Back to Journals » Cancer Management and Research » Volume 11
ELF-1 expression in nasopharyngeal carcinoma facilitates proliferation and metastasis of cancer cells via modulation of CCL2/CCR2 signaling
Authors Chen CH, Su LJ, Tsai HT, Hwang CF
Received 28 November 2018
Accepted for publication 16 April 2019
Published 6 June 2019 Volume 2019:11 Pages 5243—5254
DOI https://doi.org/10.2147/CMAR.S196355
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Chang-Han Chen,1,2 Li-Jen Su,3–5 Hsin-Ting Tsai,1 Chung-Feng Hwang6,7
1Guangdong Institute of Gastroenterology, Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Disease, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, 510020, People’s Republic of China; 2Department of Applied Chemistry, Graduate Institute of Biomedicine and Biomedical Technology, National Chi Nan University, Nantou, 54561, Taiwan; 3Department of Biomedical Sciences and Engineering, College of Health Sciences and Technology, National Central University, Taoyuan City, Taiwan; 4Education and Research Center for Technology Assisted Substance Abuse Prevention and Management, Taoyuan City, Taiwan; 5IHMed Global, Taipei City, Taiwan; 6Department of Otolaryngology, Kaohsiung Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Kaohsiung, 83301, Taiwan; 7Kaohsiung Chang Gung Head and Neck Oncology Group, Kaohsiung Chang Gung Memorial Hospital, Kaohsiung 83301, Taiwan
Background: Nasopharyngeal carcinoma (NPC) is a prevalent malignant tumor in Southeast Asia. The management of NPC has remained a challenge until now. ELF-1 is a member of the ETS family of transcription factors that regulate genes involved in cellular growth. ELF-1 expression has been reported in various cancers and is required for tumor growth and angiogenesis; however, its function in NPC remains unclear. In the present study, we characterized the role and underlying mechanism of ELF-1 in NPC.
Methods: The biological functions of ELF-1 in NPC cells such as proliferation, migration, invasion, and drug resistance were investigated using MTT, BrdU incorporation, and Transwell assays. To gain more insight into the mechanism of ELF-1 in NPC, we analyzed CCL2/CCR2 signaling by Western blotting, ELISA, siRNAs, and CCR2 antagonist.
Results: Gain-of-function of ELF-1 in TW01 and TW04 cells promoted NPC cell proliferation, BrdU incorporation, migration, invasion and cisplatin resistance. By contrast, knockdown of ELF-1 produced opposite results. Overexpression of ELF-1 enhanced the expression of CCL2 via binding to its promoter region and increased the level of the extracellular matrix protein CCL2 in cell culture medium. ELF-1 expression also modulated the downstream targets of CCL2/CCR2 signaling. Most importantly, ELF-1-induced NPC malignant phenotypes were abrogated by a CCR2 inhibitor, implying that the CCL2/CCR2 signaling axis was involved in ELF-1-mediated regulation in NPC.
Conclusion: Our data suggest that ELF-1 plays an oncogenic role in NPC development associated with the CCL2/CCR2 signaling pathway and may therefore be a potential target for NPC therapy.
Keywords: ELF-1, NPC, CCL2
Introduction
Nasopharyngeal carcinoma (NPC) occurs in certain geographic areas such as southern China and Southeast Asia, including Taiwan with an incidence of 20–50 cases per 100,000 inhabitants.1,2 Many etiological studies have revealed that NPC is positively correlated with multiple factors, such as genetics, viral infections, and environmental factors. At present, radiotherapy, chemotherapy, and chemoradiotherapy are standard treatments for NPC patients.3 Although treatment outcomes have improved with technological developments, the 5-year survival rate of NPC patients remains disappointing.4,5 Treatment failure results in local recurrence and distant metastasis, and the principle reason is advanced staging.6 Thus, to further understand the molecular mechanisms of NPC progression is crucial for the determination on therapeutic strategy of NPC.
ETS is member of an evolutionarily conserved family of transcription factor that contain a conserved ETS DNA-binding domain. ETS proteins are ubiquitously distributed in metazoans, some of which are indispensable to life.7,8 The ETS protein family participates in widespread biological processes, such as cell cycle control, proliferation, angiogenesis, and differentiation,9,10 suggesting that EST proteins play an important role in cancer progression. ELF-1 is a crucial ETS family member and expressed in specific cell cycle, and during mitotic activation and tumorigenesis.11 ELF-1 expression has been linked to hematopoiesis during embryonic development.12 ELF-1 may also act as an activator or repressor to regulate transcription of various genes in human cells. In mice, knockout endogenous of ELF-1 causes abnormalities in the development of natural killer T-cells.13–15 Pathologically, ELF-1 expression has been correlated with poor prognoses in ovarian and endometrial cancers.16,17 In melanoma, ELF is involved in tumor-induced neo-angiogenesis.18 However, ELF-1 expression is negatively associated with histological grading in breast cancer.15 Overall, these results suggest that ELF-1 could be a novel target for cancer therapy.
There is minimal information regarding the role of ELF-1 in NPC. In the present study, we characterized the biological function and potential molecular mechanism of ELF-1 during NPC tumorigenesis.
Materials and methods
Cell lines and cell culture
The TW01 and TW04 cell lines derived from primary nasopharyngeal tumours of untreated NPC patients, which provided by Chin-Tarng Lin (National Taiwan University, Taipei, Taiwan) and were cultured in an incubator (37°C, 5% CO2, and saturated humidity) with Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (FBS) and 100 mg/mL streptomycin and 100 U/mL penicillin. The culture medium was replaced every 2 days. The use of human cell lines was approved by the Institutional Review Board of Chang Gung Memorial Hospital.
RNA extraction and Q-RT-PCR
TRIzol reagent and Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) were used to extract total RNA and perform reverse transcription, respectively. Q-RT-PCR was performed using an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and an SYBR Green PCR Kit (Qiagen, Hilden, Germany). The relative mRNA expression levels were quantified using the 2−ΔΔCt method following normalization with endogenous levels of the reference mRNA GAPDH. All reactions were performed in triplicate for each sample.
Cell transfection
NPC cells were transfected with pCMV6 or pCMV6-ELF-1 using Lipofectamine 2,000 (Invitrogen) according to the manufacturer’s instructions. The transfected cells were treated with G418 (Calbiochem, La Jolla, CA, USA) for approximately 2 weeks, to select drug-resistant colonies, which were collected, expanded, and identified. The two ELF-1 siRNA sequences were 5ʹ-CACTTCAAATAGGAATCAAC-3ʹ and 5ʹ-TGCCGTAATTGTGGAACATCT-3ʹ.
Western blot analysis
Whole-cell lysates were extracted and lysed in RIPA buffer. Total protein was extracted, and protein concentration was quantified using a BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Protein was separated by SDS-PAGE and electrophoretically transferred to nitrocellulose membranes. The primary antibodies used in this study were as follows: anti-DDK (1:2,000), anti-ELF-1 (1:1,000), anti-CCL2 (1:1,000), and anti-β-actin (1:5,000; Cell Signaling Technology, USA) was used as the loading control. Immunoreactive bands were visualized using ECL detection reagent. All data analyses were repeated three times independently.
Cell proliferation and growth inhibition assays
Cell proliferation was evaluated using the MTT assay according to the manufacturer’s instructions. Briefly, we seeded approximately 1,000 cells in 200 µL medium into 96-well plates, which were cultured for the indicated days. We then added 20 μL MTT solution to the plates, and the cells were incubated for an additional 4 hrs. The optical density of each well was measured with an enzymatic reader at 490 nm. All data analyses were independently repeated three times. For the growth inhibition assays, 5,000 cells/well were plated into 96-well and cultured for 24 hrs. Next day, cisplatin concentrations from 5 μM to 40 μM or DMSO were added into each well. The MTT reagent was added after additional 48 hrs incubation at 37°C.
Transwell invasion and migration assay
The cells were then harvested, resuspended (5×104 cells/well) in 200 μl of serum-free medium, and placed in the upper compartment of a Transwell chamber with or without Matrigel. The lower compartment was filled with 500 μL of DMEM and 10% FBS. After 24 hrs of incubation, cells that penetrated through the Matrigel membrane were fixed with methanol and stained with 0.05% Crystal Violet. Six random fields of cells were counted in each well using a microscope at a magnification of ×100. All data analyses were independently repeated three times.
Chromatin immunoprecipitation (ChIP) analysis
The ChIP assay was performed according to the manufacturer’s instructions. ELF-1 and IgG antibodies were used to immunoprecipitate the protein-DNA complexes. Q-PCR was performed using primers designed to amplify the region of the CCL2 promoter. The primers were as follows: forward 5′-TGTTTACACAATCCTACAGTTCTGC-3′ and reverse 5′-TTACCTTCAGGCCACATTCC-3′.
Dual-luciferase reporter assay
Cells were transfected with pGL3 or pGL3-CCL2 containing the firefly luciferase reporter using Lipofectamine 2,000 transfection reagent and harvested 48 hrs after transfection. Luciferase activities were analyzed using the Dual-Luciferase Reporter Assay System according to the manufacturer’s protocol. Each experiment was performed in triplicate.
Statistical analysis
All analyses were performed using the GraphPad prism 5.0 software. Student’s t-test and one-way ANOVA were used for the comparison of significant differences between more than two groups. Data are presented as the mean±standard deviation. Differences were considered significant at P<0.05.
Results
ELF-1 overexpression promoted NPC cell growth
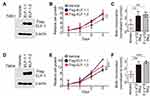
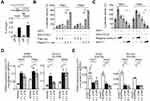
To investigate the role of ELF-1 in NPC, gain-of-function of ELF-1 in TW01 and TW04 cells was established. As shown in Figure 1A and D, the protein expression profiles of exogenous ELF-1 were determined in both cell lines by Western blotting. We then investigated if ELF-1 overexpression influenced cell growth using MTT and BrdU incorporation assays. The MTT assay showed that ELF-1-TW01 and ELF-1-TW04 transfectants exhibited significantly increased the cell proliferation compared to the vehicle control (Figure 1B and E). We further determined the effect of ELF-1 on DNA synthesis in TW01 and TW04 cells. The BrdU incorporation assay indicated that ELF-1 overexpression in NPC cells was at least 2.0-fold higher than that of vector control groups (Figure 1C and F). Collectively, these results showed that ELF-1 overexpression promoted NPC proliferation.
ELF-1 inhibition prevented NPC cell proliferation
To further validate the results of Figure 1, we knocked down the expression of ELF-1 in human NPC cell lines. Figure 2A and D showed that the protein levels of ELF-1 were significantly reduced in cells transfected with siRNA targeting ELF-1 compared with the negative control. Using the same panel, we examined whether ELF-1 knockdown affected the growth of NPC cells. The data indicated that cell proliferation and BrdU incorporation in siELF-1-TW01 and siELF-1-TW04 cells were remarkably inhibited (Figure 2B, C, E, and F), suggesting that ELF-1 expression is required for NPC proliferation.
ELF-1 expression affected NPC cell motility
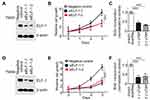
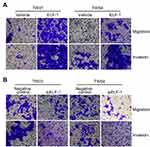
Next, the effects of ELF-1 on cell migration and invasion were investigated. Transwell assays with or without Matrigel coating were performed to examine the influence of ELF-1 on NPC cell motility. Compared to the vehicle control, high expression of ELF-1 in TW01 and TW04 cells dramatically increased the migratory and invasive abilities of NPC cells (Figures 3A, B and S1A). By contrast, compared with the negative control group, siELF-1 cells exhibited decreased migration and invasion (Figures 3C, D and S1B). Taken together, these results suggested that ELF-1 expression is important for NPC cell motility.
ELF-1 expression affected the response to cisplatin in NPC cells
To assess the role of ELF-1 in response to the chemotherapeutic drug cisplatin, we used ELF-1-overexpressing NPC cells to assess viability. Using the MTT assay, TW01 and TW04 cells expressing gain-of-function of ELF-1 were shown to exhibit increased resistance to the genotoxic drug, cisplatin in a dose-dependent manner compared with the vehicle control (Figure 3E and F). To confirm these results, we knocked down endogenous ELF-1 in NPC cells and observed the effect of ELF-1 in response to cisplatin. ELF-1-suppressed NPC cells were more sensitive to cisplatin (Figure 3G and H). Together, the results suggested that ELF-1 expression mediates the response of NPC cells to cisplatin.
CCL2 was a downstream target of ELF-1 in NPC
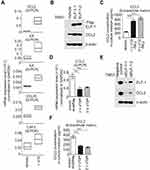
To determine the underlying mechanism involved in the ELF-1-mediated effect on the proliferation and metastasis of NPC, we used a human cytokine TaqMan Q-PCR array system to identify cytokine expression profiles associated with ELF-1 in NPC cells. Compared with the vehicle, 15 genes revealed ≥1.5-fold upregulation in ELF-1-TW01 cells; however, only 5 chemokines had over 2-fold change (Figure S2). We next verified these five gene expression profiles in gain-of-function of ELF-1-TW01 cells. Among the upregulated genes, CCL2 was significantly upregulated in ELF-1 transfectants compared with the vehicle control (Figure 4A). To confirm these results, we determined the endogenous and extracellular matrix protein levels of CCL2 in ELF-1-TW01 cells. In agreement with Q-PCR data, the endogenous and extracellular matrix protein levels of CCL2 were increased in ELF-1-TW01 stable cells compared to the vehicle (Figure 4B and C). Conversely, a significant decrease in CCL2 mRNA and protein expression was observed in the siELF-1 groups compared with the negative control (Figure 4D–F). Overall, these findings suggested that ELF-1 modulates CCL2 chemokine mRNA and protein levels in NPC cells.
ELF-1 directly targeted CCL2 and activated the CCL2/CCR2 pathway in NPC
We next performed in silico prediction based on the binding motif for ELF-1 in PROMO database, which revealed that the region of the CCL2 promoter from −392 to −404 was critical for ELF-1 recognition, indicating that CCL2 expression may be modulated by ELF-1 (Figure 5A). First, a ChIP assay was performed to show that ELF-1 directly bound to the promoter region of CCL2 in the parental TW01 and TW04 cell lines (Figure 5A). Next, we conducted a dual luciferase assay to evaluate if CCL2 was transcriptionally activated by ELF-1. TW01 and TW04 cells were co-transfected with Flag-ELF-1 and the pGL3-CCL2 promoter and their luciferase activities were assayed. As shown in Figure 5B, the luciferase activity of the CCL2 promoter was dramatically increased in NPC cells with ectopic ELF-1 expression in a dose-dependent manner. However, this effect was abolished when endogenous ELF-1 was decreased following the addition of siRNA targeting ELF-1 in TW01 and TW04 cells, compared to the corresponding negative control (Figure 5C). VEGF-A19,20 and survivin,21 two downstream targets of CCL2/CCR2 signaling, were upregulated in TW01-ELF-1 and TW04-ELF-1 transfectants (Figure 5D). By contrast, VEGF-A and survivin mRNA expression levels were decreased in NPC cells with ELF-1 knockdown (Figure 5E). Together, these results indicated that ELF-1 regulated the CCL2/CCR2 signaling pathway in NPC.
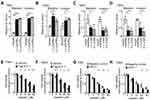
Blocking CCL2/CCR2 signaling with a CCR2 antagonist suppressed ELF-1-mediated NPC cell growth and motility
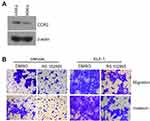
To address the regulation and apparent important role of CCL2/CCR2 in ELF-1-NPC cell proliferation, migration, and invasion, first, the endogenous protein expression profile of CCR2 in NPC cells was determined (Figure S3A). Next, ELF-1-overexpressing stable cells were treated with the CCR2 antagonist RS102895 to reduce the effects of CCL2/CCR2. Treatment of ELF-1-TW01 cells with RS102895 decreased ELF-1 induced cell growth, migration, and invasion (Figures 6A–C and S3B). Similar results were also observed in ELF-1-TW04 cells treated with a CCR2 antagonist (Figure 6D–F). Together, our data showed that CCL2 induced by ELF-1 led to increased cell growth, migration, and invasion and these effects were blocked by treatment with a CCR2 antagonist.
Discussion
The aim of this study was to determine the involvement of ELF-1 in NPC carcinogenesis. Several studies have proposed that ELF-1 expression may be involved in human cancer progression;11,17,22 however, there have been no studies characterizing the biological functions and underlying mechanisms of ELF-1 in NPC. In the present study, we showed that ELF-1 expression had significant effects on the proliferation, migration, invasion, and drug resistance of NPC cells. Remarkably, we identified a novel signaling link where ELF-1 mechanistically connected downstream proinflammatory cytokine CCL2 stimulation to CCR2 signaling activation, which contributed to cellular events, including promotion of NPC growth and motility. Together, our findings show that ELF-1 promotes NPC tumor progression via the CCL2/CCR2 signaling axis.
ELF-1 activates or represses the transcription of various genes involved in processes such as development, viral gene activation, and oncogenesis.14,23–28 Accumulating evidence have indicated that the function of ELF-1 in cancers involves both oncogenic and tumor suppressive roles.23 Several studies have reported that high ELF-1 expression is positively correlated with clinical characteristics, such as histological grading.16–18 However, in breast ductal carcinomas, ELF-1 expression is negatively correlated with tumor size.15 ELF-1 protein is localized in both the cytoplasm and nucleus. Phosphorylated ELF-1 has been shown to translocate to the nucleus to activate gene expression. Loss of the ability of T-cells to produce ELF-1 has been linked to decreased expression of T-cell receptor ζ and systemic lupus erythematosus.29 ELF-1 is also a key regulator of hematopoietic-related genes such as Scl, Fli-1, Lyl-1, Runx1, and Lmo2.12 ELF-1 has been shown to participate in blood vessel development by regulating angiogenesis-related genes such as Tie1/2, VEGFR1, angioooietin-2, and endoglin.25,30–32 Several evidences have shown that hPygo2 expression is required for ELF-1 modulation in human cancer cells, such as breast and cervical carcinoma;33 whereas dominant-active Rb overexpression inhibits ELF-1-dependent activation of hPygo2.34 Despite the function of ELF-1 as an important transcriptional regulator in both blood and endothelium, nothing is known about the biological function of ELF-1 in NPC. In the present study, we showed that ELF-1 plays an important role in NPC cell proliferation, migration, invasion, and drug resistance, suggesting that ELF-1 is a critical molecule in NPC progression.
Chemokines and their receptors are potential therapeutic targets that have been extensively investigated by pharmaceutical and biotechnology companies. CCL2, a prototypic chemokine is constitutively expressed and is important for homeostatic functions in tissues and cells. CCL2 is expressed in several cell types, including epithelial, fibroblasts, and endothelial cells, and is also a potent chemoattractant for monocytes and lymphocytes.35,36 Increased levels of CCL2 in many human cancers, such as breast, cervical, colon, and prostate have been associated with cancer progression and poor prognoses.37 In addition, CCL2 expression has been correlated with infiltration of tumor-associated macrophages, which are known to be important for tumor progression, growth, and angiogenesis.37 However, CCL2 may also have opposing anti-tumorigenic functions in the progression of melanoma by recruiting cytotoxic T lymphocytes.38 CCL2 preferentially binds the CCR2 receptor, which is expressed in various human tissues. CCL2/CCR2 signaling has also been implicated in promoting tumorigenesis and metastasis. CCL2/CCR2 signaling promotes the migration and invasion of chondrosarcoma cells by inducing MMP9 expression via Ras/Raf-1 and NF-κB activation.39 Inhibition of JAK2 expression, a downstream target of CCL2/CCR2, prevents tumor cell transmigration and decreases the induction of lung permeability, consequently attenuating metastasis.37 Although CCL2/CCR2 signaling has been investigated in many human cancers, few reports have focused on NPC. Our results demonstrated that ELF-1 bound to the promoter region of CCL2 regulated transcriptional and post-transcriptional levels of CCL2 in NPC cells. ELF-1-induced cell growth, migration, and invasion were required for CCL2/CCR2 signaling. However, blockage of CCL2/CCR2 signaling using a CCR2 antagonist reduced the ELF-1-raised effects, implying CCL2/CCR2 signaling is critical for ELF-1-mediated tumorigenesis in NPC.
Conclusions
We have provided evidence characterizing the biological role of ELF-1 in NPC. Our results showed that high expression of ELF-1 promotes cell proliferation, motility, and cisplatin resistance in NPC cell lines; conversely, inhibition of endogenous ELF-1 had the opposite effects. Furthermore, ELF-1 modulated the secretion and expression of CCL2 in NPC cells. Mechanistically, ELF-1 regulated CCL2 expression through binding to the promoter region of CCL2. ELF-1 also regulated the expression of downstream targets of CCL2/CCR2. Blockage of CCR2 signaling with a CCR2 antagonist decreased ELF-1-induced malignant phenotypes in NPC cells. Taken together, our results suggest that ELF-1/CCL2/CCR2 signaling participates in the development of NPC.
Acknowledgments
This work was supported in part by grants from the National Science Council, Taiwan (MOST-105-2314-B-182A-073-MY3 and MOST-104-2314-B-182A-076), Chang Gung Memorial Hospital (CMRPG8D1221, CMRPG8D1222, CMRPG8D1223, CMRPG8C0581, CMRPG8C0582, and CMRPG8C0583), and the Science and Technology Program of Guangzhou City (201904010356). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. An abstract of this paper was presented at the Molecular Med Tri-Con/Bio-IT World WEST Conference as a poster presentation talk with interim findings. The poster’s abstract was published in “Poster Abstracts” in Bio-IT World Conference & Expo WEST Poster Sessions.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bei JX, Li Y, Jia WH, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42(7):599–603. doi:10.1038/ng.601
2. Zhao YY, Tian Y, Liu L, et al. Inhibiting eEF-2 kinase-mediated autophagy enhanced the cytocidal effect of AKT inhibitor on human nasopharyngeal carcinoma. Drug Des Devel Ther. 2018;12:2655–2663. doi:10.2147/DDDT.S169952
3. Shen Y, Zhang S, Sun R, Wu T, Qian J. Understanding the interplay between host immunity and Epstein-barr virus in NPC patients. Emerg Microbes Infect. 2015;4(3):e20. doi:10.1038/emi.2015.20
4. Lee AW, Ma BB, Ng WT, Chan AT. Management of nasopharyngeal carcinoma: current practice and future perspective. J Clin Oncol. 2015;33(29):3356–3364. doi:10.1200/JCO.2015.60.9347
5. Zhang L, Huang Y, Hong S, et al. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: a multicentre, randomised, open-label, phase 3 trial. Lancet. 2016;388(10054):1883–1892. doi:10.1016/S0140-6736(16)31388-5
6. Zhang P, Hong H, Sun X, et al. MicroRNA-10b regulates epithelial-mesenchymal transition by modulating KLF4/Notch1/E-cadherin in cisplatin-resistant nasopharyngeal carcinoma cells. Am J Cancer Res. 2016;6(2):141–156.
7. Degnan BM, Degnan SM, Naganuma T, Morse DE. The ets multigene family is conserved throughout the metazoa. Nucleic Acids Res. 1993;21(15):3479–3484.
8. Poon GMK, Kim HM. Signatures of DNA target selectivity by ETS transcription factors. Transcription. 2017;8(3):193–203. doi:10.1080/21541264.2017.1302901
9. Findlay VJ, LaRue AC, Turner DP, Watson PM, Watson DK. Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv Cancer Res. 2013;119:1–61. doi:10.1016/B978-0-12-407190-2.00001-0
10. Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC. The ETS family of oncogenic transcription factors in solid tumours. Nat Rev Cancer. 2017;17(6):337–351. doi:10.1038/nrc.2017.20
11. Yang DX, Li NE, Ma Y, Han YC, Shi Y. Expression of Elf-1 and survivin in non-small cell lung cancer and their relationship to intratumoral microvessel density. Chin J Cancer. 2010;29(4):396–402.
12. Calero-Nieto FJ, Wood AD, Wilson NK, Kinston S, Landry JR, Gottgens B. Transcriptional regulation of Elf-1: locus-wide analysis reveals four distinct promoters, a tissue-specific enhancer, control by PU.1 and the importance of Elf-1 downregulation for erythroid maturation. Nucleic Acids Res. 2010;38(19):6363–6374. doi:10.1093/nar/gkq490
13. Walunas TL, Wang B, Wang CR, Leiden JM. Cutting edge: the Ets1 transcription factor is required for the development of NK T cells in mice. J Immunol. 2000;164(6):2857–2860.
14. Choi HJ, Geng Y, Cho H, et al. Differential requirements for the Ets transcription factor Elf-1 in the development of NKT cells and NK cells. Blood. 2011;117(6):1880–1887. doi:10.1182/blood-2010-09-309468
15. Gerloff A, Dittmer A, Oerlecke I, Holzhausen HJ, Dittmer J. Protein expression of the Ets transcription factor Elf-1 in breast cancer cells is negatively correlated with histological grading, but not with clinical outcome. Oncol Rep. 2011;26(5):1121–1125. doi:10.3892/or.2011.1409
16. Takai N, Miyazaki T, Nishida M, Shang S, Nasu K, Miyakawa I. Clinical relevance of Elf-1 overexpression in endometrial carcinoma. Gynecol Oncol. 2003;89(3):408–413.
17. Takai N, Miyazaki T, Nishida M, Nasu K, Miyakawa I. The significance of Elf-1 expression in epithelial ovarian carcinoma. Int J Mol Med. 2003;12(3):349–354.
18. Huang X, Brown C, Ni W, Maynard E, Rigby AC, Oettgen P. Critical role for the Ets transcription factor ELF-1 in the development of tumor angiogenesis. Blood. 2006;107(8):3153–3160. doi:10.1182/blood-2005-08-3206
19. Han R, Gu S, Zhang Y, et al. Estrogen promotes progression of hormone-dependent breast cancer through CCL2-CCR2 axis by upregulation of Twist via PI3K/AKT/NF-kappaB signaling. Sci Rep. 2018;8(1):9575. doi:10.1038/s41598-018-27810-6
20. Hong KH, Ryu J, Han KH. Monocyte chemoattractant protein-1-induced angiogenesis is mediated by vascular endothelial growth factor-A. Blood. 2005;105(4):1405–1407. doi:10.1182/blood-2004-08-3178
21. Zhang J, Lu Y, Pienta KJ. Multiple roles of chemokine (C-C motif) ligand 2 in promoting prostate cancer growth. J Natl Cancer Inst. 2010;102(8):522–528. doi:10.1093/jnci/djq044
22. Takai N, Ueda T, Nishida M, Nasu K, Miyakawa I. The relationship between oncogene expression and clinical outcome in endometrial carcinoma. Curr Cancer Drug Targets. 2004;4(6):511–520.
23. Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34.
24. Xiang P, Lo C, Argiropoulos B, et al. Identification of E74-like factor 1 (ELF1) as a transcriptional regulator of the Hox cofactor MEIS1. Exp Hematol. 2010;38(9):
25. Jin E, Liu J, Suehiro J, et al. Differential roles for ETS, CREB, and EGR binding sites in mediating VEGF receptor 1 expression in vivo. Blood. 2009;114(27):5557–5566. doi:10.1182/blood-2009-05-220434
26. Iljin K, Dube A, Kontusaari S, et al. Role of ets factors in the activity and endothelial cell specificity of the mouse Tie gene promoter. FASEB J. 1999;13(2):377–386.
27. Seth A, Watson DK, Blair DG, Papas TS. c-ets-2 protooncogene has mitogenic and oncogenic activity. Proc Natl Acad Sci U S A. 1989;86(20):7833–7837.
28. Leiden JM, Wang CY, Petryniak B, Markovitz DM, Nabel GJ, Thompson CB. A novel Ets-related transcription factor, Elf-1, binds to human immunodeficiency virus type 2 regulatory elements that are required for inducible trans activation in T cells. J Virol. 1992;66(10):5890–5897.
29. Juang YT, Tenbrock K, Nambiar MP, Gourley MF, Tsokos GC. Defective production of functional 98-kDa form of Elf-1 is responsible for the decreased expression of TCR zeta-chain in patients with systemic lupus erythematosus. J Immunol. 2002;169(10):6048–6055.
30. Dube A, Thai S, Gaspar J, et al. Elf-1 is a transcriptional regulator of the Tie2 gene during vascular development. Circ Res. 2001;88(2):237–244.
31. Hegen A, Koidl S, Weindel K, Marme D, Augustin HG, Fiedler U. Expression of angiopoietin-2 in endothelial cells is controlled by positive and negative regulatory promoter elements. Arterioscler Thromb Vasc Biol. 2004;24(10):1803–1809. doi:10.1161/01.ATV.0000140819.81839.0e
32. Pimanda JE, Chan WY, Donaldson IJ, Bowen M, Green AR, Gottgens B. Endoglin expression in the endothelium is regulated by Fli-1, Erg, and Elf-1 acting on the promoter and a −8-kb enhancer. Blood. 2006;107(12):4737–4745. doi:10.1182/blood-2005-12-4929
33. Andrews PG, Kennedy MW, Popadiuk CM, Kao KR. Oncogenic activation of the human Pygopus2 promoter by E74-like factor-1. Mol Cancer Res. 2008;6(2):259–266. doi:10.1158/1541-7786.MCR-07-0068
34. Tzenov YR, Andrews PG, Voisey K, et al. Human papilloma virus (HPV) E7-mediated attenuation of retinoblastoma (Rb) induces hPygopus2 expression via Elf-1 in cervical cancer. Mol Cancer Res. 2013;11(1):19–30. doi:10.1158/1541-7786.MCR-12-0510
35. Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29(6):313–326. doi:10.1089/jir.2008.0027
36. Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7(19):28697–28710. doi:10.18632/oncotarget.7376
37. O‘Connor T, Borsig L, Heikenwalder M. CCL2-CCR2 signaling in disease pathogenesis. Endocr Metab Immune Disord Drug Targets. 2015;15(2):105–118.
38. Zhang T, Somasundaram R, Berencsi K, et al. Migration of cytotoxic T lymphocytes toward melanoma cells in three-dimensional organotypic culture is dependent on CCL2 and CCR4. Eur J Immunol. 2006;36(2):457–467. doi:10.1002/eji.200526208
39. Tang CH, Tsai CC. CCL2 increases MMP-9 expression and cell motility in human chondrosarcoma cells via the Ras/Raf/MEK/ERK/NF-kappaB signaling pathway. Biochem Pharmacol. 2012;83(3):335–344. doi:10.1016/j.bcp.2011.11.013
Supplementary materials
 | Figure S2 The mRNA expression levels of chemokines in ELF-1-TW01 cell lines and the vehicle control. Fifteen genes revealed ≥1.5-fold upregulation in ELF-1-TW01 cells, compared to vehicle control. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.