Back to Journals » Clinical Interventions in Aging » Volume 16
Effects of Donor-Recipient Age Difference in Renal Transplantation, an Investigation on Renal Function and Fluid Proteome
Authors Wang X, Zu Q, Lu J, Zhang L, Zhu Q, Sun X , Dong J
Received 16 April 2021
Accepted for publication 6 July 2021
Published 27 July 2021 Volume 2021:16 Pages 1457—1470
DOI https://doi.org/10.2147/CIA.S314587
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Zhi-Ying Wu
Xinning Wang,1 Qiang Zu,1 Jinshan Lu,1 Lei Zhang,1 Qiang Zhu,1 Xuefeng Sun,2 Jun Dong1
1Department of Urology, Chinese PLA General Hospital, Beijing, People’s Republic of China; 2Department of Nephrology, Chinese PLA General Hospital, Beijing, People’s Republic of China
Correspondence: Xuefeng Sun; Jun Dong Email [email protected]; [email protected]
Introduction: Our previous study revealed that a young internal environment ameliorated kidney aging by virtue of an animal model of heterochronic parabiosis and a model of heterochronic renal transplantation. In this research, we used proteome to investigate the effects of donor-recipient age difference in clinical renal transplantation.
Methods: This study included 10 pairs of renal transplantation donors and recipients with an age difference of greater than 20 years to their corresponding recipients/donors. All recipients have received transplantation more than 3 years ago. Renal function and the serum/urine proteomes of the donors and recipients were analyzed.
Results: The renal function was similar between the young recipients and the old donors. In contrast, the renal function of the young donors was significantly superior to that of the old recipients. Furthermore, 497 and 975 proteins were identified in the serum and urine proteomes, respectively. The content of SLC3A2 in the blood was found to be related to aging, while the contents of SERPINA1 and SERPINA3 in the urine were related to immune functions after renal transplantation.
Conclusion: This study demonstrated that, in the human body, a younger internal environment could ameliorate kidney aging and provided not only clinical evidence for increasing the age limit of kidney transplant donors but also new information for kidney aging research.
Keywords: renal transplantation, aging, proteome, living donor transplantation
Background
The number of patients with end-stage renal disease (ESRD) has been increasing year by year. Renal transplantation is the best treatment for ESRD patients. However, standard criteria donors (SCDs) cannot meet the demand for renal transplantation. Although renal transplantation has advanced rapidly, the effects of the ages of the donors and recipients and the age differences between the donors and recipients on the function of the transplanted kidney remain unclear.
A number of studies have employed animal models of heterochronic parabiosis to demonstrate that a young internal environment improves the aging of the brain and heart,1,2 and enhances the regeneration capacities of muscles, liver, neural stem/progenitor cells and ovarian follicles.3–5 By virtue of an animal model of heterochronic parabiosis and a model of heterochronic renal transplantation, our previous study found that a young internal environment improved the aging of the kidney.6,7 In addition, employment of an animal model of heterochronic parabiosis with bilateral renal ischemia-reperfusion injury (IRI) revealed that a young internal environment alleviated IRI in elderly kidneys.8 These studies provide the basic research evidence for increasing the age limit of kidney transplant donors. However, there is a lack of support from clinical trials. The bioinformatics technique could provide enough information for analyzing the pathological mechanism which traditional clinical graft function assessment could not. For example, quantitative real-time PCR could provide more useful information to understand the pathogenic role of opportunistic pathogens in the urogenital tract.9 In this study, we used the proteome to find more information about the action between the internal environment and grafts.
The present study included 20 donors and recipients (10 pairs). The recipients received transplant surgery more than 3 years ago and had an age difference of greater than 20 years with their corresponding donors. The kidney functional changes in the recipients and donors were analyzed, and their serum and urine proteomes were identified. The purpose of the present study was to provide clinical evidence for increasing the age limit of kidney transplant donors and new information for research on kidney aging.
Methods
Ethics
This was a prospective, small-sample, exploratory, controlled study approved by the Ethics Committee of the People’s Liberation Army (PLA) General Hospital. The protocol followed the ethical principles in the Declaration of Helsinki (1964). All patients gave their written informed consent prior to any study procedures. After data lock, all study data and collected biological material were anonymized.
Patients
All kidney transplant surgeries and follow-ups with the donors and recipients were completed at the PLA General Hospital. All kidneys were donated voluntarily with written informed consents, and the consent was conducted in accordance with the Declaration of Istanbul. All donors and recipients underwent the following examinations: blood pressure measuring, blood routine, urine routine, renal and liver function, and ultrasound imaging of the urinary system. A total of 10 pairs of recipients and donors of a living-related kidney transplant who were direct relatives were included. The inclusion criteria were as follows: (1) no limitations on the age and sex of the donors and recipients; (2) age difference between the donors and recipients greater than 20 years; (3) donors and recipients completed 3 years of regular follow-ups; and (4) the cause of transplantation was primary glomerulonephritis. The exclusion criteria were as follows: (1) recipient developed acute rejection; (2) recipient’s original kidney disease recurred or a new kidney disease developed; (3) donor and recipient suffered concurrent hypertension, diabetes, kidney stones, kidney tumors, and urinary system infection; (4) donor and recipient with concurrent acute kidney injury, edema, pleural and peritoneal effusion, amputation, heart failure, liver disease, severe obesity, or ketoacidosis; (5) donor and recipient were currently taking high-dose steroids, cimetidine, or trimethoprim; and (6) pregnant or menstruating women.
The donors and the recipients were divided into 2 groups according to their age differences: the old donor/young recipient group, which included 5 old donors and 5 young recipients, and the young donor/old recipient group, which included 5 young donors and 5 old recipients. Eight recipients received an immunosuppressive regimen composed of prednisone, mycophenolate mofetil, and tacrolimus, one recipient received sirolimus instead of tacrolimus, and one received CsA instead of tacrolimus.
Scr levels were measured using a Roche enzymatic assay (Hitachi, Tokyo, Japan; reagents from Roche Diagnostics, Mannheim, Germany). Renal function was assessed using the estimated glomerular filtration rate (eGFR), which was calculated based on the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation.10
Sample Processing and Mass Spectrometry Analysis
Fasting venous blood and morning urine were collected from all donors and recipients for serum or urine proteomic testing. The blood was centrifuged at 5000 g for 15 min to isolate the serum, while the urine was centrifuged at 5000 g for 45 min to obtain the supernatant. The samples were stored at −80°C.
Serum Proteome Protocol
First, put the serum samples into the spin-column (Pierce™ Top 2 Abundant Protein Depletion Spin Columns, ThermoScientific) to deplete top 2 protein at the room temperature. Then loaded 5 μl serum into the column and inverse. Put the column on head-over-head shaker for 30 min at the room temperature. After that, spined column at 1000g for 2 min at RT, then got 400 μl of solution. Added 20 μl of 1M NH4HCO3 to the solution (final conc 50 mM) to adjust pH to 8.5. Heated the sample at 95°C for 2 min to denature the proteins and cooled down to RT. Added 3 μg of trypsin (Sequencing Grade Modified Trypsin, Promega) and incubated at 37°C for four hours, then the digest was ready for sRP.
Actived reverse-phase column with 3mg C18 (3 μm, Dr. Maisch GmbH) by 100μl Acetonitrile (LC/MS grade, FisherScientific). Vacuum dried peptides were dissolved in pH10 buffer (10mM Ammonium Bicarbonate, pH10 adjusted by NH4OH) and subjected to pH10 C18 reverse-phase column chromatography. Washed C18 reverse-phase column by 100μl buffer (10mM Ammonium Bicarbonate, pH10 adjusted by NH4OH) twice. Bounded peptide was eluted with step gradient of 100 µL of 6, 9, 12, 15, 18, 21, 25, 30, 35% ACN (pH10) into 1.5mL centrifuge tubes. Mixed the 6, 15, 25% ACN eluent, 9, 18, 30% ACN eluent and 12, 21, 35% ACN and pooled in to 3 pools and vacuum dried for LC-MS/MS.
Urine Proteome Protocol
Added 1mL urine into 2mL Beckman ultracentrifugation tube, centrifuged at 200,000g and room temperature for 75 min, discarded the super. Added 50ul suspension buffer (50mM Tris, 250mM sucrose, pH8.5) in each tube and laid tube down for 15min at the room temperature, then added 2.5μl 1M DTT and heated at 65 °C for 30 min. Adjusted volume to 200ul with wash buffer (10mM TEA, 100mM NaCl, pH 7.4) and centrifuged at 200,000g for 30 min, discarded the super. Added 30ul digestion buffer (30ul 50mM NH4HCO3, pH8.5), then added 500ug trypsin (Sequencing Grade Modified Trypsin, Promega) and mixed well. Then incubated at 37°C for 4h. Dried the extract with SpeedVac and it was ready for LC-MS/MS.
NanoHPLC-MS Analysis
The extracted peptides were re-suspended in 20 μL of loading solution (5% methanol containing 0.1% formic acid) and 5 μl was analyzed. Q Exactive coupled to nLC-1000 (ThermoScientific) was used. A homemade trap column (2 cm x 100 μm) and an analytical column (12 cm x 150 μm), both packed with Reprosil-Pur Basic C18 (3 µm, Dr. Maisch GmbH, Germany) were used. The 0–69 min gradient of 5–31% acetonitrile and 0.1% formic acid and the 70–75 min gradient of 95% acetonitrile and 0.1% formic acid at a flow were used. The full MS scan range was set to 300–1400 m/z and trap size for MS1 and MS2 was 3×106 and 2×105, respectively. The mass resolution for MS1 and MS2 was 120,000 and 15,000, respectively. The top 25 ions were selected for higher energy collision dissociation (HCD) with collision energy set at 27%. Dynamic exclusion was used after 1st identification with 30s exclusion duration.
Protein Identification and Label-Free Quantification
Proteome Discoverer (PD, V2.0, ThermoScientific) with Mascot (Mascot V2.3, Matrix Science) was used to search raw data against Human RefSeq database (the 2013.07.04). Mass tolerance for precursor ions was set to 20 ppm; mass tolerances of fragment ions were 0.05 Da. Carbamidomethylation of cysteine, oxidation of methionine, acetylation and butyrylation of lysine were included as variable modifications. A maximum of two missed cleavages was allowed. All assigned peptides were filtered with 1% false discovery rate (FDR) at peptide level. We only kept identifications with ≥2 unique peptides (1% FDR and ion score >20), which was stricter than 1% FDR at the protein level. All identified peptides were quantified with peak areas derived from their MS1 intensity.
Data Processing
Statistical Techniques
Continuous variables were expressed as mean±SD. Statistical analyses were performed using IBM SPSS statistics 22 software. Comparisons of continuous variables were performed by using paired Student’s t-test.
Protein Identification and Label-Free Quantification
The acquired MS/MS spectra were, respectively, searched against the target-decoy RefSeq human database (release 2013_07, containing 32,015 entries) from NCBI website (https://www.ncbi.nlm.nih.gov/refseq/) using Proteome Discoverer software version 2.0 (Thermo Fisher Scientific) with Mascot algorithm (Mascot 2.4, Matrix Science). Dynamic modifications of acetylation of the N terminus and oxidation of methionine were allowed. The precursor mass tolerance was confined within 20 ppm with fragment mass tolerance of 0.5 Dalton, and trypsin was chosen as cleavage specificity with a maximum number of allowed missed cleavages of two. The following filter was used in this study, 1% false-positive rate at protein level and each protein with ≥1 unique peptides and protein areas were used for protein quantification.11,12 After filtering the results by above filter, proteins were exported for proteomic analysis workflow described here.
Bioinformatic Analysis
All further calculations had been performed using R (version 3.4.1, http://www.r-project.org/). Before further processing, expression values were normalized for each separately. Wilcoxon–Mann–Whitney test was applied to select data sets with statistical significance (p-value≤0.05). Hierarchical clustering was done using the heatmap.2 function.
GO Functional Analysis
All differential proteins identified by two approaches were assigned their gene symbol via the DAVID database (https://david.ncifcrf.gov/). Protein classification was performed based on their functional annotations using Gene Ontology (GO) for biological process, and molecular function. When more than one assignment was available, all of the functional annotations were considered in the results.
IPA Network Analysis
All differential proteins were used for pathway analysis. For this purpose, the gene symbol was inserted into the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Mountain View, CA). This software categorizes gene products based on the location of the protein within cellular components and suggests possible biochemical, biological and molecular functions. Furthermore, proteins were mapped to genetic networks available in the Ingenuity and other databases and ranked by score. These genetic networks describe functional relationships between gene products based on known interactions in the literature. Through the IPA software, the newly formed networks were associated with known biological pathways.
Results
Patient Characteristics
All donor-recipient pairs had parent–child relationships. No significant abnormalities were detected in their blood pressure, routine urine analysis results, routine blood analysis results, and liver/renal function.
The average age difference in the old donor/young recipient group was 24 years, and the average post-transplantation time was 5 years. The average age difference in the young donor/old group was 23 years, and the average post-transplantation time was 8 years. There was no significant difference in eGFR between the young recipients and the old donors (75.54±10.3 vs 73.54±18.04, P=0.717). In contrast, the eGFR of the young donors was significantly higher than that of the old recipients (101.20±14.28 vs 64.46±18.23, p=0.041). The results are summarized in Table 1.
 |
Table 1 Characteristics and Renal Function of the Patients |
Serum Proteome
A total of 497 proteins were identified in the serum proteome, including 17 differentially expressed proteins (DEPs) in the old donor/young recipient group (Table 2) and 12 DEPs in the young donor/old recipient group (Table 3). Solute carrier 3A2 (SLC3A2) was significantly increased in old individuals in both groups.
 |
Table 2 Distinct Serum Proteins in Old Donor/Young Recipient Group |
 |
Table 3 Distinct Serum Proteins in Young Donor/Old Recipient Group |
Figures 1 and 2 show the serum proteome gene ontology (GO) results. Tables 4 and 5 illustrate the significant differential pathways, as determined by Ingenuity Pathway Analysis (IPA), in both groups. In all cases, the DEPs were mainly located in extracellular regions. In the old donor/young recipient group, the DEPs were shown to be related to physiological processes, such as immune response, coagulation reactions and acute-phase responses. In the young donor/old recipient group, the DEPs were related to lipid metabolism, glucose metabolism and other physiological processes.
 |
Table 4 IPA Pathways of Distinct Serum Proteins in Old Donor/Young Recipient Group |
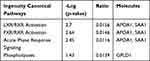 |
Table 5 IPA Pathways of Distinct Serum Proteins in Young Donor/Old Recipient Group |
 |
Figure 1 GO terms of distinct serum proteins in old donor/young recipient group. |
 |
Figure 2 GO terms of distinct serum proteins in young donor/old recipient group. |
Urine Proteome
A total of 975 proteins were identified in the urine proteome, including 12 DEPs in the old donor/young recipient group and 28 DEPs in the young donor/old recipient group (Tables 6 and 7). Alpha-1 antiproteinase (SERPINA1) and Alpha-1-Antichymotrypsin (SERPINA3) levels were significantly elevated in the transplant recipients in both groups.
 |
Table 6 Distinct Urinary Proteins in Old Donor/Young Recipient Group |
 |
Table 7 Distinct Urinary Proteins in Young Donor/Old Recipient Group |
Figures 3 and 4 show the urine proteome GO analysis results. Tables 8 and 9 illustrate the significant differential IPA pathways in both groups. In all cases, DEPs were mainly located in exosomes and external environments and were related to physiological processes, such as acute-phase responses, coagulation processes and glucose metabolism.
 |
Table 8 IPA Pathways of Distinct Urinary Proteins in Old Donor/Young Recipient Group |
 |
Table 9 IPA Pathways of Distinct Urinary Proteins in Young Donor/Old Recipient Group |
 |
Figure 3 GO terms of distinct urinary proteins in old donor/young recipient group. |
 |
Figure 4 GO terms of distinct urinary proteins in young donor/old recipient group. |
Discussion
Transplanted renal function depends on donor kidney function, IRI due to transplantation, renal damage caused by posttransplant immunological rejection and immunosuppressant toxicity, and other factors that affect renal function (such as the recurrence of the recipient’s original kidney disease and the presence of concurrent diseases). According to the exclusion criteria, the present study excluded kidney transplant recipients who suffered acute rejection and recurrence of the original kidney disease as well as donors and recipients who had concurrent diseases and factors affecting the accurate detection of serum creatinine and eGFR. Moreover, all donors underwent rigorous examinations to ensure their overall health. Therefore, renal function and degree of aging were similar between the kidneys of the donors at the time of transplantation. However, the results of the present study showed that after renal transplantation and long-term follow-up, the eGFRs of the young recipients were not significantly different from those of the old donors, while the eGFRs of the young donors were significantly higher than those of the old recipients. Due to the inevitability of IRI during transplantation and posttransplant nephrotoxic damage caused by immunosuppressants, the function of grafts couldn’t be better than the donor kidneys. This may represent an important reason why the eGFRs of young donors were significantly higher than those of the old recipients. The internal environment in which the kidneys are located affects kidney aging and function. A young internal environment reduces inflammation and apoptosis in kidneys from elderly donors and delays kidney aging. In contrast, an old internal environment increases the inflammatory and apoptotic levels in kidneys from young donors and promotes kidney aging.6,7 Appropriate interventions delay and reverse kidney aging.13 Changes in the aging status of kidneys affect renal function. A previous study showed that once the kidneys from elderly donors were transplanted into young recipients, chronic focal glomerular injury in the transplanted kidneys was repaired within months, and the kidneys maintained long-term function.14 Therefore, the aging of kidneys from young donors increases after the kidney is transplanted into old recipients, which explains why eGFR is significantly higher in young donors than in old recipients. In contrast, the aging of the kidneys from elderly donors reduces after being transplanted into young recipients, which balances the IRI developed during transplantation and the posttransplant nephrotoxic injury caused by immunosuppressants. As a result, the eGFRs of young recipients are not significantly different from those of old donors. However, there are other important reasons. The young internal environment of young recipients reduces inflammation and apoptosis in the kidneys of old recipients during transplantation, increases the level of autophagy, and alleviates renal IRI.8 In addition, young recipients highly express mature growth differentiation factor (GDF), which also promotes the repair of IRI developed during renal transplantation in old recipients.15 Therefore, although donor age is often the main reason for transplanted organ rejection,16 the age of the donor is not an independent risk factor for the reduced function of a transplanted organ.17,18
The proteomic and bioinformatic analysis showed that there were 3 significant differential IPA pathways in the serum proteome, namely, acute-phase response, farnesoid X receptor (FXR)/retinoid X receptor (RXR) activation, and liver X receptor (LXR)/RXR activation. FXR, LXR, and RXR belong to the nuclear receptor family. They regulate metabolism and transport proteins and are responsible for regulating intracellular and extracellular signaling and expression, especially glucose metabolism and lipid metabolism.19–21 Existing studies have demonstrated that high glucose promotes the senescence of renal mesangial cells,22 and that abnormal lipid metabolism is an important risk factor for cell senescence.23 Calorie restriction and pioglitazone (a drug regulating glucose metabolism) improve kidney aging.24–26 Such findings indicate that FXR/RXR activation and LXR/RXR activation may be related to kidney aging.
SLC3A2 was the only differentially expressed protein between the 2 serum proteomes. SLC3A2 is the heavy chain of cluster of CD98 and a cystine transporter responsible for maintaining the glutathione content in cells.27–29 SLC3A2 affects the first rate-limiting step in glutathione synthesis, which is the main antioxidant in cells.27,30 In addition, SLC3A2 is a key factor in integrin signaling and affects cell proliferation and division.31,32 SLC3A2 was highly expressed in the serum proteomes of the old recipients and donors, suggesting that this protein is related to kidney aging.
Analysis of the urine proteome showed that the GO term acute-phase response was included in both groups. SERPINA3 and SERPINA1 were differentially expressed in the 2 groups and were highly expressed in the urine of kidney transplant recipients. SERPINA3 functions as a protease inhibitor. It inhibits a variety of granulocyte proteases related to acute rejection responses (such as trypsin G) and has been found to be related to the occurrence of acute rejection.33–35 Fragments of SERPINA3 were found to be significantly elevated in the urine of patients suffering acute rejection.36 SERPINA1 is also a protease inhibitor and is involved in various physiological processes, such as wound healing, embryonic development, complement response, and coagulation.37,38 These findings suggest that SERPINA1 and SERPINA3 may be related to the immune response after renal transplantation and may serve as potential urinary biomarkers.
For the cell components in GO terms, the DEPs were related to the exosome, extracellular environment, particles in blood circulation, and extracellular region. Such results indicated that these changes may serve as clues for the exploration of kidney aging and renal transplantation. In addition, we observed 7 pathways that were significantly different in the 2 groups, of which 4 (acute-phase response, FXR/RXR activation, LXR/RXR activation, and atherosclerotic signal) were also identified in the blood proteome analysis. These results indicate that urine proteomics may play an important role in the study of the kidney aging and renal transplantation.
Summary
In summary, our study further demonstrated that a young internal environment might ameliorate kidney aging and improve the function of transplanted kidneys from elderly donors. Our study provided preliminary clinical evidence for increasing the age limit of renal transplant donors. However, due to the limited sample size, the findings of the present study need to be further verified by large-scale clinical studies. In addition, we found that SLC3A2 might be related to kidney aging and that SERPINA1 and SERPINA3 might be related to transplanted kidney rejection. These findings provided new clues for the investigation of related mechanisms and clinical biological marker screening.
Funding
This research was supported by the National “973” Program for Basic Research of China (NO.2013CB530800). National Natural Science Foundation of China (81870463), the fund of the State Key Laboratory of Kidney Diseases in Chinese PLA General Hospital (KF201807).
Disclosure
The authors declare no conflicts of interest.
References
1. Villeda SA, Luo J, Mosher KI, et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477(7362):90–94. doi:10.1038/nature10357
2. Loffredo FS, Steinhauser ML, Jay SM, et al. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell. 2013;153(4):828–839. doi:10.1016/j.cell.2013.04.015
3. Ruckh JM, Zhao J, Shadrach JL, et al. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell. 2012;10(1):96–103. doi:10.1016/j.stem.2011.11.019
4. Conboy IM, Conboy MJ, Wagers AJ, et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433(7027):760–764. doi:10.1038/nature03260
5. Niikura Y, Niikura T, Wang N, et al. Systemic signals in aged males exert potent rejuvenating effects on the ovarian follicle reserve in mammalian females. Aging. 2010;2(12):999–1003. doi:10.18632/aging.100255
6. Huang Q, Ning Y, Liu D, et al. A young blood environment decreases aging of senile mice kidneys. J Gerontol. 2018;73(4):421–428. doi:10.1093/gerona/glx183
7. Li D, Zhao D, Zhang W, et al. Identification of proteins potentially associated with renal aging in rats. Aging. 2018;10(6):1192–1205. doi:10.18632/aging.101460
8. Liu D, Lun L, Huang Q, et al. Youthful systemic milieu alleviates renal ischemia-reperfusion injury in elderly mice. Kidney Int. 2018;94(2):268–279. doi:10.1016/j.kint.2018.03.019
9. Sarier M, Demir M, Goktas S, et al. Results of real-time multiplex polymerase chain reaction assay in renal transplant recipients with sterile pyuria. Transplant Proc. 2017;49(6):1307–1311. doi:10.1016/j.transproceed.2017.02.051
10. Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604. doi:10.7326/0003-4819-150-9-200905050-00006
11. Lu H, Deng S, Zheng M, et al. iTRAQ plasma proteomics analysis for candidate biomarkers of type 2 incipient diabetic nephropathy. Clin Proteom. 2019;16(1):33. doi:10.1186/s12014-019-9253-1
12. Wu J, Zhang J, Wei J, et al. Urinary biomarker discovery in gliomas using mass spectrometry-based clinical proteomics. Chin Neurosurg J. 2020;6(1):11. doi:10.1186/s41016-020-00190-5
13. Kanasaki K, Kitada M, Koya D. Pathophysiology of the aging kidney and therapeutic interventions. Hypertens Res. 2012;35(12):1121–1128. doi:10.1038/hr.2012.159
14. Karatzas T, Bokos J, Katsargyris A, et al. Advanced donor age alone is not a risk factor for graft survival in kidney transplantation. Transplantat Proc. 2011;43(5):1537–1543. doi:10.1016/j.transproceed.2011.03.014
15. Zhang Y, Li Q, Liu D, et al. GDF11 improves tubular regeneration after acute kidney injury in elderly mice. Sci Rep. 2016;6:34624. doi:10.1038/srep34624
16. Kremers WK, Denic A, Lieske JC, et al. Distinguishing age-related from disease-related glomerulosclerosis on kidney biopsy: the Aging Kidney Anatomy study. Nephrol Dial Transplant. 2015;30:2034–2039. doi:10.1093/ndt/gfv072
17. Marconi L, Figueiredo A, Campos L, et al. Renal transplantation with donors older than 70 years: does age matter? Transplant Proc. 2013;45(3):1251–1254. doi:10.1016/j.transproceed.2013.02.024
18. Galeano C, Marcén R, Jimenez S, et al. Utilization of elderly kidney donors (>70 years) does not affect graft survival in the medium term. Transplant Proc. 2010;42(10):3935–3937. doi:10.1016/j.transproceed.2010.08.069
19. Ding R, Chen X, Wu D, et al. Effects of aging on kidney graft function, oxidative stress and gene expression after kidney transplantation. PLoS One. 2013;8(6):e65613. doi:10.1371/journal.pone.0065613
20. Lim WH, Clayton P, Wong G, et al. Outcomes of kidney transplantation from older living donors. Transplantation. 2013;95(1):106–113. doi:10.1097/TP.0b013e318277b2be
21. Ding L, Pang S, Sun Y, et al. Coordinated actions of FXR and LXR in metabolism: from pathogenesis to pharmacological targets for type 2 diabetes. Int J Endocrinol. 2014;2014:751859. doi:10.1155/2014/751859
22. Zhang X, Chen X, Wu D, et al. Downregulation of connexin 43 expression by high glucose induces senescence in glomerular mesangial cells. J Am Soc Nephrol. 2006;17(6):1532–1542. doi:10.1681/ASN.2005070776
23. Ademowo OS, Dias HKI, Burton DGA, et al. Lipid (per) oxidation in mitochondria: an emerging target in the ageing process? Biogerontology. 2017;18(6):859–879. doi:10.1007/s10522-017-9710-z
24. Zhang N, Li Z, Mu W, et al. Calorie restriction-induced SIRT6 activation delays aging by suppressing NF-κB signaling. Cell Cycle. 2016;15(7):1009–1018. doi:10.1080/15384101.2016.1152427
25. Cui J, Shi S, Sun X, et al. Mitochondrial autophagy involving renal injury and aging is modulated by caloric intake in aged rat kidneys. PLoS One. 2013;8(7):e69720. doi:10.1371/journal.pone.0069720
26. Yang H, Deleuze S, Zuo Y, et al. The PPARγ agonist pioglitazone ameliorates aging-related progressive renal injury. J Am Soc Nephrol. 2009;20(11):2380–2388. doi:10.1681/ASN.2008111138
27. Xu G, Pan L-X, Li H, et al. Regulation of the farnesoid X receptor (FXR) by bile acid flux in rabbits. J Biol Chem. 2002;277(52):50491–50496. doi:10.1074/jbc.M209176200
28. Cao G, Liang Y, Broderick CL, et al. Antidiabetic action of a liver x receptor agonist mediated by inhibition of hepatic gluconeogenesis. J Biol Chem. 2005;433(2):760–766. doi:10.1074/jbc.M210208200
29. Dai Z, Huang Y, Sadee W, et al. Chemoinformatics analysis identifies cytotoxic compounds susceptible to chemoresistance mediated by glutathione and cystine/glutamate transport system xc-. J Med Chem. 2020;6(8):1896–1906. doi:10.1021/jm060960h
30. Huang Y, Dai Z, Barbacioru C, et al. Cystine-glutamate transporter SLC7A11 in cancer chemosensitivity and chemoresistance. Cancer Res. 2005;65(16):7446–7454. doi:10.1158/0008-5472.CAN-04-4267
31. Cantor JM, Ginsberg MH. CD98 at the crossroads of adaptive immunity and cancer. J Cell Sci. 2012;125:1373–1382. doi:10.1242/jcs.096040
32. Poettler M, Unseld M, Braemswig K, et al. CD98hc (SLC3A2) drives integrin-dependent renal cancer cell behavior. Mol Cancer. 2013;12:169. doi:10.1186/1476-4598-12-169
33. Zhou J, Cheng Y, Tang L, et al. Up-regulation of SERPINA3 correlates with high mortality of melanoma patients and increased migration and invasion of cancer cells. Oncotarget. 2017;8(12):18712–18725. doi:10.18632/oncotarget.9409
34. Ziegler ME, Chen T, LeBlanc JF, et al. Apolipoprotein A1 and C-terminal fragment of α-1 antichymotrypsin are candidate plasma biomarkers associated with acute renal allograft rejection. Transplantation. 2011;92(4):388–395. doi:10.1097/TP.0b013e318225db6a
35. Siwy J, Zürbig P, Argiles A, et al. Noninvasive diagnosis of chronic kidney diseases using urinary proteome analysis. Nephrol Dial Transplant. 2017;32(12):2079–2089. doi:10.1093/ndt/gfw337
36. O’Riordan E, Orlova TN, Podust VN, et al. Characterization of urinary peptide biomarkers of acute rejection in renal allografts. Am J Transplant. 2007;7:930–940. doi:10.1111/j.1600-6143.2007.01733.x
37. Chandra T, Stackhouse R, Kidd VJ, et al. Sequence homology between human alpha 1-antichymotrypsin, alpha 1-antitrypsin, and antithrombin III. Biochemistry-Us. 1983;22(22):5055–5061. doi:10.1021/bi00291a001
38. Chan HJ, Li H, Liu Z, et al. SERPINA1 is a direct estrogen receptor target gene and a predictor of survival in breast cancer patients. Oncotarget. 2015;6(28):25815–25827. doi:10.18632/oncotarget.4441
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
