Back to Journals » Infection and Drug Resistance » Volume 13
Dynamic Epidemiology and Virulence Characteristics of Carbapenem-Resistant Klebsiella pneumoniae in Wenzhou, China from 2003 to 2016
Authors Zhao Y, Zhang S, Fang R, Wu Q, Li J, Zhang Y, Rocker A , Cao J, Lithgow T, Zhou T
Received 19 December 2019
Accepted for publication 3 March 2020
Published 31 March 2020 Volume 2020:13 Pages 931—940
DOI https://doi.org/10.2147/IDR.S243032
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sahil Khanna
Yajie Zhao,1,2 Siqin Zhang,1 Renchi Fang,1 Qing Wu,1 Jiahui Li,1 Yizhi Zhang,1 Andrea Rocker,3 Jianming Cao,2 Trevor Lithgow,3 Tieli Zhou1
1Department of Clinical Laboratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, People’s Republic of China; 2School of Laboratory Medicine and Life Science, Wenzhou Medical University, Wenzhou, People’s Republic of China; 3Infection and Immunity Program, Biomedicine Discovery Institute and Department of Microbiology, Monash University, Melbourne, VIC, Australia
Correspondence: Trevor Lithgow; Tieli Zhou Email [email protected]; [email protected]
Purpose: To investigate transitions in resistance mechanisms, virulence characteristics and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae (CRKP) during 2003– 2016 in a major Eastern Chinese medical center.
Patients and Methods: From a total of 2299 K. pneumoniae clinical strains collected from 2003 to 2016, 214 were found to be CRKP isolates and were selected for further study. Characterization of these was conducted by molecular detection of antibiotic resistance markers and virulence determinants, modified carbapenem inactivation method and multilocus sequence typing (MLST).
Results: In this study, the prevalence of CRKP was increasing over the 14-year period, mirroring a national trend. These CRKP strains were resistant to most of the tested, clinically relevant drugs. The majority of these CRKP strains were positive for carbapenemases, with the Klebsiella pneumoniae carbapenemase (KPC) found to be the dominant type (207/210, 98.6%). The carrier rates of virulence genes uge, entB, fimH, mrkD and ureA increased in 2016, while the ybtA, iucA and irp2 showed a relatively constant trend. From MLST data, ST11 (88.8%, 190/214) was the preponderant sequence type (ST), followed by ST15 (1.9%, 4/214) and ST656 (1.4%, 3/214). Several strains with less common STs (ST690, ST895, ST1823 and ST1384) were also detected, and these too showed high levels of antimicrobial resistance.
Conclusion: The average national rise in CRKP across China is mirrored in this in-depth analysis of a single hospital, while the prevalence of hypervirulent CRKP (such as ST15) was relatively low as of 2016. Continuous monitoring is necessary to keep track of CRKP and should include the prospect of newly emerging strains with less common STs and the prospect of detecting carbapenem-resistant, carbapenemase-negative Klebsiella pneumoniae.
Keywords: Klebsiella pneumoniae, carbapenem-resistant, antimicrobial resistance, ST11, epidemiology
Introduction
Klebsiella pneumoniae is currently one of the important opportunistic pathogens in hospitals where it can cause numerous infections, including urinary tract infections, pneumonias, bacteremia, liver abscesses and surgical wound infections.1,2 From the first report (1997) and outbreaks (early 2000s) of carbapenem-resistant K. pneumoniae (CRKP), they have been detected as spreading worldwide.3,4 In 2005, the China Antimicrobial Surveillance Network (CHINET) was established to provide a nationwide monitoring of hospitals responsible for the treatment of a total of 960 million people (http://www.carss.cn/). From the report of CHINET, an upward trend in the appearance of CRKP was observed from 2.4% in 2005 to 13.4% in 20145, which is of grave concern given that carbapenems were considered as the last-line antibiotics for treatment of infections caused by K. pneumonia. This increased detection of CRKP is being seen elsewhere in the world too.1,2,6,7
The lineage of K. pneumoniae shows carbapenem-resistance is by expression of the K. pneumoniae carbapenemase (KPC),8–10 and plasmids that enable KPC-expression are spreading globally.8,11 Molecular epidemiological analyses show the emergence and dissemination of a dominant sequence type 11 (ST11) which is resistant to various antibiotics, especially to carbapenems.6,10,12-15 ST11 has been demonstrated as the predominant clone of KPC-producing K. pneumoniae in Asia13,16 and now in Europe.10 However, another sequence type, ST15, is emerging as a new global threat because it is hypervirulent in addition to being multi-drug resistant.10,17 Hypervirulent isolates causing community-acquired infections were first reported in the 1980s in Taiwan.18–21 They have been shown to be highly invasive, in that they usually cause severe life-threatening infections which are associated with high morbidity and mortality in both hospital and community.20,22,23 Hypervirulence of K. pneumoniae was associated with hypermucoviscosity and diagnostic virulence genes such as rmpA and magA.24 The emergence of hypervirulent lineages carrying antimicrobial resistance genes including carbapenemases is now being increasingly reported: with limited treatment measures and high mortality, they are considered a “super-bug”.18–21
With reports of hypervirulent CRKP increasing rapidly in China,25–29 we sought to deep dive into the details of this emergence using microbiological characterization and patient metadata acquired in a single large hospital in Wenzhou, China from 2003 to 2016. In this study, we identified the resistance mechanism and virulence characteristics of 214 isolates of CRKP from amongst 2299 total isolates of K. pneumoniae associated with infection, and the proportion of isolated CRKP was increasing and reaches up to 20% in the final year of the study (2016). Mirroring the international trends of this period, the vast majority of CRKP strains were found to encode the carbapenemase KPC (98.6%) and the dominant lineage of CRKP was ST11 (88.8%).
Materials and Methods
Clinical Isolates and Identification and Antimicrobial Susceptibility Profiling
The retrospective study was conducted at the First Affiliated Hospital of Wenzhou Medical University, a 3263-bed tertiary hospital serving as the largest health-care centers in the southern province of Zhejiang, China. From 2003 to 2016, a total of 2674 clinical K. pneumoniae isolates were collected from various clinical specimens. Bacterial identification and antimicrobial susceptibility test (ASTs) were performed through the VITEK® automated microbiology analyzer (BioMerieux, Lyons, France). All the isolates were stored in frozen condition at −80°C with 30% glycerol. Any isolate of K. pneumoniae which was non-susceptible to at least one carbapenem was defined as CRKP.30 The minimum inhibitory concentration (MIC) of carbapenem (imipenem, meropenem and ertapenem) of these CRKP was rechecked using the agar dilution method. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as controls for the antimicrobial testing.
Clinical Data Collection
Clinical data including age, gender, hospitalization, clinical diagnosis, antimicrobial therapy, invasive treatments and outcomes were extracted from Electronic Medical Records. Informed consent was obtained from the patients involved in the current study.
Modified Carbapenem Inactivation Method (mCIM)
The modified carbapenem inactivation method (mCIM) was performed based on the CLSI standard (CLSI, 2016) in order to identify the carbapenem producing among the isolates which were not susceptible to one or more carbapenems. A 1-μL loopful experimental isolates were emulsified in 2 mL Tryptic Soy Broth (TSB) and a 10-μg meropenem disk was added to each tube. After 4 h incubation at 37°C, remove the meropenem disk and place on the Mueller Hinton agar (MHA) plate with E. coli ATCC 25922 inoculation. The plate was incubated at 37°C for 18–24 h. K. pneumoniae Kp 1 (which was positive for blaKPC-2) and E. coli ATCC 25922 were used as positive and negative controls, respectively.
Detection of Carbapenem-Resistance Genes and Virulence Factors
DNA was extracted from K. pneumoniae strains by the boiling method31 and used as the template in polymerase chain reactions (PCR). Screening made use of primers specific for the genes encoding carbapenemases (KPC, plasmid-encoded metallo-β-lactamase gene (IMP) and the New Delhi metallo-β-lactamase (NDM)), virulence genes (including rmpA, rmpA2, iucA, irp2, magA, iroD, iroN, kfuBC, wcaG, alls, ybtA, ureA, uge, wabG, entB, fimH and mrkD)32–34 and capsular serotype-specific genes diagnostic for capsule types K1, K2, K5, K20, K54 and K57.35
High levels of capsulation are considered representative of, and potentially contributing to, hypervirulence.36 The extent of capsulation can be diagnostically measured using the string test.36,37 Isolates with a viscous string >5 mm in the string test were considered positive for the hypermucoviscous phenotype as previously described.36
Multilocus Sequence Typing (MLST)
Oligonucleotides representing the primer sequences provided by Institute Pasteur MLST (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html) were used to amplify fragments of seven housekeeping genes (gapA, pgi, rpoB, mdh, phoE, infB and tonB). The sequences from these seven loci define the sequence type (ST) as documented in the MLST online database. Novel STs were submitted to the database for new designations. The classification of clonal complex (CC) was analyzed by eBURST v3 (http://eburst.mlst.net).38 The same clonal complex included at least four STs that share six identical alleles. STs which did not belong to any CC were defined as singletons.
Statistical Analysis
Data were analyzed through the statistical package SPSS for Windows version 22.0 (SPSS Inc., Chicago, IL, USA). The Chi-square test and Fisher’s exact test were used for statistical analysis, p-value of <0.05 was considered as statistically significant, and all tests were two-tailed.
Results
The Collection Characterizations of the CRKP Isolates
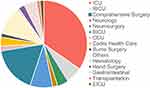
Over the course of 14 years (2003 to 2016), 2334 K. pneumoniae isolates were collected at the First Affiliated Hospital of Wenzhou Medical University (Table 1). Of these, 214 isolates non-repetitive K. pneumoniae were identified as carbapenem-resistant using a VITEK® automated microbiology analyzer. The major clinical specimens harboring CRKP were sputum (102/214, 47.7%), followed by blood (50/214, 23.4%), urine (22/214, 10.3%) and shunt fluid (11/214, 5.1%) (Figure 1A and C). The major wards of which isolates from retrieved were intensive care unit (ICU) (69/214, 32.2%), brain intensive care unit (BICU) (37/214,17.3%), neurosurgery (19/214, 8.9%) and respiratory intensive care unit (RICU) (14/214, 6.5%), which covered 65.0% of these strains (Figure 1B and D).
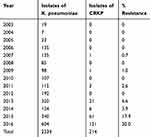 |
Table 1 Collection Summary of CRKP from 2003 to 2016 |
To gain a better understanding of the antimicrobial resistance and virulence characteristics we analyzed the 214 CRKP isolates further.
Molecular Typing Based on MLST
The 214 CRKP belonged to 18 different STs, including a novel ST2740 (Figure 2). The ST11 CRKP was emerging in 2011, and the proportion was increasing over time. Ultimately, ST11 became the predominant ST, accounting for 93.4% (113/121) of isolates in 2016. Over all CRKP, the main sequence type was ST11 (88.8%, 190/214), followed by ST15 (1.9%, 4/214), ST656 (1.4%, 3/214), ST23 (0.9%, 2/214) and ST 893 (0.9%, 2/214). Single isolates were also recovered in 13 other STs (Figure 2). Across all these types, ST11, ST690, ST895 and ST1384 belonged to the same CC (90.2%, 193/214) and the remaining 14 STs were separated into CC singletons by eBURST software. The proportion of ST11 increased constantly over time, from 66.7% (2011) to 93.3% (2016) (Figure 2), indicating its dominance. In contrast to this finding, only four ST15 CRKP were isolated, but since these all occurred in 2016, future monitoring is warranted to determine how this hypervirulent form of K. pneumoniae develops in the next few years.
Antimicrobial Susceptibility Profiles of CRKP Isolates
All the 214 CRKP strains (100%) were resistant to nearly all β-lactam antibiotics tested including aztreonam (ATM), ceftazidime (CAZ), ceftriaxone (CRO), piperacillin-tazobactam (TZP) and ampicillin-sulbactam (SAM) except for the fourth-generation cephalosporin cefepime (FEP) (Figure 3A). However, by 2013, even resistance against FEP was seen to increase to 91% (Figure 3B). A trend of increasing resistance was also observed for the fluoroquinolones ciprofloxacin (CIP) and levofloxacin (LVX), rising from 40% in 2007 to 98% in 2016, and trimethoprim-sulfamethoxazole (SXT) reaching 60% in 2016. In 2014/2015 high resistance rates were observed for the gentamicin (GEN, 83% ~ 85%) and tobramycin (TOB, 83% ~ 89%), but these rates decreased to approximately 62% and 40% by 2016, respectively (Figure 3B and C).
The Major Carbapenem-Resistance Mechanism of CRKP
To identify the genetic mechanism of carbapenem resistance, all 214 CRKP isolates were analyzed for the presence of carbapenemase genes. Indeed, carbapenemase carriage was the main factor leading to resistance with 98.1% of the isolates (210/214) being carbapenemase-positive by PCR (209/214) or mCIM (1/214). Among these isolates, most carried blaKPC-2 (207/210, 98.6%) and among these isolates, two strains (ST15) isolated in 2016 harbored both blaKPC-2 and blaIMP genes. Two isolates (ST2740, 2013 and ST15, 2016) only carried blaIMP.
Of the remaining five isolates, only one strain (F1384) was positive in mCIM, but negative for blaKPC, blaIMP and blaNDM, indicating the expression of an unidentified carbapenemase. The other four (F844, FK688, FK1934 and FK2820) were PCR-negative for the bla genes and were confirmed to be negative for carbapenemase expression using mCIM (data not shown), indicating alternative resistance mechanisms contribute to carbapenem resistance as well.
Virulence Factors and Clinical Characteristic Analysis
In order to explore the virulence characteristics of those CRKP and their dynamics over time, 50 CRKP isolated from blood specimens were selected. These patients were divided into two groups according to the clinical outcomes (Cured & Clinical improvement, Unhealed & In-hospital mortality). All the outcomes were defined based on clinical diagnosis and patient status before they discharged. The data for strains isolated from 2003 to 2015 were combined to provide a base-line because of the small annual number identified in these years. Seventeen virulence genes and K-serotype-specific alleles were identified by PCR and sequencing, and the carrier rates of virulence factors grouped by the differing clinical outcomes and years were documented (Table 2).
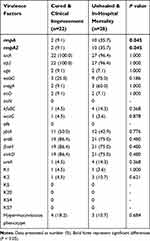 |
Table 2 The Carrier Rates of Virulence Factors in CRKP from Blood Samples Among Different Outcome Groups |
Among the 50 isolates, rmpA/A2 showed significant difference on different clinical outcomes (P = 0.045), and the carrier rates in the groups which patients were failed for treatment (10/28, 35.7%) were higher than the patients with improving quality of the care (2/22, 9.1%), indicating a negative impact of rmpA/A2 on treatment effect. The other virulence genes which usually were regarded as associated with hypervirulence such as iucA (encoding aerobactin synthetases), ybtA and irp2 (encoding yersiniabactin synthetases) were relatively constant across different groups. Unhealed & In-hospital mortality group exhibited the highest carrier rate of wabG (9/28, 75.0%), magA (3/28, 60.0%), kfuBC (4/28, 14.3%) and ureA (4/28, 14.3%), which may relate to the bacterial virulence. Some capsule types common in China, such as K5, K20, K54, K57, were not identified in this study (Table 2). The capsule types K1 and K2 which are associated with virulence were detected in some isolates. Among these isolates, the carrier rate of K1 was similar in two groups (4.5%: 3.6%), while more isolates from untreated patients were positive for K2 (3/28, 10.7%). Seven strains (14.0%) were positive for string test and presented a hypermucoviscous phenotype, but these were mostly associated with improving outcomes (4/22, 18.2%). All strains were found to be negative for several virulence-related genes such as alls and iroN (Table 2).
As documented in Table 3, kfuBC, entB, fimH and mrkD varied across different years (P = 0.020, 0.000, 0.000, 0.000, respectively). Among them, the 2007–2015 group showed a significant difference with the highest carrier rates of all the genes mentioned above. The carrier rates of virulence genes uge (from 4.2% to 11.5%) and ureA (from 4.2% to 15.4%) increased in 2016. The prevalence of entB, fimH and mrkD increased in 2016 to reach 100%. ybtA (~44.0%), iucA (~97.9%), irp2 (~98.1%) were constant during this period. K1 and K2 remained a stable tendency among these years. The virulence genes which were associated with In-hospital mortality (wabG, magA, kfuBC and ureA) showed a dynamic change from 2007 to 2016, for example, wabG and ureA had a slight increasing trend while the magA and kfuBC diminished in 2016.
 |
Table 3 The Carrier Rates of Virulence Factors in CRKP from Blood Samples Isolated Across 2007 to 2016 |
Discussion
The emergence of carbapenem-resistant Enterobacteriaceae (CRE) strains is a major burden for public health-care systems,39,40 with long-term surveillance studies identifying emerging high-risk strains of CRKP.41,42 Here, we analyzed a 14-year time-window for trends in resistance phenotypes, virulence factors and sequence profiles of CRKP isolates from a major hospital in Wenzhou. This study enabled an in-depth look at a single major hospital within the time-frame of the national surveillance averages gathered through CHINET.5
In this hospital, CRKP was first isolated in 2007, and thereafter the proportion of CRKP among the total K. pneumoniae isolates increased from 0.7% (2007) to 20.0% in 2016. The dominant cause in carbapenem-resistance in this time period was via strains carrying carbapenemase resistance genes, as 98.6% of all CRKP were carbapenemase-positive. Similar trends are evident in surveillance studies in China5 and Europe,10 with a likely cause being selective pressure due to the widespread use of carbapenems.43,44 KPC-2 identified in 98.6% of CRKP strains in our study also represents the predominant carbapenemase in Asia.45 In our hospital, prescription of carbapenems (meropenem, imipenem and imipenem/cilastatin sodium) was introduced early in the twenty-first century and used as the last-line antibiotics in treatment for patients with impaired immune function. However, five strains were negative for the bla gene detection and only one strain was positive for the carbapenemase production via mCIM experiment, which means the other four CRKP (F844, FK688, FK1934 and FK2820) have other mechanisms for carbapenem resistance. The possible explanations are showed below: (i) there exist other carbapenemase genes such as other β-lactamase, AmpC enzyme or ESBLs,46 (ii) there were mutations in or loss of outer membrane porins (especially for the OmpK35 and OmpK36),47 or (iii) there exists the active drug efflux. Among these four strains, FK2820 lacked the OmpK36 porins in the previous study (data not published), which may cause the carbapenem resistance. And the relative detections of other stains could be performed in a further study.
Worryingly, all CRKP isolated in 2016 were resistant to all β-lactam and β-lactam/β-lactamase inhibitor combinations tested. In addition, increasing resistance rates were observed for fluoroquinolones and trimethoprim-sulfamethoxazole (SXT) in these strains, yielding multi-drug resistant strains that are increasingly difficult to treat. Similar effects have been reported previously.48 As for SXT, a fluctuant pattern was observed from 2013 to 2015, indicating strictly drug application strategy was performed during this period but the rate rebounded in 2016 according to the selective pressure in the clinic. On a positive note, the prevalence of resistance to aminoglycosides decreased, from around 85% in 2015 (84% for GEN and 86% for TOB) to less than 65% in 2016 (62% for GEN and 40% for TOB). This can potentially be explained by the adjustment of antibiotic usage strategies in infection treatment: in this hospital, clinical use of GEN/TOB was reduced due to the toxicity in the inner ear and kidney. There are insufficient data that are yet to establish a causal link, but the data are consistent with changes in drug-resistance profiles that show population structures can change towards sensitivity to existing antimicrobials, and that withdrawal of drug use may play a role in this.
The major ST within the CRKP isolates identified in our study was ST11 (93.4%), consistent with previous investigations showing that ST11 rose to be the predominant clone of CRKP in Asia.49 Of the remaining 17 ST, ST15 (FK 2836, FK 3006 and FK 3020) emerged in 2016 and is of growing concern given its characterization as both multidrug resistant and hypervirulent,10 which lead to the unfavorable prognosis of two patients and in-hospital mortality of one patient.
Previously, the phenotype “hypermucoviscous” was defined as a representation for hypervirulence in K. pneumoniae, which related to poor prognosis.50,51 Our study adds weight to the growing understanding18,52 that there is no simple relationship between the capsule phenotype and virulence: the hypermucoviscous strains identified in this study were predominantly associated with the “cured & improvement” group. The rmpA/A2 (responsible for upregulation of the synthesis of capsular polysaccharide),53 consistent with the emergence of the isolates with hypermucoviscous type, suggesting these CRKP got more capsule later. The isolates from patients who failed the treatment seem to have higher carrier rates of rmpA/A2 (P= 0.045) compared to the clinical improving group. These two genes were regarded as one of the most important virulence factors due to the activation of cps transcription,53 which were consistent with the results of our study. K1 and K2 have a strong association with hypervirulence of K. pneumoniae,54 and genes associated with these serotypes were prevalent in the “Unhealed & In-hospital mortality” group of isolates, but so too were other genes designated as promoting virulence, including iron acquisition relative genes (iucA, irp2, iroD, kfuBC),18,55 capsule production genes (wabG, magA)32 and ureA (nitrogen source in host).56 When analyzed across the 14-year time-window, virulence profiles were seen to change. Although the carrier rates of some virulence genes such as irp2, magA, iroD, ybtA, wcaG and kfuBC illustrated slight decrease, other genes such as entB, fimH and mrkD presented an obvious increased trend from 58.3% (14/24) to 100% (26/26) in 2016, suggesting the possible virulence enhancement among CRKP. Interestingly, the carrier rates of siderophores genes such as iucA (~97.9%) and irp2 (~98.1%) among all these strains were relatively high. These genes were usually regarded as the markers of hypervirulence,16,18,26,55 but our data showed that these virulence genes presented in very early year in the same strains, which hinted the possibly wrong markers we used to identify the hypervirulent K. pneumoniae in the clinical and better gene markers should be discussed for monitoring the clinical outcomes in the further study.
What’s more, due to the iro, iuc and rmpA/A2 loci were usually found to exist on virulence plasmid, the relatively stable carrier rates of these genes (rmpA/A2, iucA, iroN) in our study over time indicated the potential transmission of virulence plasmid like pLVPK,57,58 which might need be focused on in the further study.
Conclusion
In conclusion, the resistance mechanism, virulence factors and molecular epidemiology of CRKP during 2003 to 2016 were explored. The primary carbapenemase resistance mechanism (KPC-production) and ST type revealed the potential dissemination of ST11 CRKP in hospital. In addition to ST11, attention should also be paid to some less common STs with high degrees of antimicrobial resistance. Furthermore, the virulence-associated gene profile across 14 years suggested the potential transmission of virulence plasmids and effective strategies should be constantly applied in order to push back against this latent threat to the future.
Ethics and Consent Statement
No samples were collected specifically for this research; only anonymized clinical residual samples collected during routine hospital procedures were used for this study. This study has been designed in accordance with the Declaration of Helsinki (2013) (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/) and been approved by the First Affiliated Hospital of Wenzhou Medical University.
Disclosure
Dr Andrea Rocker reports grants from National Health and Medical Research Council of Australia, during the conduct of the study. The authors report no other conflicts of interest in this work.
References
1. Paczosa M, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80:629–661.
2. Navon-venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–275. doi:10.1093/femsre/fux013
3. MacKenzie FM, Forbes KJ, Dorai-john T, Amyes SG, Gould IM. Emergence of a carbapenem-resistant Klebsiella pneumoniae. Lancet. 1997;350:783. doi:10.1016/S0140-6736(05)62567-6
4. Arnold RS, Thom KA, Sharma S, Phillips M, Kristie Johnson J, Morgan DJ. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J. 2011;104:40–45. doi:10.1097/SMJ.0b013e3181fd7d5a
5. Hu FP, Guo Y, Zhu DM, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin Microbiol Infect. 2016;22(Suppl 1):S9–S14. doi:10.1016/j.cmi.2016.01.001
6. Pitout J, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59:5873–5884. doi:10.1128/AAC.01019-15
7. Tzouvelekis L, Markogiannakis A, Psichogiou M, Tassios P, Daikos G. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25:682–707. doi:10.1128/CMR.05035-11
8. Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–696. doi:10.1016/j.tim.2014.09.003
9. Chen S, Hu F, Xu X, et al. High prevalence of KPC-2-type carbapenemase coupled with CTX-M-type extended-spectrum beta-lactamases in carbapenem-resistant Klebsiella pneumoniae in a teaching hospital in China. Antimicrob Agents Chemother. 2011;55:2493–2494. doi:10.1128/AAC.00047-11
10. David S, Reuter S, Harris SR, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4(11):1919–1929. doi:10.1038/s41564-019-0492-8
11. Zhan L, Wang S, Guo Y, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol. 2017;7:182. doi:10.3389/fcimb.2017.00182
12. Bi W, Liu H, Dunstan RA, et al. Extensively drug-resistant Klebsiella pneumoniae causing nosocomial bloodstream infections in China: molecular investigation of antibiotic resistance determinants, informing therapy, and clinical outcomes. Front Microbiol. 2017;8:1230. doi:10.3389/fmicb.2017.01230
13. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66:307–312. doi:10.1093/jac/dkq431
14. DSouza R, Pinto NA, Hwang I, et al. Molecular epidemiology and resistome analysis of multidrug-resistant ST11 Klebsiella pneumoniae strain containing multiple copies of extended-spectrum β-lactamase genes using whole-genome sequencing. New Microbiol. 2017;40:38–44.
15. Lee C, Lee J, Park K, Kim Y, Jeong B, Lee S. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. doi:10.3389/fmicb.2016.00895
16. Struve C, Roe C, Stegger M, et al. Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio. 2015;6:e00630. doi:10.1128/mBio.00630-15
17. Caneiras C, Lito L, Melo-cristino J, Duarte A. Community- and hospital-acquired Klebsiella pneumoniae urinary tract infections in portugal: virulence and antibiotic resistance. Microorganisms. 2019;7(5):138. doi:10.3390/microorganisms7050138
18. Catalán-nájera J, Garza-ramos U, Barrios-camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8:1111–1123. doi:10.1080/21505594.2017.1317412
19. Chew KL, Lin RTP, Teo JWP. Klebsiella pneumoniae in Singapore: hypervirulent infections and the carbapenemase threat. Front Cell Infect Microbiol. 2017;7:515. doi:10.3389/fcimb.2017.00515
20. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46. doi:10.1016/S1473-3099(17)30489-9
21. Shon AS, Russo TA. Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol. 2012;7:669–671. doi:10.2217/fmb.12.43
22. Gu D, Lv H, Sun Q, Shu L, Zhang R. Emergence of tet(A) and blaKPC-2 co-carrying plasmid from a ST11 hypervirulent Klebsiella pneumoniae isolate in patient’s gut. Int J Antimicrob Agents. 2018;52:307–308. doi:10.1016/j.ijantimicag.2018.06.003
23. Yu F, Lv J, Niu S, et al. Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (sequence type 258 [ST258] and ST11) and hypervirulent (ST23, ST65, ST86, and ST375) strains. J Clin Microbiol. 2018;56. doi:10.1128/JCM.00731-18
24. Shankar C, Veeraraghavan B, Nabarro LEB, Ravi R, Ragupathi NKD, Rupali P. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 2018;18:6. doi:10.1186/s12866-017-1148-6
25. Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58:225–232. doi:10.1093/cid/cit675
26. Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann Clin Microbiol Antimicrob. 2019;18:4. doi:10.1186/s12941-018-0302-9
27. Wei DD, Wan LG, Deng Q, Liu Y. Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in Mainland China. Diagn Microbiol Infect Dis. 2016;85:192–194. doi:10.1016/j.diagmicrobio.2015.03.012
28. Xu M, Fu Y, Fang Y, et al. High prevalence of KPC-2-producing hypervirulent Klebsiella pneumoniae causing meningitis in Eastern China. Infect Drug Resist. 2019;12:641–653. doi:10.2147/IDR.S191892
29. Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60:709–711. doi:10.1128/AAC.02173-15
30. Braykov NP, Eber MR, Klein EY, Morgan DJ, Laxminarayan R. Trends in resistance to carbapenems and third-generation cephalosporins among clinical isolates of Klebsiella pneumoniae in the United States, 1999-2010. Infect Control Hosp Epidemiol. 2013;34:259–268. doi:10.1086/669523
31. Feizabadi MM, Delfani S, Raji N, et al. Distribution of blaTEM, blaSHV, blaCTX-M genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran, Iran. Microbial Drug Resist. 2010;16:49–53. doi:10.1089/mdr.2009.0096
32. Candan E, Aksöz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62:867–874. doi:10.18388/abp.2015_1148
33. Luo Y, Wang Y, Ye L, Yang J. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect. 2014;20:O818–O824. doi:10.1111/1469-0691.12664
34. Zhang S, Yang G, Ye Q, Wu Q, Zhang J, Huang Y. Phenotypic and genotypic characterization of isolated from retail foods in China. Front Microbiol. 2018;9:289. doi:10.3389/fmicb.2018.00289
35. Wasfi R, Elkhatib W, Ashour H. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;6:38929. doi:10.1038/srep38929
36. Liu Y, Li B, Zhang Y, et al. Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agents Chemother. 2014;58:5379–5385. doi:10.1128/AAC.02523-14
37. Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. 2015;37:107–112. doi:10.1016/j.ijid.2015.06.023
38. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi:10.1128/JB.186.5.1518-1530.2004
39. Li Y, Sun QL, Shen Y, et al. Rapid increase in prevalence of carbapenem-resistant Enterobacteriaceae (CRE) and emergence of colistin resistance gene mcr-1 in CRE in a hospital in Henan, China. J Clin Microbiol. 2018;56. doi:10.1128/JCM.01932-17
40. Xu Y, Gu B, Huang M, et al. Epidemiology of carbapenem resistant Enterobacteriaceae (CRE) during 2000-2012 in Asia. J Thorac Dis. 2015;7:376–385. doi:10.3978/j.issn.2072-1439.2014.12.33
41. Demiraslan H, Cevahir F, Berk E, Metan G, Cetin M, Alp E. Is surveillance for colonization of carbapenem-resistant gram-negative bacteria important in adult bone marrow transplantation units? Am J Infect Control. 2017;45:735–739. doi:10.1016/j.ajic.2017.01.006
42. Richter SS, Marchaim D. Screening for carbapenem-resistant Enterobacteriaceae: who, When, and how? Virulence. 2017;8:417–426. doi:10.1080/21505594.2016.1255381
43. Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3:15–21. doi:10.1177/2049936115621709
44. Pavez M, Vieira C, de Araujo MR, et al. Molecular mechanisms of membrane impermeability in clinical isolates of Enterobacteriaceae exposed to imipenem selective pressure. Int J Antimicrob Agents. 2016;48:78–85. doi:10.1016/j.ijantimicag.2016.04.016
45. Li H, Zhang J, Liu Y, et al. Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn Microbiol Infect Dis. 2014;78:63–65. doi:10.1016/j.diagmicrobio.2013.10.002
46. Miao M, Wen H, Xu P, et al. Genetic diversity of Carbapenem-Resistant Enterobacteriaceae (CRE) clinical isolates from a tertiary hospital in Eastern China. Front Microbiol. 2018;9:3341. doi:10.3389/fmicb.2018.03341
47. Du J, Cao J, Shen L, et al. Molecular epidemiology of extensively drug-resistant Klebsiella pneumoniae outbreak in Wenzhou, Southern China. J Med Microbiol. 2016;65:1111–1118. doi:10.1099/jmm.0.000338
48. Hagiya H, Aoki K, Akeda Y, et al. Nosocomial transmission of carbapenem-resistant Klebsiella pneumoniae elucidated by single-nucleotide variation analysis: a case investigation. Infection. 2017;45:221–225. doi:10.1007/s15010-017-0986-3
49. Jiang Y, Wei Z, Wang Y, Hua X, Feng Y, Yu Y. Tracking a hospital outbreak of KPC-producing ST11 Klebsiella pneumoniae with whole genome sequencing. Clin Microbiol Infect. 2015;21:1001–1007. doi:10.1016/j.cmi.2015.07.001
50. Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199:697–705. doi:10.1084/jem.20030857
51. Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45:284–293. doi:10.1086/519262
52. Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56. doi:10.1128/JCM.00776-18
53. Hsu C, Lin T, Chen Y, Chou H, Wang J. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology. 2011;157:3446–3457. doi:10.1099/mic.0.050336-0
54. Zhang Y, Zhao C, Wang Q, et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. 2016;60:6115–6120. doi:10.1128/AAC.01127-16
55. de Cassia Andrade Melo R, de Barros EM, Loureiro NG, et al. Presence of fimH, mrkD, and irp2 virulence genes in KPC-2-producing Klebsiella pneumoniae isolates in Recife-PE, Brazil. Curr Microbiol. 2014;69:824–831. doi:10.1007/s00284-014-0662-0
56. Mathers A, Peirano G, Pitout J. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28:565–591. doi:10.1128/CMR.00116-14
57. Harada S, Aoki K, Ishii Y, et al. Emergence of IMP-producing hypervirulent Klebsiella pneumoniae carrying a pLVPK-like virulence plasmid. Int J Antimicrob Agents. 2019;53:873–875. doi:10.1016/j.ijantimicag.2019.05.007
58. Lam MMC, Wyres KL, Judd LM, et al. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018;10:77. doi:10.1186/s13073-018-0587-5
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.