Back to Journals » International Journal of Nanomedicine » Volume 12
Development of gold nanoparticles coated with silica containing the antibiofilm drug cinnamaldehyde and their effects on pathogenic bacteria
Authors Ramasamy M , Lee JH, Lee J
Received 19 January 2017
Accepted for publication 10 March 2017
Published 6 April 2017 Volume 2017:12 Pages 2813—2828
DOI https://doi.org/10.2147/IJN.S132784
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Mohankandhasamy Ramasamy, Jin-Hyung Lee, Jintae Lee
School of Chemical Engineering, Yeungnam University, Gyeongsan, Republic of Korea
Abstract: Emerging resistance to antibiotics is a mounting worldwide health concern and increases the need for nonantibiotic strategies to combat infectious diseases caused by bacterial pathogens. In this study, the authors used the antibiofilm activity of the naturally occurring antimicrobial cinnamaldehyde (CNMA) conjugated to the surface of gold nanoparticles (GNPs) to deliver CNMA efficiently and eradicate biofilms of Gram-negative organisms (enterohemorrhagic Escherichia coli O157:H7, and Pseudomonas aeruginosa), Gram positive (methicillin-sensitive Staphylococcus aureus organisms, and methicillin-resistant Staphylococcus aureus) bacteria. CNMA-GNPs containing 0.005% (v/v) of CNMA were found to inhibit biofilm formation efficiently. The distributions of nanoparticles in biofilm cells and their biofilm disruption activities, including distorted cell morphology, were determined by transmission electron microscopy. In addition to their antibiofilm activities, CNMA-GNPs attenuated S. aureus virulence and protected Caenorhabditis elegans (C. elegans) worms. Here, the authors report the antibiofilm effects of CNMA-GNPs and suggest that they could be used to treat pathogenic bacterial infections in vivo.
Keywords: antibiofilm, biocompatible, Caenorhabditis elegans, cinnamaldehyde, gold nanoparticles, Staphylococcus aureus
Introduction
Infectious diseases are leading causes of death worldwide and have high morbidity and mortality rates.1,2 Antimicrobial resistance and lack of effective broad-spectrum antibiotics exacerbate this situation. More than 80% of infections arise from microbial biofilms,3 that is, organized colonies bound by secreted extracellular polymeric substances (EPSs) on abiotic or biotic surfaces, which are responsible for persistent disease and biomedical device-related infections. Furthermore, EPSs protect individual cells from conventional antibiotics, nutrient limitations, and immunologic host defense systems.4 Antimicrobial resistance usually occurs due to the failure of antibiotics to penetrate biofilms.5,6 Many harmful biofilms found on medical devices or wound surfaces are difficult to eradicate using simple surface disinfectants, and thus, their control and treatment are urgently required. Clearly, developing novel antibacterial agents that penetrate EPSs and destroy pathogenic biofilms are required to address these infections. In the present study, nanotechnology was used to develop a new means of effectively combating biofilm infections.
The production of antimicrobial agents embedded in nanomaterials provides a potential platform to prevent payload degradation and circumvent other limitations, such as, poor delivery of water-insoluble drugs, unsatisfactory drug uptake, excessive drug efflux, and acquired resistance of infectious microbes.7–9 Because of their unique optical, electrical, and physicochemical characteristics, nanomaterials and functionalized nanomaterials have attracted the attentions of researchers for the synthesis, biolabeling, imaging, diagnosis, and therapeutic delivery of antibacterial agents.10,11 Moreover, the conjugation of antimicrobial drugs with nanomaterials protects these drugs from surrounding environments and helps ensure the delivery of high concentrations payload drugs at targeted sites.12,13 Because of their inertness, biocompatibility, and ease of functionalization, gold nanoparticles (GNPs) are viewed as effective nanocarriers and have been utilized in this study.14,15
Increasing antibiotic and silver resistance by bacteria are the leading causes to select alternative antibacterials that will stop the spread of resistant genes. Recently, antimicrobial effects of essential oils have been applied widely, especially against multidrug resistant microorganisms.16,17 The antibacterial mechanism of essential oil is complex, and so the development of resistance by the bacteria is more difficult. In particular, these “green” antimicrobials are cheap, biocompatible, and potent biofilm inhibitors.18,19 Cinnamaldehyde (CNMA), which is present in cinnamon bark, is a major antimicrobial component and has also been found to have diverse biological effects, which include anticancer effects.20–22 Mechanistically, CNMA, decreases metabolic activity, coagulates cytoplasmic materials and limits the replication rate in P. aeruginosa, inhibits swimming motility in E. coli, inhibits quorum sensing, interferes with the autoinducer-2 (AI-2) system, and decreases biofilm formation in S. aureus at subinhibitory concentrations. Therefore, the use of CNMA at low concentrations could exert antibiofilm activity, which could prevent bacteria colonization.23 However, the clinical use of CNMA is made difficult due to its instability, poor aqueous solubility, poor dispersability, toxicity, and because it is difficult to deliver it to target sites at required concentrations.24–26 The delivery of CNMA encapsulated in nanocarriers could overcome these problems, but the antibiofilm activity of CNMA conjugated on GNPs has not been previously explored.
Herein, we describe the conjugation of CNMA on an inorganic nanomaterial and its treatment efficiency against bacterial biofilms. CNMA incorporated in the silica coating and attached on GNPs had potent antimicrobial activity on biofilms. Biofilm inhibition by CNMA-GNPs and their antibacterial effects were visualized by confocal laser scanning microscopy (CLSM) and transmission electron microscopy (TEM). Furthermore, CNMA-GNPs showed enhanced biocompatibility and therapeutic effects in infected C. elegans.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains used were enterohemorrhagic Escherichia coli (E. coli) O157:H7 [ATCC 43895, EDL933], Pseudomonas aeruginosa (P. aeruginosa PAO1), methicillin-sensitive Staphylococcus aureus (MSSA 6538) [ATCC 6538], and methicillin-resistant Staphylococcus aureus (MRSA) strain [ATCC BAA-1707]. Bacterial experiments were performed at 37°C in Luria–Bertani (LB) medium for E. coli, P. aeruginosa, and S. aureus MSSA strains and in LB medium containing 0.2% glucose for S. aureus MRSA strain. For phenotypic assays, stationary phase cells were reinoculated into LB medium at an initial turbidity of 0.05 (OD600). All reagents and materials used in this study were purchased from Sigma-Aldrich (St Louis, MO, USA) and used as received.
Synthesis of cinnamaldehyde-conjugated gold nanoparticles (CNMA-GNPs)
Initially, GNPs were synthesized as previously described with minor modifications.27 Next, Tween 80 was suspended in a mixture of ethanol and propanediol, 20 μL CNMA, and 0.1 M tetraethyl orthosilicate (TEOS) were added consecutively and vortexed for 10 min. The cleaned GNPs (0.1%) were then added dropwise into this mixture with vigorous vortexing until a clear dispersion was obtained and then the reaction was continued for an additional 30 min. Further, the prepared nanodispersion was sonicated for 30 min and allowed to rest overnight at 4°C prior to use. Finally, differential centrifugal sedimentation was utilized to separate CNMA-loaded GNPs from uncoated GNPs. The same procedure without CNMA was used to produce silica-coated gold nanoparticles (Si-GNPs).
Characterizations of nanoparticles
The absorbance spectra of the synthesized nanoparticles were recorded between 180 and 700 nm using an UV-Visible spectrophotometer (UV-1800, Shimadzu, Kyoto, Japan). After appropriate dilutions, the impact of surface CNMA conjugation on GNPs was investigated and compared.
A Zetasizer Nano ZS dynamic light-scattering (DLS) analyzer (Malvern Instruments, Malvern, UK) was used to determine average particle sizes (nm) and zeta potentials (mV) of the nanoparticles. Samples were diluted 1,000 times with ultrapure water and results were obtained by averaging 15 runs. Samples were sonicated prior to DLS measurements.
Infrared spectra were recorded on a Perkin-Elmer Spectrum Two® instrument by attenuated total reflectance Fourier transform infrared spectroscopy (ATR-FT-IR) using a spectrometer with a resolution of 4 cm−1 (Perkin-Elmer Inc., Norwalk, CT, USA). A minimum of sixteen scans were collected and processed using Spectrum 10™ software (Perkin-Elmer Inc.).
The crystal structures of nanoparticles-thin film were determined using an X-ray powder diffractometer (XRD, PANalytical, Almelo, the Netherlands) and Cu Kα radiation (λ=0.1518 nm) at 40 kV and 30 mA in the range of 10°–90° and compared with those of standard compounds in the Joint Committee on Powder Diffraction Standards (JCPDS) databank.
For X-ray photoelectron spectroscopy (XPS), thin films were made by drop-casting nanoparticle dispersions on silicon wafers and drying at 80°C to remove residual solvents. XPS measurements were performed using the ESCALAB 250 XPS System (Thermo Fisher Scientific, Ashford, UK) featuring monochromatized Al Kα X-rays (hν =1,486.6 eV); for calibration purposes, the binding energy of the C 1s peak was set at 284.6 eV. After Shirley-type background subtraction, chemically distinct species of core-level spectra were resolved using Gaussian–Lorentzian shape lines and a nonlinear least-squares fitting procedure using CasaXPS software.
Morphologies and surface modifications of synthesized samples were investigated by high-resolution transmission electron microscopy (HR-TEM, Tecnai G2 F20, FEI, Hillsboro, OR, USA) at an accelerating voltage of 200 kV. Initially, aqueous dispersions of samples were drop cast onto 300-square mesh carbon coated TEM grids and dried under a UV lamp.
Amounts of CNMA in nanodispersions were determined by UV-Vis spectroscopy at 290 nm. The standard calibration curve obtained was linear in the range of 0.00002%–0.002% (v/v) with a correlation coefficient of R2=0.999. The amount of CNMA conjugated was calculated from the ratio of the amount of CNMA on GNPs to the total amount of CNMA added to dispersion.
In vitro CNMA release was analyzed using a dialysis method. As-prepared CNMA-GNPs were placed in the dialysis membrane tubing (Spectra/Por; MWCO 3,500 Da, Rancho Dominguez, CA, USA), which was then immersed in sodium acetate buffer (0.05 M) having either pH 5.0 or pH 7.4. Samples were withdrawn at predetermined time intervals and the amounts of CNMA released was quantified by UV-Vis absorbance at 290 nm.28
Biofilm formation assay
Biofilm formation assays were performed on 96-well polystyrene plates (SPL Life Sciences, Korea), as previously reported.29 Briefly, overnight cultures were adjusted to an OD600 of 0.05 in LB medium and cocultured with CNMA or GNP dispersions without or with CNMA for 24 h without shaking at 37°C. Cell growth was assessed by measuring absorbance at OD620 using a Thermo Scientific Multiskan EX (Thermo Fisher Scientific, Vantaa, Finland). Plates were rinsed with copious water to remove planktonic cells and stained with 300 μL of crystal violet (0.1%, v/v) for 20 min at room temperature. Plates were then emptied, washed with water, blotted onto tissue paper towels, and dried. The crystal violet was then extracted using ethanol (95%, v/v), and total biofilm formation was measured at OD570. Each experiment was performed using three independent cultures. Results are the averages and standard deviations (SD) of three replicate tests. The significance of biofilm inhibition was determined using the Student’s t-test and significance was accepted for P-values of <0.05.
Evaluation of biofilm growth by confocal microscopy
CLSM was performed using a Nikon Eclipse Ti (Tokyo, Japan) microscope on MSSA and MRSA biofilms in the presence of CNMA, Si-GNPs, or CNMA-GNPs.30 In addition, biofilm formation by both strains was compared when they were grown in medium alone in 96-well polystyrene plates. Bacteria were stained with carboxyfluorescein diacetate succinimidyl ester (Invitrogen, Molecular probes Inc., Eugene, OR, USA) and visualized at a magnification of 20× using an Ar laser with excitation and emission wavelengths of 488 nm and 500–550 nm, respectively. All confocal images of the same strains were captured under the same conditions. Colored 2D sections were reconstructed into 3D images of biofilms using NIS-Elements C version 3.2 (Nikon Eclipse).
Ultrastructural analysis of nanoparticle treated MSSA 6538 biofilms
Synthesized nanoparticle-treated and untreated biofilms of MSSA 6538 were examined by TEM. Initially, overnight grown cells in 96-well plates were incubated with nanodispersions for 24 h without shaking at 37°C. For prefixation, cells were then treated with aldehyde mixture (glutaraldehyde 2.5% and formaldehyde 2%) and kept overnight at 4°C. After washing, cells were harvested by sonication to disrupt biofilms, immediately postfixed using 2% osmium tetroxide overnight at 4°C, and washed with 0.2 M phosphate buffer. Cell blocks were made using 2% agarose in sterile tubes, which were then sliced to the desired size. Specimens were dehydrated in an ethanol series and embedded in an Epon resin mixture (Electron Microscopy Sciences, Hatfield, PA, USA), which was treated at different temperatures to ensure complete polymerization. Thin sections were obtained using a MT-X ultramicrotome (Boeckeler Instruments, Tucson, AZ, USA) and loaded onto TEM copper grids. Finally, sections were stained with 1% uranyl acetate followed by lead citrate to remove any traces of moisture. Microscopy was performed using an H-7600 electron microscope (Hitachi Ltd., Tokyo, Japan) at 120-keV.
Survival assay using C. elegans
We assessed and evaluated the effects of CNMA-GNPs at 0.005% v/v on C. elegans especially with respect to survival against infection. As a toxicity assay, the impact of CNMA-GNPs on the viability of nematodes was also evaluated after ingestion of nanoparticles at different times. Adult populations of fer-15; fem-1 worms (n=20) were infected with lawns of MSSA 6538 on NGM plates which were preincubated with or without CNMA-GNPs. Infection-free C. elegans plates were prepared after feeding E. coli OP50 (considered a control strain) as a common food source. Nematode plates were incubated at 25°C, and C. elegans viability was monitored for about 3 weeks or until complete reduction of the infected nematode population. To count live worms, plates were gently tapped. Three independent experiments (n=60) were performed. Results are expressed as percentage survival versus nontreated worms.
Microscopic assessment of nanoparticle internalization
To investigate nano–bio interactions, starved C. elegans were fed on the lawns of nanodispersions mixed MSSA 6538 suspensions on NGM plates for 24 h. Animals were monitored closely at 1 h intervals and then transferred and imaged (with or without anaesthetization) using the iRiS™ Digital Cell Imaging System (Logos Biosystems, Anyang, Korea) equipped with an inverted Plan Achromatic 20× or 40× Ph objective lens. Worms were imaged using a monochrome CMOS camera at 20 or 40×. iRiS™ LED filter cubes (RFP filter) with excitation and emission wavelengths of 530/40 nm and 605/55 nm, respectively, were utilized to capture the phase contrast images of nanoparticles in C. elegans.
Results and discussion
Fabrication and characterizations of CNMA-GNPs
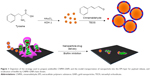
This study describes the processes of synthesizing CNMA-GNPs to be used as an antibiofilm agent as sketched in Figure 1. Initially, GNPs were synthesized by adding gold chloride solution to a tyrosine-KOH mixture, which was then boiled for 3 min to form tyrosine-capped GNPs. The functional phenolic group of tyrosine acted as a reducing group and facilitated the formation of GNPs surface functionalized with oxidized tyrosine molecules, which acted to stabilize GNPs in aqueous solution. Surface amines on GNPs were then reacted with the aldehyde groups of CNMA in the presence of ethanol to form imine linkage. Further surface functionalizations with Tween 80 and TEOS enabled CNMA loading in the silica layer to produce CNMA-GNPs nanodispersions (Figure 1). Because silica is biocompatible,31 it was chosen to stabilize conjugated CNMA on GNPs and to protect the incorporated CNMA. TEM was used to evaluate the core size, while DLS was used to measure the hydrodynamic sizes of nanoparticles. GNPs, Si-GNPs, and CNMA-GNPs were found to have average hydrodynamic particle sizes of 20±6, 155±30, and 326±48 nm, respectively. DLS measurements of Si-GNPs and CNMA-GNPs revealed they were significantly larger than GNPs, which was possibly explained by surface functionalization and CNMA incorporation. The mean Zeta potential of GNPs was −36±1 mV, and after surface modifications it increased to −32±1 for Si-GNPs and −45±1 mV for CNMA-GNPs. The high negative surface charge of CNMA-GNPs was attributed to the incorporation of CNMA and the silica coating, which substantially increased the stabilities of CNMA-GNP nanodispersions.
The optical properties of prepared nanodispersions were investigated by UV-visible spectroscopy as they exhibited unique absorption bands at specific wavelengths. Surface plasmon resonance (SPR) is sensitive to nanoparticle size, shape, interparticle distance, and the surrounding microenvironment.32 Surface-modified GNPs (Figure 2A) showed alterations in their optical properties and exhibited two or three bands with bathochromic shifts in spectra. Si-GNPs exhibited a minimal shift to 530 nm typical of a thin surface coating of silica, whereas CNMA-GNPs showed a larger absorption shift to 540 nm. This shift indicated that GNPs had been coated with silica and incorporated CNMA. Most importantly, CNMA-GNPs exhibited a peak at 290 nm that exactly matched a peak in the spectrum of CNMA,33 which confirmed the successful integration of CNMA into the surfaces of GNPs. The presence of individual well-resolved bands with shifts in the SPRs of GNPs, and appearance of the sharp CNMA peak, provided direct evidence of the formation of CNMA-GNPs with high stability.
The formation of tyrosine-functionalized GNPs and their further surface functionalizations with silica and CNMA were investigated by ATR-FT-IR spectroscopy (Figure 2B). The peaks observed for GNPs at 1,444, 1,224, and 1,076 corresponded to the stretching vibrations of –C=O–, –NH2, and –COO– groups. The appearance of symmetrical NH3+ bending vibration at 1,510 cm−1 indicated strong binding of amine group on GNPs.34 A Si-O-Si asymmetric stretching band at 980–1,190 cm−1 was observed for Si-GNPs and CNMA-GNPs but not for GNPs. The presence of signature peaks at 1,671 and 1,613 cm−1 in nanocomposites indicated binding of CNMA to GNPs.22 CNMA-GNPs had distinctive signatures at 688 (CH=CH bending), 748 (–CH bending) and 1,458 cm−1 (–CH stretching), corresponding to the phenyl group of CNMA. A signal at 1,744 cm−1 represented the presence of a carbonyl or carboxylate group of nanodispersions.35 These results confirmed the presence of a silica coating and the incorporation of CNMA onto GNPs.
Figure S1 shows the crystalline natures of GNPs as determined by XRD, before and after surface modifications. For GNPs, diffraction patterns at 38.2°, 44.5°, 64.5°, and 78.5° corresponded to the (111), (200), (220), and (311) planes of a face centered cubic structure. These lattice constants were similar to standard Au values as per JCPDS 4-0784 and 4-0783.36 In contrast, the 2θ peaks of monomeric Au in the Si-GNPs and CNMA-GNPs were weak, which could have been due to strong surface coating effects of silica and CNMA. The sharp peaks observed indicated good crystallinity, which concurred with TEM findings. These results further confirmed GNPs had been surface functionalized.
TEM images (Figure 2C) revealed the sizes, shapes, and surface modifications of GNPs. As shown, GNPs were spherical and had a uniform particle size distribution. In addition, Si-GNPs showed a dark Au core with a distinguishable thin silica coating which appeared in aggregation as compared to GNPs. On the other hand, CNMA-GNPs were coated in a thin silica layer with uniform dispersion. CNMA-GNPs exhibited distinct interfacial and spatial arrangements of surface coating in nanodispersions that revealed the presence Au core, silica coating, and incorporated CNMA (Figure 2C). TEM image of CNMA-GNPs showed the uniform distribution of silica-coated GNPs that might contain CNMA attached on their surfaces. Notably, the monodispersed CNMA-GNPs comprised of high surface area that potentiates its interaction, penetration, and distribution with the biological systems.
XPS analysis was performed to investigate the surface make-up of CNMA-GNPs (Figure S2). A high-resolution wide-scan spectrum showed the existence of C 1s, O 1s, N 1s, Si 2p, and Au0 4f7/2 and 4f5/2 in the survey spectrum of CNMA-GNPs (Figure S2A). The binding energies of C–C, C–O, C–N, and C=O were 284.8, 285.9, 286, and 288.5 eV, respectively, representing the chemical states of C 1s (Figure S2B). The higher intensity of C–N was attributed to bond formation between GNPs and CNMA in nanocomposites.37 However, no change was observed in the chemical state of O 1s at 532 eV (Figure S2C). Figure S2D shows the spectra of N 1s, which contained two components at 398.5 and 399 eV that were attributed to neutral amino groups and amide N atoms.37,38 The N 1s spectrum was of lower intensity than other spectra, possibly due to near complete reaction of NH2 groups with CNMA.39 The existence of a strong Si-O peak at 103 eV indicated that GNP surfaces were coated with silica (Figure S2E). Figure S2F shows two distinct binding energies at 83.8 and 87.4 eV, which were in good agreement with Au(III),40 and minor deconvulsions indicating Au-NH bonding. Au0 4f7/2 and 4f5/2 spectra were of reduced intensity, presumably because of the presence of surface silica and CNMA. High-resolution spectra of elements and of derivative peaks with specific binding energies confirmed successful surface conjugation on GNPs.
The release kinetics of CNMA from CNMA-GNPs at pH 5.0 and 7.4 is depicted in Figure S3. In vitro release of CNMA-GNPs was studied to analyze the fate of CNMA in 1) acidic pH, present in the biofilm environment 41 and 2) physiological pH, matching to body fluids. Initially, a short burst release of CNMA was observed at both pH values up to 4 h, followed by sustained CNMA release with increased time periods. Due to the presence of Schiff base system, CNMA should get hydrolyzed from CNMA-GNPs, resulting in faster release. The higher CNMA release found at pH 5.0 could be attributed to the ionic interactions of free aldehyde groups of CNMA with the liberated protons under lower pH conditions. The increased payload release in acidic pH enhanced the efficacy of CNMA-GNPs to deliver the CNMA at biofilm environment.
Antibiofilm activities of CNMA-GNPs
The effects of free CNMA, Si-GNPs, and CNMA-GNPs on pathogenic biofilms at 0%–0.025% v/v concentrations, which was equivalent to either CNMA loadings or GNPs, were investigated (Figure 3). Four pathogenic strains of E. coli O157:H7, P. aeruginosa, MSSA 6538, and MRSA, were chosen for this purpose. Each bacteria formed different level of biofilms according to their nature. Because of this phenomenon, E. coli O157:H7 and P. aeruginosa formed less biofilm compared to MSSA and MRSA, which were supplemented with glucose in LB media. At the concentrations examined, free CNMA and Si-GNPs failed to exert sufficient antibiofilm activity (Figure 3A and B). CNMA alone showed considerable antibiofilm activity (Figure 3A) and reduced growth (Figure S4A) against E. coli O157:H7 and MSSA 6538 at 0.025% v/v, the concentration of which was as high as CNMA-GNPs. On the other hand, as shown in Figures 3B and S4B, Si-GNPs had no inhibitory effect on bacterial biofilm or cell growth, respectively, indicating Si surface-functionalized GNPs were nontoxic. Only CNMA-GNPs effectively and dose-dependently inhibited biofilm formation by all four bacteria (Figure 3C), including respective cell growths (Figure S4C). In particular, CNMA-GNPs at a loading of only 0.005% v/v significantly inhibited biofilm formation (>50%), and this might be due to the existed CNMA (Figure 3C). In contrast, P. aeruginosa biofilms were much more resistant to CNMA-GNPs at this concentration, possibly because of the ability of this bacterium to change its outer membrane.23 However, P. aeruginosa biofilm and cell growth were completely inhibited by CNMA-GNPs at 0.025% v/v. To corroborate antibiofilm results, we examined biofilm formation using E. coli O157:H7, P. aeruginosa, MSSA 6538, and MRSA using CLSM. In all the strains, CNMA-GNPs diffused into the EPSs matrix and completely disrupted biofilms (Figure 3D), whereas neither CNMA nor Si-GNPs had any impact. Ultrasmall size and higher surface area enabled CNMA-GNPs to permeate through the biofilms, thus causing antibiofilm activity than free CNMA which could not pass through the typical layers of biofilms. Even though CNMA is lipophilic, it could not cross the resilient layers of EPS present in the biofilms and showed less activity in the confocal microscopy images than CNMA-GNPs. This observation indicates CNMA-GNPs permeated biofilm layers and delivered CNMA through nanodispersion to eradicate sessile bacteria.
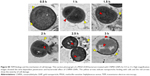
Recently, metal nanoparticles have proven to be efficient against pathogenic bacteria by disturbing membrane integrity and creating pores that cause leakage of cytoplasmic materials and ultrastructural changes of bacterial lysis.42–44 Therefore, TEM was employed to visualize the impacts of CNMA-GNPs on the ultrastructure of MSSA 6538 biofilms. As shown in Figure 4A and A′, colony biofilms grown without treatment displayed well-conserved morphological characters, such as closely packed cells, typical cytoplasmic membranes, and distinctive cell wall. Normal and magnified views of biofilm cells (Figure 4B and B′) exposed to Si-GNPs showed similar morphological features. Notably, the lack of damage to cells with attached Si-GNPs indicated they were not harmful to cells. In stark contrast, CNMA-GNPs exhibited antibiofilm activity as indicated by reduced cell density and scattered cellular debris (Figure 4C and C′). Some morphological changes were also observed, such as the swelling and disintegration of bacterial cells. Furthermore, the EDS spectra (Figure 4D) shows the characteristic elemental peaks of C, O, Au, and Si, which confirms the successful attachment of CNMA-GNPs on bacteria.
Figure S5 shows that CNMA-GNPs interacted with bacterial cell walls and exerted antimicrobial activity within a few hours. Since nanodispersed materials have small sizes, they are more likely to contact bacteria and be attached due to their lipophilic attraction of CNMA in CNMA-GNPs. After exposure to CNMA-GNPs for 0.5–1 h, signs of cell damage, such as, premature cell division and cell wall thickening were observed, and after exposure for 1.5–2 h, interacted CNMA-GNPs caused severe cell damage, and exposure for 2.5–3 h caused the complete disintegration of bacterial cells.
To the best of our knowledge, this is the first report to describe the preparation and silica coating of CNMA-conjugated GNPs to produce water dispersible CNMA-GNPs with enhanced antimicrobial/antibiofilm activities against various bacteria, especially enterohemorrhagic E. coli O157:H7, P. aeruginosa, and MRSA. A previous study described, the antibiofilm activity of silica nanoparticle stabilized CNMA capsules.39 However, an extremely high concentration of CNMA (5%, v/v)23,39 was needed for effective antibacterial activity, which was much higher than the CNMA-GNP concentrations found to be needed in this study. Si et al45 reported that in the absence of a stabilizing agent, CNMA was completely dissolved into the media and only had slight antibacterial activity.
The mechanism responsible for the antibiofilm activity of CNMA has not been fully elucidated. Our results suggest nanodispersions containing CNMA could have acted by inhibiting quorum sensing,46 by attaching cell walls due to lipophilic nature of CNMA,47 by interacting with cytoplasmic contents,48 by releasing CNMA, or by causing protein precipitation.49 Importantly, CNMA-GNPs were markedly more effective than free CNMA, which supports the notion that nanodispersions increase interactions with biofilms.50
As demonstrated earlier,51 we also claimed that the diminutive sizes of CNMA-GNPs might allow them to invade protective layers of EPS and kill bacteria. Furthermore, the acidic pH in biofilm environments could hydrolyze nanodispersions and promote the sustained release of CNMA.41,52 Notably, CNMA-GNPs were more effective than CNMA alone against the tested bacteria, including multidrug resistant strains, which supports the hypothesis that nanodispersions offer a valuable treatment alternative to traditional antibiotics.
Survival of infected C. elegans
Caenorhabditis elegans have been shown to be an excellent model for studies on the in vivo pathogenicity of S. aureus.53,54 Since S. aureus infects and kills nematodes,53 and in this study, we evaluated the in vivo effects including toxicity of nanoparticles with and without CNMA using C. elegans. It was observed (Figure 5) that CNMA-GNPs significantly prolonged the viability of infected nematodes, indicating that CNMA-GNPs reduce the virulence of S. aureus. On the other hand, Si-GNPs failed to prevent nematode death due to the highly virulent S. aureus. Additionally, the effect of CNMA-GNPs on C. elegans survival was comparable with its effect on the common food: E. coli OP50. These observations suggest CNMA-GNPs disinfected the nematode environment and showed twofold increase in worm survivability. This significant C. elegans survival from S. aureus infection after treatment with CNMA-GNPs might be because of the loaded CNMA. CNMA acts as a quorum sensing inhibitor and probably interferes with the cellular processes of S. aureus, including swarming motility, and might possibly affect the bacterial dissemination in the host.30,55 Additionally, the prolonged survival rates of the nematodes shown in Figure 5 indicate that CNMA-GNPs have no significant biological toxicity under a long incubation period. The observed no toxicity of CNMA-GNPs prominently suggested that the silica coating does not allow the biological system of the nematode to interact with nanoparticle core directly, thus resulting less toxicity.31
Internalizations and excretion of CNMA-GNPs in C. elegans
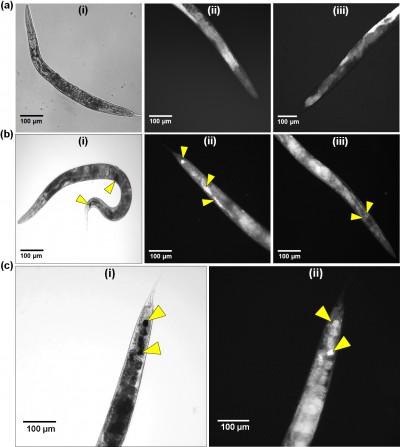
To determine nanoparticle uptake, internalization, and excretion by the nematode, C. elegans were exposed to CNMA-GNPs at a CNMA loading of 0.005% v/v for 24 h and then imaged. Figure 6 shows the phase contrast and dark field images of nematodes treated without (Figure 6A) or with CNMA-GNPs (Figure 6B and C). The phase contrast images of whole nematode (Figure 6A [a]) and corresponding dark field images of the head and tail portion (Figure 6A [b and c]) showed no changes in the contrast signals of the internal structures of the nematode, whereas, the phase contrast image in Figure 6B (a) shows the presence of accumulated CNMA-GNPs (black color) in the intestine and excretory organs. Dark field images in Figure 6B (b and c) depict CNMA-GNPs as bright spots. CNMA-GNPs were internalized by C. elegans, as monodispersed nanoparticles, which were observed in the pharynx, whereas aggregated particles were visible as bright spots in the intestine and anus regions (yellow arrows). In addition, the magnified C. elegans tail view shown in Figure 6C (a and b) demonstrates contrast signal enhancement by CNMA-GNPs within the nematode. Furthermore, when animals were fed with CNMA-GNPs, they retained the ingested particles for more than 12 h. In a previous study when C. elegans was deprived of food, no or few particles were excreted.56 The retention of particles in the intestines of treated worms indicates CNMA-GNPs were biostable in the worm’s digestive system, which can even degrade bacteria in ~2 min.57 As similar report of Mohan et al,58 after resumption of feeding the worms with E. coli OP50, the CNMA-GNPs were pushed down by the gut lumen and were excreted out of nematode within an hour. A video showing the excretion of CNMA-GNP aggregates by C. elegans fed with bacteria can be found in Supplementary materials (Video S1). Particles in the pharynx of C. elegans reached the digestive system apparently unaffected, similar to the available literature reports.58,59 In another study, the presence of digestive enzymes and acidic environment of the digestive system of C. elegans were found to dissolve metal nanoparticles and thus have toxic effects on C. elegans.60,61 However, in the case of CNMA-GNPs, the silica coating must be digested before the gold core is accessible to the intestinal environment, which suggests that the silica coating on CNMA-GNPs protects the bare GNPs and thus decreases potential toxicity issues.
Conclusion
In summary, we report the development of a biocompatible antimicrobial nanodrug delivery system (CNMA-GNPs). The surface-engineered gold nanoparticles produced successfully eradicated pathogenic biofilms, including those of antibiotic-resistant strains. Furthermore, CNMA-GNPs effectively eliminated pathogenic infection, and thus promoted C. elegans viability. Future interdisciplinary studies will be carried out to understand the governing molecular interactions between CNMA-GNPs and bacteria or biofilms, which will offer the possibility to rationalize the novel strategies to target infectious wounds. Furthermore, combining CNMA-GNPs with photothermal therapy in clinical models is highly expected to produce advanced nanomedicine for the treatment of chronic infected wounds.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) funded by the Korea government (MSIP) (#2015R1A2A2A01004542 to Jintae Lee) and by the Priority Research Center Program through the NRF funded by the Ministry of Education (#2014R1A6A1031189).
Disclosure
The authors report no conflicts of interest in this work.
References
World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. Geneva: World Health Organization; 2014:257. Available from: http://www.who.int/drugresistance/documents/surveillancereport/en/. Accessed December 10, 2016. | ||
Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362(19):1804–1813. | ||
Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2(2):114–122. | ||
Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):1487. | ||
Lewis K. Persister cells and the riddle of biofilm survival. Biochemistry (Moscow). 2005;70(2):267–274. | ||
Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39. | ||
Zhu X, Radovic-Moreno AF, Wu J, Langer R, Shi J. Nanomedicine in the management of microbial infection – Overview and Perspectives. Nano Today. 2014;9(4):478–498. | ||
Huh AJ, Kwon YJ. “Nanoantibiotics”: a new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J Control Release. 2011;156(2):128–145. | ||
Pelgrift RY, Friedman AJ. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv Drug Deliv Rev. 2013;65(13–14):1803–1815. | ||
Jain KK. Applications of nanobiotechnology in clinical diagnostics. Clin Chem. 2007;53(11):2002–2009. | ||
Aravinthan A, Govarthanan M, Selvam K, et al. Sunroot mediated synthesis and characterization of silver nanoparticles and evaluation of its antibacterial and rat splenocyte cytotoxic effects. Int J Nanomedicine. 2015;10:1977. | ||
Taylor E, Webster TJ. Reducing infections through nanotechnology and nanoparticles. Int J Nanomedicine. 2011;6:1463. | ||
Leid JG, Ditto AJ, Knapp A, et al. In vitro antimicrobial studies of silver carbene complexes: activity of free and nanoparticle carbene formulations against clinical isolates of pathogenic bacteria. J Antimicrob Chemother. 2012;67(1):138–148. | ||
Ashraf S, Pelaz B, del Pino P, et al. Gold-based nanomaterials for applications in nanomedicine. Top Curr Chem. 2016;370:169–202. | ||
Daraee H, Eatemadi A, Abbasi E, Fekri Aval S, Kouhi M, Akbarzadeh A. Application of gold nanoparticles in biomedical and drug delivery. Artif Cells Nanomed Biotechnol. 2016;44(1):410–422. | ||
Hemaiswarya S, Kruthiventi AK, Doble M. Synergism between natural products and antibiotics against infectious diseases. Phytomedicine. 2008;15(8):639–652. | ||
Kalemba D, Kunicka A. Antibacterial and antifungal properties of essential oils. Curr Med Chem. 2003;10(10):813–829. | ||
Kavanaugh NL, Ribbeck K. Selected antimicrobial essential oils eradicate Pseudomonas spp. and Staphylococcus aureus biofilms. Appl Environ Microbiol. 2012;78(11):4057–4061. | ||
Nostro A, Roccaro AS, Bisignano G, et al. Effects of oregano, carvacrol and thymol on Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Med Microbiol. 2007;56(4):519–523. | ||
Balaguer MP, Lopez-Carballo G, Catala R, Gavara R, Hernandez-Munoz P. Antifungal properties of gliadin films incorporating cinnamaldehyde and application in active food packaging of bread and cheese spread foodstuffs. Int J Food Microbiol. 2013;166(3):369–377. | ||
Koppikar SJ, Choudhari AS, Suryavanshi SA, Kumari S, Chattopadhyay S, Kaul-Ghanekar R. Aqueous cinnamon extract (ACE-c) from the bark of Cinnamomum cassia causes apoptosis in human cervical cancer cell line (SiHa) through loss of mitochondrial membrane potential. BMC Cancer. 2010;10:210. | ||
Wani KD, Kadu BS, Mansara P, et al. Synthesis, characterization and in vitro study of biocompatible cinnamaldehyde functionalized magnetite nanoparticles (CPGF Nps) for hyperthermia and drug delivery applications in breast cancer. PloS One. 2014;9(9):e107315. | ||
Zodrow KR, Schiffman JD, Elimelech M. Biodegradable polymer (PLGA) coatings featuring cinnamaldehyde and carvacrol mitigate biofilm formation. Langmuir. 2012;28(39):13993–13999. | ||
Chen H, Davidson PM, Zhong Q. Impacts of sample preparation methods on solubility and antilisterial characteristics of essential oil components in milk. Appl Environ Microbiol. 2014;80(3):907–916. | ||
Gomes C, Moreira RG, Castell-Perez E. Poly (DL-lactide-co-glycolide) (PLGA) nanoparticles with entrapped trans-cinnamaldehyde and eugenol for antimicrobial delivery applications. J Food Sci. 2011;76(2):N16–N24. | ||
Stammati A, Bonsi P, Zucco F, Moezelaar R, Alakomi HL, von Wright A. Toxicity of selected plant volatiles in microbial and mammalian short-term assays. Food Chem Toxicol. 1999;37(8):813–823. | ||
Dubey K, Anand BG, Badhwar R, et al. Tyrosine-and tryptophan-coated gold nanoparticles inhibit amyloid aggregation of insulin. Amino Acids. 2015;47(12):2551–2560. | ||
Akbarzadeh A, Zarghami N, Mikaeili H, et al. Synthesis, characterization, and in vitro evaluation of novel polymer-coated magnetic nanoparticles for controlled delivery of doxorubicin. Nanotechnol Sci Appl. 2012;5:13–25. | ||
Lee JH, Cho MH, Lee J. 3-Indolylacetonitrile decreases Escherichia coli O157:H7 biofilm formation and Pseudomonas aeruginosa virulence. Environ Microbiol. 2011;13(1):62–73. | ||
Cho HS, Lee JH, Cho MH, Lee J. Red wines and flavonoids diminish Staphylococcus aureus virulence with anti-biofilm and anti-hemolytic activities. Biofouling. 2015;31(1):1–11. | ||
Ramasamy M, Das M, An SSA, Yi DK. Role of surface modification in zinc oxide nanoparticles and its toxicity assessment toward human dermal fibroblast cells. Int J Nanomedicine. 2014;9:3707–3718. | ||
Moores A, Goettmann F. The plasmon band in noble metal nanoparticles: an introduction to theory and applications. New J Chem. 2006;30(8):1121–1132. | ||
Guo K, Chen Y. Simple and rapid detection of aromatic amines using a thin layer chromatography plate. Anal Methods. 2010;2(8):1156–1159. | ||
Selvakannan P, Swami A, Srisathiyanarayanan D, et al. Synthesis of aqueous Au core-Ag shell nanoparticles using tyrosine as a pH-dependent reducing agent and assembling phase-transferred silver nanoparticles at the air-water interface. Langmuir. 2004;20(18):7825–7836. | ||
Chen H, Hu X, Chen E, et al. Preparation, characterization, and properties of chitosan films with cinnamaldehyde nanoemulsions. Food Hydrocoll. 2016;61:662–671. | ||
Shin Y, Bae IT, Arey BW, Exarhos GJ. Facile stabilization of gold-silver alloy nanoparticles on cellulose nanocrystal. J Phys Chem C Nanomater Interfaces. 2008;112(13):4844–4848. | ||
Techane SD, Gamble LJ, Castner DG. X-ray photoelectron spectroscopy characterization of gold nanoparticles functionalized with amine-terminated alkanethiols. Biointerphases. 2011;6(3):98–104. | ||
Khan SA, Gambhir S, Ahmad A. Extracellular biosynthesis of gadolinium oxide (Gd2O3) nanoparticles, their biodistribution and bioconjugation with the chemically modified anticancer drug taxol. Beilstein J Nanotechnol. 2014;5(1):249–257. | ||
Duncan B, Li X, Landis RF, et al. Nanoparticle-stabilized capsules for the treatment of bacterial biofilms. ACS Nano. 2015;9(8):7775–7782. | ||
Movahed SK, Fakharian M, Dabiri M, Bazgir A. Gold nanoparticle decorated reduced graphene oxide sheets with high catalytic activity for Ullmann homocoupling. RSC Adv. 2014;4(10):5243–5247. | ||
Harrison JJ, Ceri H, Turner RJ. Multimetal resistance and tolerance in microbial biofilms. Nat Rev Microbiol. 2007;5(12):928–938. | ||
Seil JT, Webster TJ. Antimicrobial applications of nanotechnology: methods and literature. Int J Nanomedicine. 2012;7:2767–2781. | ||
Moritz M, Geszke-Moritz M. The newest achievements in synthesis, immobilization and practical applications of antibacterial nanoparticles. Chem Eng J. 2013;228:596–613. | ||
Krishnamoorthy K, Premanathan M, Veerapandian M, Kim SJ. Nanostructured molybdenum oxide-based antibacterial paint: effective growth inhibition of various pathogenic bacteria. Nanotechnology. 2014;25(31):315101. | ||
Si W, Gong J, Chanas C, et al. In vitro assessment of antimicrobial activity of carvacrol, thymol and cinnamaldehyde towards Salmonella serotype typhimurium DT104: effects of pig diets and emulsification in hydrocolloids. J Appl Microbiol. 2006;101(6):1282–1291. | ||
Shafreen B, Mohmed R, Selvaraj C, Singh SK, Karutha Pandian S. In silico and in vitro studies of cinnamaldehyde and their derivatives against LuxS in Streptococcus pyogenes: effects on biofilm and virulence genes. J Mol Recognit. 2014;27(2):106–116. | ||
Khan MS, Ahmad I. In vitro antifungal, anti-elastase and anti-keratinase activity of essential oils of cinnamomum-, syzygium- and cymbopogon-species against Aspergillus fumigatus and Trichophyton rubrum. Phytomedicine. 2011;19(1):48–55. | ||
Helander IM, Alakomi HL, Latva-Kala K, et al. Characterization of the action of selected essential oil components on Gram-negative bacteria. J Agric Food Chem. 1998;46(9):3590–3595. | ||
Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–899. | ||
Gaidhani S, Singh R, Singh D, et al. Biofilm disruption activity of silver nanoparticles synthesized by Acinetobacter calcoaceticus PUCM 1005. Mater Lett. 2013;108:324–327. | ||
Ikuma K, Decho AW, Lau BL. When nanoparticles meet biofilms – interactions guiding the environmental fate and accumulation of nanoparticles. Front Microbiol. 2015;6:591. | ||
Li XN, Yeh YC, Giri K, et al. Control of nanoparticle penetration into biofilms through surface design. Chem Commun (Camb). 2015;51(2):282–285. | ||
Sifri CD, Begun J, Ausubel FM, Calderwood SB. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun. 2003;71(4):2208–2217. | ||
Moy TI, Conery AL, Larkins-Ford J, et al. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem Biol. 2009;4(7):527–533. | ||
Kim YG, Lee JH, Kim SI, Baek KH, Lee J. Cinnamon bark oil and its components inhibit biofilm formation and toxin production. Int J Food Microbiol. 2015;195:30–39. | ||
Lim SF, Riehn R, Ryu WS, et al. In vivo and scanning electron microscopy imaging of upconverting nanophosphors in Caenorhabditis elegans. Nano Lett. 2006;6(2):169–174. | ||
McGhee J. The C. elegans intestine. In: WormBook, ed. The C. elegans Research Community, WormBook; 2007. Available from: http://www.wormbook.org | ||
Mohan N, Chen CS, Hsieh HH, Wu YC, Chang HC. In vivo imaging and toxicity assessments of fluorescent nanodiamonds in Caenorhabditis elegans. Nano Lett. 2010;10(9):3692–3699. | ||
Avery L, You YJ. C. elegans feeding. In: WormBook: The Online Review of C. elegans Biology. Pasadena, CA: Worm Book; 2012:1. | ||
Chauhan VM, Orsi G, Brown A, Pritchard DI, Aylott JW. Mapping the pharyngeal and intestinal pH of Caenorhabditis elegans and real-time luminal pH oscillations using extended dynamic range pH-sensitive nanosensors. ACS Nano. 2013;7(6):5577–5587. | ||
Gonzalez-Moragas L, Yu SM, Carenza E, Laromaine A, Roig A. Protective Effects of Bovine Serum Albumin on Superparamagnetic Iron Oxide Nanoparticles Evaluated in the Nematode Caenorhabditis elegans. ACS Biomater Sci Engi. 2015;1(11):1129–1138. |
Supplementary materials
Interaction of CNMA-GNPs with planktonic MSSA 6538
The ability of CNMA-GNPs to affect the maturation of planktonic bacteria was investigated by TEM. Cultures in the logarithmic phase were exposed to 0.005% (v/v) CNMA containing GNPs for 0–3 h at 37°C with shaking at 250 rpm. After exposure, bacteria were washed with PBS buffer by centrifugation at 4000 × g for 10 min at 4°C and then fixed for 12 h at 4°C with aldehyde mixture (glutaraldehyde 2.5% and formaldehyde 2%). Cells were then washed with PBS, post-fixed for 30 min with 1% osmium tetroxide, dehydrated in a graded acetone series, and embedded in Epon resin mixture (Hatfield, USA). Ultrathin sections were obtained using a MT-X ultramicrotome (Tucson, USA), stained with uranyl acetate and lead citrate, and observed under a H-7600 electron microscope (Tokyo) at 120 keV.
  | Figure S1 X-ray diffraction patterns of GNPs (black), Si-GNPs (red) and CNMA-GNPs (blue). |
  | Figure S3 In vitro release profile of CNMA from CNMA-GNPs in different pH buffered saline at 37°C. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.