Back to Journals » International Journal of General Medicine » Volume 13
Deficient Expression of DGCR8 in Human Testis is Related to Spermatogenesis Dysfunction, Especially in Meiosis I
Authors Babakhanzadeh E, Khodadadian A, Nazari M, Dehghan Tezerjani M , Aghaei SM, Ghasemifar S, Hosseinnia M, Mazaheri M
Received 25 March 2020
Accepted for publication 6 May 2020
Published 15 May 2020 Volume 2020:13 Pages 185—192
DOI https://doi.org/10.2147/IJGM.S255431
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Emad Babakhanzadeh,1,2,* Ali Khodadadian,1,* Majid Nazari,1 Masoud Dehghan Tezerjani,1 Seyed Mohsen Aghaei,1 Sina Ghasemifar,1 Mehdi Hosseinnia,3 Mahta Mazaheri1,4
1Department of Medical Genetics, Shahid Sadoughi University of Medical Sciences, Yazd, Iran; 2Medical Genetics Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran; 3Department of Biology, Faculty of Science, University of Guilan, Rasht, Iran; 4Mother and Newborn Health Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran
*These authors contributed equally to this work
Correspondence: Mahta Mazaheri Email [email protected]
Introduction: DiGeorge syndrome critical region gene 8 (DGCR8) contributes to miRNA biogenesis, and defects in its expression could lead to defects in spermatogenesis.
Methods: Here, we assess gene and protein expression levels of DGCR8 in the testicular biopsy specimens obtained from men with obstructive azoospermia (OA, n = 19) and various types of non-obstructive azoospermia (NOA) including maturation arrest (MA, n = 17), Sertoli cell-only syndrome (SCOS, n = 20) and hypospermatogenesis (HYPO, 18). Also, samples of men with NOA were divided into two groups based on successful and unsuccessful sperm recovery, NOA+ in 21 patients and NOA− in 34 patients.
Results: Examinations disclosed a severe decrease in DGCR8 in samples with MA and SCOS in comparison to OA samples (P < 0.001). Also, the results showed DGCR8 has significantly lower expression in testis tissues of NOA− group in comparison to NOA+ group (p< 0.05). Western blot analysis confirmed that the DGCR8 protein was not expressed in SCOS samples and had a very low expression in MA and HYPO samples.
Discussion: The results of this survey showed that DGCR8 is an important gene for the entire spermatogenesis pathway. Moreover, DGCR8 gene plays an important role in the diagnosis of NOA subgroups, and also the expression changes in it might contribute to SCOS or MA phenotypes. This gene with considering other related genes can also be a predictor of sperm retrieval.
Keywords: DGCR8, obstructive azoospermia, non-obstructive azoospermia, spermatogenesis
Introduction
Azoospermia is one of the major worldwide problems,1,2 and commonly categorized into two groups, including obstructive azoospermia (OA) and non-obstructive azoospermia (NOA).3–6 OA, are mainly because of male genital system blockage, although they usually have normal spermatogenesis in most cases. NOA, routinely described as a testicular failure, in which condition that spermatozoa is completely absent in ejaculation due to spermatogenesis defects, NOA subtypes are categorized into three major classes, Sertoli cell-only syndrome (SCOS), meiotic arrest (MA), and hypospermatogenesis (HYPO).7 While SCO defined as a severe spermatogenesis defect and Sertoli cells line the seminiferous tubules. MA is briefly called incomplete maturation of germ cells. Hypospermatogenesis is referred to as abnormally diminished and defected sperm production, the spermatogenesis process in most cases is stopped at either spermatocyte (SC) or round spermatid stage.8–10
Despite the efforts of many researchers to clarify the basic molecular mechanisms of male infertility, the etiology of almost 75% of the cases remains obscure and thought to be genetic.11 This indicates a lack of a comprehensive understanding of the underlying molecular mechanisms and genetic factors involved in infertility. Unfortunately, due to the lack of suitable biomarkers and the limited data available for evaluation of sperm recovery, sometimes a useless surgery is performed on patients. Therefore, it seems necessary to introduce new and more favorable methods, as well as the introduction of appropriate genes, serum markers, and targeted biomarkers; also the development of more sensitive and noninvasive techniques for the detection of spermatogenesis in testicular azoospermic individuals, which in some OA patients can reduce the request for a diagnostic testicular biopsy. In NOA patients, a reliable diagnostic test can have many benefits, such as prepare a more detailed valuation of histopathological subtypes, prognosis of testicular sperm extraction (TESE) outcome, and simplify good planning for assisted reproductive technology (ART).
In recent years, abundant evidence has shown the key role of miRNAs in different kinds of biological processes such as cell growth, apoptosis, differentiation, and spermatogenesis,12,13 it is reported that more than 30% of genes are regulated by miRNAs and they are conserved in different species.14 Furthermore, some of miRNAs show high levels of expression in certain stages of spermatogenesis.15,16 DGCR8 plays a crucial role in the biogenesis and maturation of miRNAs.17 The biogenesis of miRNA is a two-step process, miRNAs genes are generally transcribed by RNA pol II, termed primary miRNA transcripts (pri-miRNA). In a first step, the pri-miRNAs are processed in the nucleus by a set of proteins includes DGCR8 and DROSHA, to form into ~70-nt miRNA stem-loop precursor (pre-miRNA), which is a faulty structure. The pre-miRNAs are then transmitted to the cytoplasm by a complex of Exportin 5 and Ran-GTP. In the cytoplasm, where the pre-miRNAs undergo cleavage by cytoplasmic RNase III enzyme, DICER1, to give rise to double-stranded RNA of ~22-nt long, called the miRNA:miRNA* duplex. This duplex is then separated by a helicase and the miRNA strand is loaded into ribonucleoprotein complex known as the miRNA-induced silencing complex (miRISC), holding members of the Argonaute protein family, and this complex can bind to partial or complementary sequences of their target mRNAs, in most cases, inhibit the expression of target genes by posttranscriptional mechanism and promoting mRNA decay, whereas the miRNA* is usually degraded.18–20 In this regard, several experiments of knockout mouse models have shown the role of Dgcr8 in mice infertility. For instance, conditional deletion of Dgcr8 in Sertoli cells or early stages of germ cell maturation in mice lead to adult infertility or subfertility cause of spermatogenesis failure.21 In Dgcr8 -deleted testis, the spermatogenesis process was stopped at an any early stage of proliferation or differentiation.22
As discussed above, regarding the importance of DGCR8 in male infertility, we designed the study in azoospermic individuals to measure expression levels of the DGCR8 gene at mRNA level using quantitative real-time polymerase chain reaction (RT-qPCR), and assess the protein level of DGCR8 by immunohistochemistry and Western blot techniques.
Patients and Methods
Patients
Seventy-four NOA and OA men admitted to the Abortion Research Centre, Yazd Reproductive Sciences Institute, were entered into this study. All subjects were undergoing bilateral testicular tissue micro-dissection (mTESE) operations to attain spermatozoa for intracytoplasmic sperm injection (ICSI). The study was approved by the local Ethics Committee and written informed consent was obtained from all subjects. Preoperative tests included karyotyping and Y chromosome microdeletion analysis, and the levels of serum follicle-stimulating hormone (FSH), luteinizing hormone (LH) and testosterone. Patients were not receiving hormone therapy and all had primary infertility. No history of TESE and cryptorchidism was reported for any of the participants. Patients with cystic fibrosis, chromosomal abnormalities and Y chromosome microdeletion were omitted from the study. Subjects with normal spermatogenesis were included as the control group. Following TESE, testicular samples were split into three; one was fixed in Bouin’s solution for histological examination and two other sections instantly frozen in liquid nitrogen for RNA extraction and Western blot assay. Histological examination was carried out with hematoxylin and eosin (H&E) and interpreted by a trained pathologist to classify the samples with normal spermatogenesis (OA, n = 19), lack of germ cells (SCOS, n = 20), declined number of spermatozoa (HYPO, n = 18), and incomplete maturation of germ cells (MA, n = 17). Also, samples of men with NOA were divided into two groups based on successful and unsuccessful sperm recovery, NOA+ in 21 patients and NOA− in 34 patients, Table 1.
 |
Table 1 Number of Specimens Included in NOA+ and NOA− Groups |
RNA Isolation and cDNA Synthesis
Frozen testis tissues were homogenized, and total RNA extracted using the miRNeasy Micro Kit (Qiagen, Germany), according to the manufacturer’s protocol. Then, on-column DNase digestion and in-solution DNase digestion were accomplished to eliminate DNA contamination. The purity and concentration of RNA were specified by spectrophotometer (NanoDrop, Thermo Fisher) and OD 260/280. Complementary DNA (cDNA) of DGCR8 was synthesized using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, C.N: 4368813, 4368814, 4374966, and 4374967) according to the manufacturer’s instructions.
Real-Time Quantitative PCR (RT-qPCR)
RT-RT-qPCR was performed on a Real-Time PCR System (Applied Biosystem, ABI, Step One Plus, USA) by 1.0 μL of produced cDNA, 10 μL of the SYBR Green master mix (AB Applied Biosystems), 1 μL of each primer and 7.0 μL of DNase/RNase-free water for the gene expression profile. β-actin was used as the reference gene and DGCR8 also as the target gene, and with designed primers listed in Table 2. The reactions were performed with initial denaturation at 95°C for 8 min, followed by 40 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 30 sec, extension at 72°C for 30 sec, and a final extension at 72°C for 10 min. The relative gene expression was calculated by using 2−∆∆Ct quantitative method.
 |
Table 2 Real-Time RT-PCR Primers Applied in This Experiment |
Immunohistochemistry
Bouin’s fixed paraffin-embedded biopsies at 5-μm thickness deparaffinized in fresh xylene for 10 minutes. Then rehydrated in a series of decreasing ethanol concentrations. Slides were put into a microwave container and exposed to Citrate buffer (pH 6.0) and heated for 10 minutes. Slides were cooled in the Citrate buffer for 35 minutes. Slides were washed four times with 1x TBS for 3 minutes. For quelling endogenous peroxidase function, slides were incubated with 3% H2O2 for 10 minutes. Use 5% BSA in 1x TBS for blocking for 1 hour. Incubate sections with primary antibody against DGCR8 (ProteinTech, 10996-1-AP) protein diluted 1:100 in 1x TBS for overnight at 4°C. Following primary antibody, incubated slides were washed three times with 1x TBS for 3 minutes. Then subsequently incubated with secondary goat anti-human-IgG-(H-L) antibody conjugated with horseradish peroxidase (HRP) (ProteinTech, SA00001-17) (diluted 1:150 in 1x TBS) at room temperature (RT) for 30 minutes. As a negative control, a section on each slide was incubated with 1x TBS instead of primary antibody in each experiment. Then, the sections were stained with Liquid DAB at RT for 10 min. And rinse sections gently with sufficient distilled water. Following staining, slides were counterstained with haematoxylin (30 s at 25–30 °C) and transferred slides into a 1% HCl, 95% ethanol solution for 10 seconds; and instantly transferred to distilled water. Slides were immersed sequentially into 60%, 80%, 95% and 100% ethanol baths for 5 minutes each. Ultimately, slides were submerged in xylene (5 min at 25–30 °C) and mounted on microscope slides for evaluation by light microscopy.
Western Blot
Western blot examination was conducted as formerly explained.23 Briefly, equal amounts of proteins (35mg) obtained from testis samples were separated with 12% SDS-PAGE and electrotransferred onto nitrocellulose paper. The membranes were blocked with (2–5%) non-fat dry milk in 1x TBST (10 mM/L Tris-HCl, pH 8.0; 150 mM/L NaCl, 0.1% Tween 20) for 1 hour at 25–30°C, and then incubated with primary DGCR8 (1:300; ProteinTech, 10996-1-AP), ACTB (1:300; ABIN4284408) antibody at 4°C overnight, then with secondary goat anti-human-IgG-(H-L) antibody conjugated with horseradish peroxidase (HRP) (1:500; ProteinTech, SA00001-17) for 1 hour at 25°C. At the end, the immunoreactive signals were determined via the ECL kit (Thermo Scientific).
Statistical Analysis
Statistical comparisons between the groups were determined using a ONE WAY ANOVA followed by Dunnett’s multiple comparison post-test. Also, statistical comparisons between two groups (NOA+, NOA−) were determined by independent samples t-test. P-values less than 0.05 were deemed to be statistically significant. All analyses were executed using GraphPad Prism 6 software.
Results
Clinical Characteristics of Patients
No significant differences were indicated in age, LH, FSH, and testosterone serum concentrations between the four groups (OA [n = 19], HYPO [n = 18], MA [n = 17], and SCOS [n = 20]) (Table 3). Also, the results showed no significant differences in these parameters between the successful and unsuccessful TESE groups (NOA+ and NOA− respectively) (Table 4).
 |
Table 3 The Clinical Features of Patient Groups |
 |
Table 4 The Clinical Features of Patients with NOA+ and NOA− Groups |
Expression Analysis of DGCR8 Gene
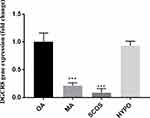
There was a significant difference in the expression of the DGCR8 gene in the four groups. As shown in Figure 1, the expression of DGCR8 was significantly lower in the MA (n = 17) and the SCOS (n = 20) than the positive control (OA, n = 19) (P < 0.001, Dunn’s post-test). While, HYPO samples did not show a significant difference with the control group (Figure 1). Also, the results of the RT-qPCR analysis between NOA+ and NOA− testicular samples showed a significantly lower expression of DGCR8 gene in testis tissues of NOA− group in comparison to NOA+ group (Figure 2). The significance was calculated at p<0.05 level.
Immunohistochemical Analysis of DGCR8 Protein
To analyze the role of DGCR8 in men with azoospermia, we examined DGCR8 expression by IHC in testicular biopsies from men with OA (n = 3), HS (n = 3), MA (n = 3), and SCO (n = 3). The DGCR8 protein was seen in spermatocytes (with a higher intensity than other stages), round spermatids, and spermatozoa. While relatively weak staining was observed in spermatogonia. IHC results of DGCR8 expression in the testis tissues displaying the negative staining of this protein in Sertoli cells. This protein also has low expression in MA and HYPO specimens (Figure 3).
Western Blot Analysis of DGCR8 Protein
To confirm the results of RT-RT-qPCR Western blotting and IHC, we performed the Western blotting. As shown in Figure 4, DGCR8 protein was not expressed in SCOS samples and had a very low expression in MA and HYPO samples.
 |
Figure 4 Western blotting test results for protein expression of DGCR8 in HP, MA, SCOS, and OA samples compared to control group. |
Discussion
Sperm production has been reported in approximately 60% of men with NOA.24 If Testicular tissue micro-dissection (mTESE) procedure is able to retrieve sperm, men with NOA could have their biological offspring via intracytoplasmic sperm injection (ICSI). Undoubtedly the only reliable predictor of sperm recovery in mTESE is histopathological examination. Failure to sperm retrieval may bring unfortunate outcomes for couples who will not want to undergo sperm or embryo donation. The second surgery after the first mTESE is good manner for detection of the possible spermatogenic locus for these couples. So it is indispensable to specify reliable factors that can be applied to anticipate the sperm retrieval rate (SRR) of mTESE.25–27
To the best of our knowledge, our work was the first that appraise the expression of DGCR8 gene in NOA and OA tissue samples. The results of this experiment indicated no significant differences in age, LH, FSH, and testosterone serum concentrations between the four parameters. Also, the results showed no significant differences in these parameters between the successful and unsuccessful mTESE groups (NOA+ and NOA− respectively). The results also showed that the DGCR8 gene expression in MA and SCOS testicular tissues were significantly lower than those of samples with OA (control group), while HYPO samples had no significant difference with the control group.
It seems logical that finding sperm in a small number of testicular tissue samples of SCOS and MA by RT-RT-qPCR technique. Because there have been many studies in which patients with SCOS and MA histology have succeeded in recovering sperm. Likely demonstrate the presence of cryptic foci of spermatogenesis in these specimens or mixed patterns in NOA subtypes (HS with SCO and MA with SCO).28 Success rates of sperm recovery by mTESE depend on histopathological subtypes and are appraised to be 79% for HS, 47% for MA, and 24% for SCO.29
Yeon Sun Kim et al reported that a defect in the Dgcr8 gene leads to infertility in mice.30 In another study, researcher observed that DGCR8 germ cell deficient in mice result in accumulative errors in meiotic and post-meiotic stages as well as non-functional and atypical spermatozoa that ended up with complete infertility.31 Ce´line Zimmermann et al in Dgcr8 mutant mice reported that dysfunction of the normal Dgcr8 gene could lead to spermatid without function or azoospermia.21
Immunohistochemistry results of DGCR8 expression in the testis tissues displaying the negative staining of this protein in Sertoli cells. Also, this protein has low expression in MA and HYPO specimens. Immunohistochemical staining in the control specimens represented that DGCR8 was expressed in stages from early meiosis spermatocytes up to postmeiotic spermatids. Secondary spermatocytes in histological studies either absent or hardly detectable, due to they do not have much time to enter the next stage. In contrast, DGCR8 was hardly identifiable in spermatogenic cells and spermatids of the HYPO and MA samples. These results are indicative of this fact that the DGCR8 is likely play a more significant role in the meiotic and post-meiotic stages and it points out that the DGCR8 is more likely to be transcribed in the germ cells. Because SCOS samples have no germ cells, the expression of the DGCR8 was not detected.
Interestingly, the results of the RT-RT-qPCR technique showed a clear difference with the immunohistochemical results, in the HYPO samples, compared with the control samples. There was no significant difference in the RT-RT-qPCR technique, but in the immunohistochemistry assay, there was a difference in the intensity of staining. Some experiments illustrate that mRNA levels of a gene cannot always anticipate protein levels. Schwa Nhausser et al.32 Found that the main regulators of the cell cycle and growth have a poor relationship between mRNAs and protein levels. Western blot analysis confirmed that the DGCR8 protein was not expressed in SCOS samples and had a very low expression in MA and HYPO samples. The results suggest that the DGCR8 transcript may be affected by post-transcriptional regulation (eg miRNAs) and that its expression in HYPO samples is reduced.
In sum, it can be guessed that the expression of DGCR8 follows a three-step pattern. First, it helps to mitotic division in spermatogonium, and then plays an important role in the meiosis stage, which seems to be more effective than the other stages, it has also been reported that the removal of the DGCR8 causes a halt in meiosis I due to disarrange spindles and fault in chromosomal alignment,22,33 third in the final stages of spermatogenesis in elongated spermatids.
Our results also showed a significant difference in the expression of the DGCR8 among those whose sperm was successfully recovered and those that were not (NOA+ and NOA− groups, respectively), recoveries had much higher expression than unrecovered ones. This result suggests that the use of DGCR8 along with other predictor genes which identified in researchers’ study can be a potential sperm recovery biomarker. Probably, patients showing high levels of DGCR8 expression have a greater chance of successful sperm recovery than those have low expression. We did not notice any significant difference in levels of clinical hormonal parameters consist of testosterone, FSH and LH among NOA+ and NOA− individuals. This finding brings this doubt closer to reality, which uses DGCR8 as a predictive biomarker for sperm retrieval.
The results of this study showed that DGCR8 is an important gene for the entire spermatogenesis pathway. Moreover, the DGCR8 gene plays an important role in the diagnosis of NOA subgroups, also expression changes in it might contribute to SCOS or MA phenotypes. This gene can also be a predictor of sperm retrieval. However, more studies with more patients are indispensable for confirming the results of this experiment.
Abbreviations
DGCR8, DiGeorge syndrome critical region gene 8; NOA, non-obstructive azoospermia; OA, obstructive azoospermia; HYPO, hypospermatogenesis; MA, maturation arrest; SCOS, Sertoli cell-only syndrome.
Ethics Approval and Consent to Participate
The study was conducted in accordance with the Declaration of Helsinki. Moreover, the study was approved by the local Ethics Committee (IR.SSU.MEDICINE.REC.1396.220) and written informed consent was obtained from all subjects.
Acknowledgments
This experiment was funded by a grant from the Shahid Sadoughi University of Medical Sciences of Yazd. The authors would like to thank to Dr. S. Mirjalili, Dr. Montazeri, and Mr. Heydarian for their valuable support.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
All authors declared no potential conflicts of interest with respect to this work.
References
1. Dubé E, Hermo L, Chan PT, Cyr D. Alterations in gene expression in the caput epididymides of nonobstructive azoospermic men. Biol Reprod. 2008;78(2):342–351.
2. Vij SC, Sabanegh E, Agarwal A. Biological therapy for non-obstructive azoospermia. Expert Opin Biol Ther. 2018;18(1):19–23. doi:10.1080/14712598.2018.1380622
3. Donoso P, Tournaye H, Devroey P. Which is the best sperm retrieval technique for non-obstructive azoospermia? A systematic review. Hum Reprod Update. 2007;13(6):539–549. doi:10.1093/humupd/dmm029
4. Jarvi K, Lo K, Fischer A, et al. CUA guideline: the workup of azoospermic males. Can Urol Assoc J. 2010;4(3):163. doi:10.5489/cuaj.10050
5. Lee JY, Dada R, Sabanegh E, Carpi A, Agarwal A. Role of genetics in azoospermia. Urology. 2011;77(3):598–601. doi:10.1016/j.urology.2010.10.001
6. Ma M, Yang S, Zhang Z, et al. Sertoli cells from non-obstructive azoospermia and obstructive azoospermia patients show distinct morphology, Raman spectrum and biochemical phenotype. Hum Reprod. 2013;28(7):1863–1873. doi:10.1093/humrep/det068
7. Wosnitzer M, Goldstein M, Hardy MP. Review of azoospermia. Spermatogenesis. 2014;4(1):e28218. doi:10.4161/spmg.28218
8. Devroey P, Liu J, Nagy Z, et al. Pregnancies after testicular sperm extraction and intracytoplasmic sperm injection in non-obstructive azoospermia. Hum Reprod. 1995;10(6):1457–1460. doi:10.1093/HUMREP/10.6.1457
9. Hauser R, Botchan A, Amit A, et al. Multiple testicular sampling in non-obstructive azoospermia–is it necessary? Hum Reprod. 1998;13(11):3081–3085. doi:10.1093/humrep/13.11.3081
10. Silber SJ, Van Steirteghem AC, Devroey P. Sertoli Cell Only Revisited. Oxford University Press; 1995.
11. Okada H, Tajima A, Shichiri K, Tanaka A, Tanaka K, Inoue I. Genome-wide expression of azoospermia testes demonstrates a specific profile and implicates ART3 in genetic susceptibility. PLoS Genet. 2008;4(2):e26. doi:10.1371/journal.pgen.0040026
12. Hayashi K, de Sousa Lopes SMC, Kaneda M, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3(3):e1738. doi:10.1371/journal.pone.0001738
13. Kotaja N. MicroRNAs and spermatogenesis. Fertil Steril. 2014;101(6):1552–1562. doi:10.1016/j.fertnstert.2014.04.025
14. Lewis BP, Shih I-H, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798.
15. Dabaja AA, Mielnik A, Robinson BD, et al. Possible germ cell-Sertoli cell interactions are critical for establishing appropriate expression levels for the Sertoli cell-specific MicroRNA, miR-202-5p, in human testis. Basic Clin Androl. 2015;25(1):2.
16. Lian J, Tian H, Liu L, et al. Downregulation of microRNA-383 is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation by targeting IRF1. Cell Death Dis. 2010;1(11):e94.
17. Guo WT, Wang Y. Dgcr8 knockout approaches to understand microRNA functions in vitro and in vivo. Cell Mol Life Sci. 2019;76(9):1697–1711. doi:10.1007/s00018-019-03020-9
18. Guo H, Ingolia NT, Weissman JS, Bartel DPJN. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835.
19. Huang XM, Tian QN, Bao ZX, et al. Cloning and identification of microRNAs in earthworm (Eisenia fetida). Biochem Genet. 2012;50(1–2):1–11.
20. Long JE, Chen HX. Identification and characteristics of cattle microRNAs by homology searching and small RNA cloning. Biochem Genet. 2009;47(5–6):329–343. doi:10.1007/s10528-009-9234-6
21. Zimmermann C, Romero Y, Warnefors M, et al. Germ cell-specific targeting of DICER or DGCR8 reveals a novel role for endo-siRNAs in the progression of mammalian spermatogenesis and male fertility. PLoS One. 2014;9(9):e107023.
22. Modzelewski AJ, Hilz S, Crate EA, et al. Dgcr8 and Dicer are essential for sex chromosome integrity during meiosis in males. J Cell Sci. 2015;128(12):2314–2327.
23. He Y, Luo M, Yi M, et al. Identification of a testis-enriched heat shock protein and fourteen members of hsp70 family in the swamp eel. PLoS One. 2013;8(6):e65269. doi:10.1371/journal.pone.0065269
24. Schlegel PN. Nonobstructive azoospermia: a revolutionary surgical approach and results. Semin Reprod Med. 2009;27(2):165-170.
25. PLAS E, RIEDL CR, ENGELHARDT PF, MÜHLBAUER H, PFLÜGER H. Unilateral or bilateral testicular biopsy in the era of intracytoplasmic sperm injection. J Urol. 1999;162(6):2010–2013. doi:10.1016/S0022-5347(05)68089-5
26. Tsujimura A, Matsumiya K, Miyagawa Y, et al. Prediction of successful outcome of microdissection testicular sperm extraction in men with idiopathic nonobstructive azoospermia. J Urol. 2004;172(5):1944–1947. doi:10.1097/01.ju.0000142885.20116.60
27. Vernaeve V, Verheyen G, Goossens A, Van Steirteghem A, Devroey P, Tournaye H. How successful is repeat testicular sperm extraction in patients with azoospermia? Hum Reprod. 2006;21(6):1551–1554. doi:10.1093/humrep/del012
28. Lin YM, Kuo PL, Lin YH, Teng YN, Lin JSN. Messenger RNA transcripts of the meiotic regulator BOULE in the testis of azoospermic men and their application in predicting the success of sperm retrieval. Hum Reprod. 2005;20(3):782–788. doi:10.1093/humrep/deh647
29. Schlegel PN, Palermo GD, Goldstein M, et al. Testicular sperm extraction with intracytoplasmic sperm injection for nonobstructive azoospermia. Urology. 1997;49(3):435–440.
30. Kim YS, Kim H-R, Kim H, et al. Deficiency in DGCR8-dependent canonical microRNAs causes infertility due to multiple abnormalities during uterine development in mice. Sci Rep. 2016;6:20242. doi:10.1038/srep20242
31. Eisenberg I, Kotaja N, Goldman-Wohl D, Imbar T. microRNA in human reproduction. Adv Exp Med Biol. 2015;888:353–387.
32. Schwanhäusser B, Busse D, Li N, et al. Corrigendum: global quantification of mammalian gene expression control. Nature. 2013;495(7439):126.
33. Tang F, Kaneda M, O’Carroll D, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21(6):644–648.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.