Back to Journals » Breast Cancer: Targets and Therapy » Volume 9
CYP2D6 polymorphisms may predict occurrence of adverse effects to tamoxifen: a preliminary retrospective study
Authors Wickramage I , Tennekoon KH , Ariyaratne MAY, Hewage AS, Sundralingam T
Received 3 November 2016
Accepted for publication 2 February 2017
Published 6 March 2017 Volume 2017:9 Pages 111—120
DOI https://doi.org/10.2147/BCTT.S126557
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Pranela Rameshwar
Video abstract presented by Ishani Wickramage.
Views: 1739
Ishani Wickramage,1 Kamani Hemamala Tennekoon,1 Merenchi Arachchige Yasantha Ariyaratne,2 Asanka Sudeshini Hewage,1 Tharmini Sundralingam,1
1Institute of Biochemistry, Molecular Biology and Biotechnology (IBMBB), University of Colombo, Colombo, Sri Lanka; 2National Cancer Institute, Maharagama, Sri Lanka
Introduction and aims: Tamoxifen is an adjuvant drug effective in treating hormone receptor – positive breast cancer. However, 30%–50% of patients relapse and many develop adverse effects, such as hot flashes and fatty liver. Allelic variations altering the activity of cytochrome P450-2D6 enzyme affect response to tamoxifen by modulating metabolism of tamoxifen into its pharmacologically active metabolite endoxifen. Although association between CYP2D6 polymorphisms and recurrence of breast cancer in patients on tamoxifen had been reported, little evidence exists on association between these polymorphisms and adverse effects to tamoxifen. This study explored the association between CYP2D6 polymorphisms and tamoxifen effects, hitherto not studied in Sri Lanka.
Methods: A retrospective preliminary study was carried out on 24 breast cancer patients on tamoxifen for minimally 3 months attending National Cancer Institute, Maharagama, Sri Lanka. They were not on CYP2D6-inhibiting drugs, chemotherapy or other endocrine therapy, and had no conditions that could occur as adverse effects to tamoxifen before starting the therapy. Their blood samples were collected, DNA was extracted and genotyped using SNaPshot Multiplex sequencing based single-nucleotide polymorphism (SNP) assay.
Results: SNP/allele frequencies detected: 1846G>A (confirmatory of *4 null allele)=8.3%; 2549delA (confirmatory of *3 null allele)=50%; 100C>T (suggestive of *10 reduced functional allele, in addition to other alleles)=0%; combination of 2988G>A, -1584C and 2850C>T (strongly suggestive of *41 or other reduced functional allele)=4.8%. Occurrence of heterozygous 2988G>A SNP with -1584C and 2850C>T was significantly higher among those with ultrasound-diagnosed fatty liver following the commencement of tamoxifen therapy (P=0.029). Adverse effects occurred at a significantly higher frequency among postmenopausal women (P=0.041). Three patients who developed recurrence of breast cancer had no association with SNPs tested.
Conclusions: CYP2D6 SNP combination 2988G>A, -1584C and 2850C>T, strongly suggestive of *41 reduced functional allele, is likely to be useful in predicting occurrence of adverse effect fatty liver in breast cancer patients on tamoxifen, thereby alternative treatment can be considered and lifestyle modifications implemented. Larger sample studies are recommended with the measurement of tamoxifen and metabolite levels. Alternative therapy should be considered for postmenopausal patients.
Keywords: fatty-liver, 2988G>A, CYP2D6*41, intermediate-metabolizer, SNP
Introduction
Breast cancer is the commonest cancer among females in Sri Lanka, accounting for 27% of all cancers in women.1 In 2010 in Sri Lanka, 2401 women were diagnosed with breast cancer (crude incidence rate=23.1, age standardized rate=23.0) and 659 women died due to breast cancer (crude death rate=6.3, age standardized rate=6.4).1 One in 40 females in Sri Lanka has a life-time risk of developing breast cancer.1
Estrogen receptor- and progesterone receptor-positive (PgR+) breast cancers depend on these hormones for their growth and replication.2 Most breast cancers (75%) are estrogen receptor positive (ER+), and out of those, 65% are also PgR+.3 Tamoxifen acts as an antagonist on ERs in breast tissue4 and blocks the effects of estrogen on proliferation of breast cancer cells.5 It is effective in ~70% of ER+ and/or PgR+ breast cancers.3 However, 30%–50% of patients on tamoxifen therapy have been known to relapse.6
Tamoxifen is a pro drug. It must be metabolized into its abundant and pharmacologically active metabolite endoxifen (4-hydroxy-N-desmethyltamoxifen) to elicit its optimal action.7 Tamoxifen is a selective estrogen receptor modulator (SERM).4 It shows ER agonist, antagonist or mixed activity in different tissues.5
Many patients on tamoxifen develop adverse effects to this drug. They are frequently reported by patients as a reason for discontinuation of therapy, which would otherwise be potentially lifesaving.8 The estrogenic or antiestrogenic actions on non-breast tissues account for some of the undesirable adverse effects of tamoxifen.
The commonest adverse effect of tamoxifen is hot flashes.9 Fisher et al found that ~80% of women receiving tamoxifen experienced hot flashes.9 Hot flashes are sudden and transient episodes of heat and sweating with possible palpitations and anxiety.10 Their pathophysiology is unclear and appears to involve thermoregulatory and neurochemical disruptions.9 Estrogen appears to stabilize these thermoregulatory disruptions,10 but poor correlation of frequency and severity of hot flashes with plasma estrogen levels suggest that estrogen does not have an exclusive role in the pathophysiology.8 The antiestrogenic action of tamoxifen seems to be responsible for the development of hot flashes.
Tamoxifen acts as an agonist on endometrial tissue.4 It stimulates the proliferation of endometrial cells and causes endometrial thickening.5 It also increases the risk of endometrial cancer.9 Possibly through the same agonist action on the uterus, it is also known to cause adverse effects, such as uterine fibroids, post-menopausal vaginal bleeding and menstrual changes.11
Fatty liver or hepatic steatosis is the deposition of fat within hepatocytes.12 Approximately 43% of patients on tamoxifen therapy are known to develop fatty liver.13 Tamoxifen-induced hepatic steatosis is commonly of macrovesicular type14 and is found to occur in the majority of patients within the first 2 years of therapy.14 A study done by Cole et al revealed that increased de novo fatty acid synthesis is the primary event leading to tamoxifen-induced fatty liver in the mouse.13 This was explained by the marked decrease in the phosphorylation, and thereby the activation of acetyl-coenzyme A carboxylase, which catalyzes the committed step in fatty acid synthesis.13 In vitro studies suggest that the upregulation of sterol regulatory element-binding protein-1c and its downstream lipogenesis target genes may be the key mechanism of tamoxifen-induced hepatic steatosis.15 The evidence for the effect of tamoxifen on fatty acid β-oxidation is conflicting. Although Cole et al13 and Gudbrandsen et al16 found β-oxidation of fatty acids to be unchanged among rodents, several others found that β-oxidation was inhibited with the use of tamoxifen.12,17,18 The remaining fatty acids were found to be converted to triglycerides resulting in hepatic steatosis.12
The risk factors associated with the development of tamoxifen-induced liver injury are the same factors associated with traditional nonalcoholic fatty liver disease (NAFLD), namely diabetes mellitus, hypertension, hyperlipidemia and obesity.14 Tamoxifen-induced macrovesicluar steatosis is thought to occur mainly by worsening of underlying NAFLD12 and also by increasing the development of fatty liver and retarding fatty liver improvement.19
Serum aminotranferase levels are known to increase with a number of liver abnormalities known to occur with long-term tamoxifen therapy, such as fatty liver, steatohepatitis, liver fibrosis, cirrhosis and rarely, acute liver injury, which is considered to probably occur due to an idiosyncratic reaction to a tamoxifen metabolite rather than estrogenic effects.20 Elevated liver enzymes usually return to normal within 6 months of stopping tamoxifen therapy.14 It is unclear which particular metabolites of tamoxifen play specific roles in causing liver abnormalities.20 Increased transaminase levels seen in tamoxifen-induced hepatic steatosis is attributed to the inhibition of fatty acid β-oxidation, which causes inhibition of the electron transport chain resulting in the formation of reactive oxygen species (ROS).12 ROS interact with nonesterified polyunsaturated fatty acids (PUFA) to produce lipid peroxidation products, which cause hepatic damage.12 However, only half of the patients who develop tamoxifen-associated hepatic steatosis show increase in liver transaminase levels.14
Metabolism of tamoxifen depends on hepatic cytochrome P450 enzymes (CYPs). Tamoxifen is N-methylated to N-desmethyltamoxifen, predominantly by CYP3A4/5.21 Tamoxifen is also converted to 4-hydroxytamoxifen, principally by CYP2D6.3 Both these metabolites are oxidized to form endoxifen.3 Conversion of N-desmethyltamoxifen to endoxifen is almost exclusively catalyzed by CYP2D6.21 Although 4-hydroxytamoxifen and endoxifen have equivalent antiestrogenic potencies and have higher affinity to the ER than tamoxifen or N-desmethyltamoxifen, endoxifen plasma concentrations are 6- to 12-fold higher than 4-hydroxytamoxifen.21 Therefore, endoxifen is considered the main, abundant, pharmacology-active metabolite of tamoxifen responsible for its activity.3,21 Out of the two pathways of tamoxifen metabolism, the formation of N-desmethyltamoxifen is the major route of tamoxifen’s metabolism to endoxifen.21 Therefore, it is evident that as the main pathway of the formation of endoxifen from tamoxifen largely depends on CYP2D6, alterations in this enzyme would considerably affect the activity of tamoxifen.
Groups of single-nucleotide polymorphisms (SNPs) occurring in CYP2D6 gene result in numerous allelic variations. CYP2D6 is a highly polymorphic gene and more than 100 alleles have been reported to date.22 The nucleotide changes reported to date responsible for these alleles are published in The Human Cytochrome P450 (CYP) Allele Nomenclature Database,22 and those of the alleles selected to be studied in this research are described in Table 1. Many CYP2D6 alleles (and some SNPs alone) alter the level of activity of the resulting CYP2D6 enzyme. Some alleles render the resulting enzyme inactive (eg, *3 and *4).22 Some alleles (eg, *10, *17 and *41) reduce the level of activity of the enzyme.22 Alleles such as *1 and *2 result in normally functional enzyme.22 On the basis of CYP2D6 allele combinations, 4 metabolizer phenotypes have been identified. They are poor metabolizers (PM), intermediate metabolizers (IM), extensive metabolizers (EM) and ultra-rapid metabolizers (UM).2 Bearers of two inactive alleles are classified as PM.7 Those who carry one or two decreased activity alleles and those who carry one inactive or decreased activity allele in combination with a functional allele are categorized as IM.7 Those with more than two copies of functional alleles are called UM.7 People with normal enzyme activity are EM.3
  | Table 1 The CYP2D6 SNPs selected to be analyzed in this study Note: Nucleotide variations in bold are the major SNPs/alterations responsible for the phenotype of the corresponding allele.22 Abbreviation: SNPs, single-nucleotide polymorphisms. |
The CYP2D6 allele variations and the resulting metabolizer status phenotype show significant interethnic differences. PM phenotype is predominantly found among Caucasians with a high prevalence of 5%–10%, whereas <1% of the East Asians showed this phenotype.23 CYP2D6*4 null allele is most prevalent among Caucasians.23 1846G>A SNP exclusively found in the *4 allele22 is confirmatory of the presence of this allele. Another null functional allele contributing to the PM phenotype, *3, has a deletion of an adenine nucleotide at its 2549th position, which is specific to this allele.22 CYP2D6*10 reduced-functional allelic variation and the IM phenotype are most commonly found in Asians.23 CYP2D6*41 is also a reduced functional allele and a variant of CYP2D6*2.24 They both contain 2850C>T, but only CYP2D6*41 has −1584C in its promoter region instead of G.24 In addition, CYP2D6*41 has the intron 6 SNP 2988G>A, which has been unequivocally identified by Toscano et al as the causative mutation responsible for the low expression of this allele, with strong evidence for its modulating activity in alternative splicing events.24 2988G>A is also detected in two other alleles: CYP2D6*69 which is a reduced functional allele and CYP2D6*91 which causes a splicing defect with as yet unreported activity level of the resulting enzyme.22 Therefore, although the presence of 2988G>A SNP is not confirmatory of CYP2D6*41 allele, the presence of this SNP alone can be considered to reduce the activity of the resulting enzyme.
To date, two studies have been published in Sri Lanka on CYP2D6. Weerasuriya et al detected the CYP2D6*4 (CYP2D6B) null functional allele at an allele frequency of 9% and did not detect the null functional CYP2D6*3 (CYP2D6A) allele tested.25 In 2013, Tharanga et al detected the following alleles CYP2D6*3 (null functional), *10 (poorly functional), *2 (functional) and *4 (null functional) at the respective allele frequencies of 44.9%, 39.3%, 38.1% and 12.2%.26
Although variable associations between CYP2D6 polymorphisms and recurrence of breast cancer in patients on tamoxifen have been reported throughout the world, little evidence exists on association between these polymorphisms and adverse effects to tamoxifen. Genetic functional polymorphisms of CYP450c17 gene, which codes for the P450c17α enzyme responsible for regulating serum estrogen, has been found to possibly influence the individual susceptibility to develop tamoxifen-induced hepatic steatosis.27 However, this enzyme does not alter the metabolism of tamoxifen. The genetic polymorphisms with direct effect on tamoxifen metabolism and their association with the development of adverse effects to tamoxifen need to be studied further.
This study explored the association between CYP2D6 polymorphisms and tamoxifen effects, hitherto not studied in Sri Lanka.
Methods
Ethical clearance for this study was obtained from the Ethical Review Committee, Faculty of Medicine, University of Colombo, Sri Lanka (EC-15-024).
A preliminary retrospective study was carried out on 24 breast cancer patients during May to July 2015, who have been on tamoxifen 20 mg daily for minimally 3 months.
All study participants recruited were breast cancer patients who were registered to obtain treatment at National Cancer Institute, Maharagama, Sri Lanka. Patients attended the hospital clinic at frequencies such as once a month, once in 3 months or once in 6 months, depending on the progress of their treatment. Each patient had a file maintained under her name in the clinic, which contained her bed head tickets of hospital admissions, records of clinical assessments by doctors during clinic visits, investigation reports and drug charts.
Study participants were recruited on a day of their routine clinic visit. Prior to recruitment, patients were assessed for exclusion criteria. Those who were concurrently on other medicines known to inhibit CYP2D6, cytotoxic chemotherapy and other endocrine therapy, as recorded on the patients’ drug charts in the hospital records, were not recruited. Also, those who showed evidence of having conditions before the date of commencement of the tamoxifen therapy, which could also arise as adverse effects of tamoxifen, were excluded from the study. Presence of hot flashes, menstrual changes and postmenopausal vaginal bleeding prior to starting tamoxifen were excluded by examining the clinicians’ records of clinical assessment of the patient and confirmed by interviewing the patient prior to recruitment. Diagnoses of fatty liver, uterine fibroids and endometrial thickening were excluded by examining the hospital records of investigations performed by doctors treating the patient, performed for the initial assessment of breast cancer and in preparation for surgery for the treatment of breast cancer. Blood reports of serum alanine transaminase (ALT) and aspartate transaminase levels were checked to exclude abnormal liver functions prior to commencement of the tamoxifen therapy.
Selected patients who met these eligibility criteria were then recruited to the study after obtaining informed written consent. The study participants were then interviewed using an interviewer administered questionnaire, and further assessment of hospital records was carried out.
Patients’ clinic files were checked for the records following commencement of the tamoxifen therapy. Diagnoses of fatty liver and uterine fibroids were obtained from the investigative reports of abdominal ultrasound scans performed by doctors treating the patient. Information about hot flashes, menstrual changes and postmenopausal vaginal bleeding were obtained by viewing the clinic records of the doctors’ clinical assessment of the patient during clinic visits and confirmed by interviewing the study participant by the investigators. Endometrial thickening was assessed using investigation records of trans-vaginal ultrasound scans performed by the doctors treating the patient. Blood reports were assessed for liver function tests.
Several CYP2D6 SNPs were selected to be studied in these patients based on two features of the resulting alleles. The SNPs selected were the major alterations responsible for alleles which reduced the level of activity of the resulting CYP2D6 enzyme and alleles that have been found or suspected to be present among Sri Lankans based on previous studies. The participants’ venous blood samples were collected. DNA was extracted using modified Miller’s protocol. Polymerase chain reaction amplified DNA samples were genotyped using SNaPshot Multiplex sequencing based SNP assay.
SNaPshot Sample reaction mix was prepared using ABI PRISM® SNaPshot™ Multiplex kit. Applied BiosystemsTM 3500 Dx Series Genetic Analyzer machine was used for capillary electrophoresis and the results were analyzed using GeneMapper software. All laboratory procedures were carried out at Institute of Biochemistry, Molecular Biology and Biotechnology, University of Colombo, Sri Lanka. IBM Statistical Package for the Social Sciences (SPSS) statistics software version 21 was used to statistically analyze the data. As the sample size was small, two-sided Fisher’s exact test was used to determine the association between genotype and clinical response. The level of significance was set at P<0.05.
Results
The CYP2D6 SNPs 1846G>A, 2549delA, 100C>T, 2988G>A, 2850C>T and −1584C>G were selected to be analyzed in this study, as shown in Table 1. The demographic and clinical details of the study participants are shown in Table 2.
  | Table 2 The demographic and clinical details of the study participants Abbreviations: M, menopausal; m, months; +, present; −, absent; P, premenopausal; y, years. |
Table 3 shows the frequency of CYP2D6 SNPs detected among the study participants. The response of each participant to tamoxifen in developing either a recurrence of breast cancer or adverse effects is shown in Table 4. This table also elaborates the CYP2D6 SNPs detected in each participant.
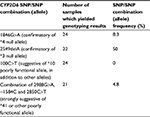  | Table 3 Frequency of CYP2D6 gene SNPs among study participants Abbreviation: SNPs, single nucleotide polymorphisms. |
The associations between the presence of CYP2D6 SNP combinations/alleles and the development of adverse effects to tamoxifen are shown in Table 5.
As SNPs 1846G>A and 2549delA are specific to the alleles CYP2D6*4 and CYP2D6*3, respectively, the presence of these SNPs were considered confirmatory of these alleles. 2988G>A together with 2850C>T and −1584C was considered to be strongly suggestive of CYP2D6*41. As explained earlier, the presence of SNP 2988G>A alone was taken to be highly predictive of reduced metabolism of the tamoxifen.
All SNPs detected were present in heterozygosity, except for one patient who had homozygous TT genotype for the 2850C>T SNP. There was no association between developing recurrence of breast cancer and the SNPs tested. SNP/allele frequencies detected were as follows: 1846G>A (confirmatory of *4 null allele)=8.3%; 2549delA (confirmatory of *3 null allele)=50%; 100C>T (suggestive of *10 poorly functional allele, in addition to other alleles)=0%; combination of 2988G>A, −1584C and 2850C>T (strongly suggestive of *41 or other poorly functional allele)=4.8%.
Adverse effects occurred at a significantly higher frequency among postmenopausal women (P=0.041).
Four patients had fatty liver diagnosed following the commencement of tamoxifen therapy. Only one of these patients (S19) showed increased ALT levels. None of the four patients had diabetes mellitus or hypertension. Only one patient (S19) had diagnosed hyperlipidemia during tamoxifen therapy.
Occurrence of heterozygous 2988G>A SNP together with 2850T and −1584C was significantly higher among those with ultrasound-diagnosed fatty liver following the commencement of tamoxifen therapy (P=0.029; Table 5). 1846G>A SNP was detected among patients who experienced increased ALT enzymes (P=0.312), hot flashes (P=0.437) and increased endometrial thickness (P=0.437), but these findings were not statistically significant (Table 5). 2988G>A SNP together with 2850T and −1584C was also nonsignificantly seen among patients who had increased ALT levels (P=0.186), hot flashes (P=0.271) and increased endometrial thickness (P=0.271; Table 5).
Discussion
Those who are heterozygous for a reduced functional CYP2D6 allele are classified as IM.7 Therefore, the patients in our study who were heterozygous for 2988G>A SNP together with 2850T and −1584C, strongly suggestive of *41 or other poorly functional allele, can be classified as IM. Bearers of one inactive allele such as CYP2D6*4 in combination with a functional allele are also categorized as IM.7 Therefore, all four patients in our study in whom the 1846G>A SNP was detected in heterozygosity are IM.
Our study showed a significant association between the presence of heterozygous 2988G>A SNP together with 2850T and −1584C, strongly suggestive of *41 or other poorly functional allele, thus making these patients IM, and the development of fatty liver in patients after starting tamoxifen therapy. Through the examination of hospital records, the patients were found to have had normal hepatic status prior to starting tamoxifen. Our study established the temporal relationship between the commencement of tamoxifen therapy and occurrence of fatty liver. However, the causation cannot be proven from our study.
CYP2D6 almost exclusively catalyzes the last step of the major pathway of metabolism of tamoxifen, conversion of N-desmethyltamoxifen to endoxifen.21 Also, CYP2D6 is largely involved in its minor metabolic pathway, conversion of tamoxifen to 4-hydroxytamoxifen, which is subsequently oxidized to endoxifen.3,21 Individuals with reduced activity of CYP2D6 enzymes such as IM have lower levels of endoxifen than their wild-type counterparts. Therefore, it can be postulated that the occurrence of adverse effect fatty liver may have a positive association with the low levels of the pharmacologically active metabolite endoxifen. As documented in several studies, tamoxifen causes mitochondrial dysfunction.12,17,18 When hepatic mitochondrial β-oxidation is inhibited, the accumulated fatty acids are converted to triglycerides, which result in hepatic steatosis.12 Ribeiro et al found that endoxifen has less harmful effects on mitochondria when compared with tamoxifen.28 Therefore, those with the lower levels of endoxifen are more likely to suffer mitochondrial dysfunction and develop fatty liver. These findings may explain the significantly higher levels of fatty liver detected among the IM in our study. We recommend further studies with the measurement of levels of tamoxifen and its metabolites, especially endoxifen, to confirm the association between CYP2D6 polymorphisms and hepatic adverse effects of tamoxifen.
Our study also detected the heterozygous SNP 1846G>A (CYP2D6*4) and combination of heterozygous 2988G>A, 2850T and −1584C in patients who developed elevation of ALT levels, hot flashes and endometrial thickening following the commencement of tamoxifen therapy. However, the association between these SNPs and potential tamoxifen adverse effects did not reach statistical significance.
Mitochondrial damage and the resultant inhibition of the electron transport chain result in the formation of ROS, which react with PUFA to produce lipid peroxidation products, which damage the liver.12 This could lead to the elevation of ALT levels. As endoxifen causes less mitochondrial damage than tamoxifen, those with lower levels of endoxifen, that is, IM when compared with EM are more likely to have elevated ALT levels. This was detected in our study, albeit at a nonsignificant level.
Hot flashes because of tamoxifen therapy are hypothesized to occur because of the potent antiestrogen action of endoxifen.21 It was also found by Briest et al that those with CYP2D6 alleles which reduced the metabolism of tamoxifen, who thereby had less endoxifen, were less likely to develop hot flashes.21 A contradictory observation was made in our study that those with IM phenotypes, with heterozygous *4 or potential *41 alleles, had a statistically nonsignificantly higher likelihood of developing hot flashes. A larger sample will probably reinstate the nonsignificance of this association. Women who reported hot flashes have been found less likely to suffer a breast cancer recurrence,21 possibly because these women had higher levels of endoxifen, which is the pharmacologically active metabolite of tamoxifen crucial for its therapeutic effect.3 None of the three patients in our study who had recurrences while on tamoxifen reported hot flashes. However, the sample size in this study is too small to arrive at strong conclusions.
Being an SERM, tamoxifen has estrogen agonist actions on the uterus4 and causes proliferation and thickening of the endometrium.5 Tamoxifen and endoxifen have similar estrogenic effects on the uterus.29 Therefore, metabolizer status of tamoxifen is not expected to have an association with endometrial effects. However, our study detected a nonsignificant association between intermediate metabolizer phenotypes, with heterozygous *4 or potential *41 alleles, and endometrial thickening. As earlier, a larger sample will perhaps magnify this nonsignificance.
Similar to the findings of other studies,9 this study found a statistically significant higher frequency of occurrence of adverse effects among postmenopausal women when compared with premenopausal women. Tamoxifen is considered the adjuvant drug of choice in premenopausal women.3 The risk:benefit ratio for high-risk women who use tamoxifen as a chemopreventive drug is most favorable during the premenopausal period30 because tamoxifen is not known to increase the incidence of blood clots, stroke or endometrial cancer in premenopausal women.9 Postmenopausal women have been found to receive higher benefit from aromatase inhibitors in the adjuvant setting.3 Therefore, the higher occurrence of adverse effects of tamoxifen among postmenopausal women found in this study also favors the use of alternative therapy, such as aromatase inhibitors for postmenopausal patients.
CYP2D6 2988G>A polymorphism together with 2850T and −1584C is likely to be useful in predicting the occurrence of adverse effect fatty liver in breast cancer patients on tamoxifen, and thereby alternative treatment can be considered and lifestyle modifications implemented. Alternative therapy besides tamoxifen should be considered for postmenopausal patients.
Limitations
The biggest limitation in this study is the small sample size. We recommend further studies with larger samples and measurement of levels of tamoxifen and its metabolites to further assess the association between CYP2D6 polymorphisms and adverse effects of tamoxifen.
Disclosure
The authors report no conflicts of interest in this work.
References
National Cancer Control Program. Cancer Incidence Data Sri Lanka 2010. 12th pub. Colombo: National Cancer Control Program, Ministry of Health, Nutrition and Indigenous Medicine; 2016. | ||
Zafra-Ceres M, de Haro T, Farez-Vidal E, et al. Influence of CYP2D6 polymorphisms on serum levels of tamoxifen metabolites in Spanish women with breast cancer. Int J Med Sci. 2013;10(7):932–937. | ||
Hoskins JM, Carey LA, McLeod HL. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer. 2009;9(8):576–586. | ||
Rang HP, Dale MM, Ritter JM, et al. Anticancer drugs. In: Rang and Dale’s Pharmacology. 7th ed. Spain: Elsevier Inc; 2012:673–688. | ||
Brunton LL, Parker KL, editors. Goodman and Gilman’s Manual of Pharmacology and Therapeutics. 7th ed. New York, NY: McGraw-Hill; 2008. | ||
Clarke R, Leonessa F, Welch JN, Skaar TC. Cellular and molecular pharmacology of antiestrogen action and resistance. Pharmacol Rev. 2001;53(1):25–71. | ||
Dean L. Tamoxifen therapy and CYP2D6 Genotype. medical genetics summaries [Internet]. [Created October 7, 2014. Last Update: May 3, 2016]. Avialbale from: http://www.ncbi.nlm.nih.gov/books/NBK247013. Accessed September 6, 2016. | ||
Klingman L, Younus J. Management of hot flashes in women with breast cancer. Curr Oncol. 2010;17(1):81–86. | ||
Fisher B, Costantino JP, Wickerham DL, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90(18):1371–1388. | ||
Fisher WI, Johnson AK, Elkins GR, et al. Risk factors, pathophysiology, and treatment of hot flashes in cancer. CA Cancer J Clin. 2013;63(3):167–192. | ||
Joint Formulary Committee. British National Formulary. London: BMJ Group and Pharmaceutical Press; 2015:167–192. | ||
Patel V, Sanyal AJ. Drug-induced steatohepatitis. Clin Liver Dis. 2013;17(4):533–546. | ||
Cole LK, Jacobs RL, Vance DE. Tamoxifen induces triacylglycerol accumulation in the mouse liver by activation of fatty acid synthesis. Hepatology. 2010;52(4):1258–1265. | ||
Rabinowich L, Shibolet O. Drug induced steatohepatitis: an uncommon culprit of a common disease. Biomed Res Int. 2015;2015:168905. | ||
Zhao F, Xie P, Jiang J, Zhang L, An W, Zhan Y. The effect and mechanism of tamoxifen-induced hepatocyte steatosis in vitro. Int J Mol Sci. 2014;15(3):4019–4030. | ||
Gudbrandsen OA, Rost TH, Berge RK. Causes and prevention of tamoxifen-induced accumulation of triacylglycerol in rat liver. J Lipid Res. 2006;47(10):2223–2232. | ||
Farrell GC. Drugs and steatohepatitis. Semin Liver Dis. 2002;22(2):185–194. | ||
Larosche I, Letteron P, Fromenty B, et al. Tamoxifen inhibits topoisomerases, depletes mitochondrial DNA, and tiggers steatosis in mouse liver. J Pharmacol Exp Ther. 2007;321(2):526–535. | ||
Pan HJ, Chang HT, Lee CH. Association between tamoxifen treatment and the development of different stages of nonalcoholic fatty liver disease among breast cancer patients. J Formos Med Assoc. 2016;115(6):411–417. | ||
U.S. National Library of Medicine [Internet]. Drug record: tamoxifen. LiverTox, clinical and research information on drug-induced liver injury; [updated 2016 June 28]. Available from: http://livertox.nih.gov/Tamoxifen.htm. Accessed September 6,2016. | ||
Briest S, Stearns V. Tamoxifen metabolism and its effect on endocrine treatment of breast cancer. Clin Adv Hematol Oncol. 2009;7(3):185–192. | ||
Human Cytochrome P450 (CYP) Allele Nomenclature Committee. CYP2D6 allele nomenclature. Available from: http://www.cypalleles.ki.se/cyp2d6.htm. Accessed September 6, 2016. | ||
De Souza JA, Olopade OI. CYP2D6 genotyping and tamoxifen: an unfinished story in the quest for personalized medicine. Semin Oncol. 2011;38(2):263–273. | ||
Toscano C, Klein K, Blievernicht, et al. Impaired expression of CYP2D6 in intermediate metabolizers carrying the *41 allele caused by the intronic SNP 2988G>A: evidence for modulation of splicing events. Pharmacogenet Genomics. 2006;16(10):755–765. | ||
Weerasuriya K, Jayakody RL, Smith CA, Wolf CR, Tucker GT, Lennard MS. Debrisoquine and mephenytoin oxidation in Sinhalese: a population study. Br J Clin Pharmacol. 1994;38(5):466–470. | ||
Tharanga TDP, Jinadasa CMV, Risama MF, Galappatthy P, Jayakody RL, Dissanayake VHW. Genetic variants in the cytochrome P450 2D6 gene in the Sri Lankan population. Indian J Hum Genet. 2013;19(4):392–396. | ||
Ohnishi T, Ogawa Y, Saibara T, et al. CYP17 polymorphism as a risk factor of tamoxifen-induced hepatic steatosis in breast cancer patients. Oncol Rep. 2005;13(3):485–489. | ||
Ribeiro MP, Santos AE, Custodio JB. Mitochondria: the gateway for tamoxifen-induced liver injury. Toxicology. 2014;323:10–18. | ||
Schweikart KM, Eldridge SR, Safgren SL, et al. Comparative uterotrophic effects of endoxifen and tamoxifen in ovariectomized Sprague-Dawley rats. Toxicol Pathol. 2014;42(8):1188–1196. | ||
Jordan VC. Tamoxifen (ICI46,474) as a targeted therapy to treat and prevent breast cancer. Br J Pharmacol. 2006;147 (Suppl 1):S269–S276. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.