Back to Journals » OncoTargets and Therapy » Volume 11
Crizotinib presented with promising efficacy but for concomitant mutation in next-generation sequencing-identified ROS1-rearranged non-small-cell lung cancer
Authors Zeng L, Li Y, Xiao L, Xiong Y, Liu L, Jiang W, Heng J, Qu J, Yang N, Zhang Y
Received 14 June 2018
Accepted for publication 17 September 2018
Published 15 October 2018 Volume 2018:11 Pages 6937—6945
DOI https://doi.org/10.2147/OTT.S176273
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Liang Zeng,* Yizhi Li,* Lili Xiao,* Yi Xiong, Li Liu, Wenjuan Jiang, Jianfu Heng, Jingjing Qu, Nong Yang, Yongchang Zhang
Department of Medical Oncology, Lung Cancer and Gastrointestinal Unit, Hunan Cancer Hospital/The Affiliated Cancer Hospital of Xiangya School of Medicine, Changsha 410013, China
*These authors contributed equally to this work
Introduction: Data of standard tyrosine kinase inhibitor (TKI) treatment outcome in next-generation sequencing (NGS)-identified ROS1-rearranged non-small-cell lung cancer (NSCLC) were rare. Thus, it is practical and necessary to evaluate the efficacy and influential factors of crizotinib in real-world practice.
Patients and methods: A total of 1,466 NSCLC patients with positive targeted NGS test results from September 2015 to January 2018 were enrolled in this real-world retrospective study. Twenty-two patients had ROS1 rearrangement detected by NGS. The efficacy and safety of crizotinib were evaluated. Subgroups of concomitant mutations, brain metastasis, and fusion variants were also analyzed.
Results: Among all the patients, the occurrence rate of ROS1 rearrangement was 1.5% (22 of 1,466). Ten ROS1 fusion partners were detected, and the most common variant was CD74, which accounted for 50% (11 of 22). Five patients were found to carry dual ROS1 fusion partners, and 23% (5 of 22) of patients were detected with concomitant mutations, including TP53&PIK3CA&mTOR mutation, TP53&CDKN2A mutation, TP53&BRCA2 mutation, ALK missense mutation (p.R311H), and MET amplification. Among 22 patients with ROS1-rearranged NSCLC, 20 patients were diagnosed at stage IV, and 19 patients received crizotinib treatment. The average follow-up period was 16 months. The overall response rate (ORR) of crizotinib in unselected crizotinib-treated patients was 89%, and the median progression-free survival time (mPFS) was 13.6 months. It was shown that NSCLC patients with exclusive ROS1 rearrangement had a longer PFS than those carrying concomitant mutations (15.5 vs 8.5 months, P=0.0213). There were no newly occurring intolerant adverse events in this study.
Conclusion: Crizotinib is highly effective in NGS-identified ROS1-rearranged advanced NSCLC in real-word clinical practice, and the data are consistent with previous clinical trials applying fluorescence in situ hybridization/real-time PCR for ROS1 companion diagnosis. Concomitant mutations may not be rare and may deteriorate the PFS of crizotinib in patients with ROS1-rearranged NSCLC.
Keywords: ROS1, next-generation sequencing, crizotinib, concomitant mutation, NSCLC, efficacy, TP53, safety
Introduction
Lung cancer is the primary cause of cancer-associated mortality worldwide, and non-small-cell lung cancer (NSCLC) accounts for 85% of lung cancer.1 A few oncogenic driver mutations have been established in NSCLC, especially in adenocarcinoma, such as EGFR mutation, anaplastic lymphoma kinase (ALK) rearrangements, ROS1 rearrangements, and BRAF V600E mutations.2–6 Targeting these mutations facilitates treatment decisions and the clinical management of NSCLC patients and significantly improves the prognosis of NSCLC patients.7–9 The ROS1 rearrangement was first reported in 2007, and it occurs in 1%–2% unselected NSCLC patients and 5% of NSCLC patients without EGFR mutations and ALK rearrangements.10–13 Similar to the ALK rearrangement, the ROS1 rearrangement was commonly found in young age group, nonsmokers, and patients with the adenocarcinoma histology.14–16 ROS1 is a receptor of tyrosine kinase in the insulin receptor family.10,17 The kinase domains of ALK and ROS1 rearrangement proteins are highly homogenous, sharing 77% amino acid identity within the ATP-binding sites.10,18 Therefore, the ALK inhibitor crizotinib was supposed to be effective for NSCLC patients with ROS1 rearrangement. On the basis of robust data of efficacy and safety observed in previous multicenter studies,10,14,19,20 crizotinib is recommended to treat NSCLC patients with ROS1 rearrangement. The US Food and Drug Administration (FDA) and China FDA approved crizotinib for the treatment of advanced NSCLC patients with ROS1 rearrangement in 2015 and 2018, respectively. In previous crizotinib efficacy studies, fluorescence in situ hybridization (FISH) and real-time (RT)-PCR were the predominant ROS1 identification methods, and FISH represents the gold standard for determining ROS1 positivity in clinical trials.21,22 Compared with FISH and RT-PCR, next-generation sequencing (NGS) has the advantage of the ability to detect and find specific and novel ROS1 fusion partners, the frequencies of ROS1 rearrangement, tumor mutational burden, and other concomitant mutations.23 This year, the National Comprehensive Cancer Network (NCCN) guidelines (version 1.2018) added NGS as a standard method to detect ROS1 fusion in NSCLCs to facilitate crizotinib treatment.24 To the best of our knowledge, no efficacy data of crizotinib in ROS1-rearranged NSCLC patients identified by NGS have been reported. Thus, we conducted this real-world retrospective study to verify the efficacy of crizotinib in ROS1-rearranged NSCLC identified by NGS as well as to explore the impact of potential genetic or clinical influential factors.
Patients and methods
Patients
This study was conducted in a group of NSCLC patients who received targeted NGS testing (Burning Rock Dx; Burning Rock Biotech Ltd, Guangzhou China; 56 gene or 168 gene panel) from September 2015 to January 2018 (time close to crizotinib launch) at the Hunan Cancer Hospital, Changsha, China. Patients with NSCLC identified with ROS1 rearrangement by targeted NGS were enrolled. A total of 22 patients with NSCLC were identified with ROS1 rearrangement using targeted NGS, and 19 of them received crizotinib treatment in their clinical course (Table 3). NGS-detected samples were tumor tissues (n=19), malignant plural effusions (n=1) embedded in paraffin samples, and plasma (n=1). Pathological diagnosis and staging were carried out according to the staging system of the 2009 International Association for the Study of Lung Cancer (version 8). All the eligible patients’ clinical data included patient age, gender, smoking status, histological type, Eastern Cooperative Oncology Group (ECOG) performance status (PS), and staging (Table 1). The clinical responses were evaluated according to the response evaluation criteria in solid tumors (RECIST) version 1.113. Progression-free survival (PFS) was measured from the first day of crizotinib administration until tumor progression or death. Approval was obtained from the ethics committee of Hunan Cancer Hospital.
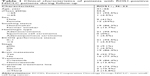  | Table 1 Clinical characteristics of patients with ROS1-positive NSCLC patients during follow-up |
Ethical statement
This project did not use the tissue samples, and all the medical records of patients were only for publication, not for any private business. So, patient consent was not required by the Ethics Committee in this project. This project was approved by ClinicalTrials.gov (NO NCT03646994) and Ethics Committee of Hunan Cancer Hospital (NO 2018230).
Specimen validation
Formalin-fixed tissues from patients’ tumor biopsies were used for targeted NGS in 19 patients. In addition, all samples were reviewed by pathologists to confirm the tumor histology. One patient’s plasma and one patient’s malignant plural effusion were used for NGS for lack of tumor tissue.
Next-generation sequencing
DNA was profiled using a commercially available capture-based targeted sequencing panel (Burning Rock Biotech Ltd, Guangzhou, China), targeting 56 or 168 genes. The concentration of the DNA samples was measured with the Qubit dsDNA assay. Fragments of 200–400 bp sizes were selected with beads (Agencourt AMPure XP kit; Beckman-Coulter, Brea, CA, USA), followed by hybridization with the capture probe baits, hybrid selection with magnetic beads, and PCR amplification. Then, a bioanalyzer high-sensitivity DNA assay was used to assess the quality and size range. Available indexed samples were sequenced on a Nextseq (Illumina, San Diego, CA, USA) with pair-end reads. In 19 NSCLC patients identified by targeted NGS in our follow-up, 5 patients were identified by the 168-gene panel and 14 patients were identified by the 56-gene panel. Panels were selected according to patients’ clinical characteristics and financial situation.
The 56 genes include ALK, BRAF, EGFR, ERBB2, KRAS, MET, RET, ROS1, AKT1, ARAF, CCND1, CDK4, CKD6, CKNNB1, DDR2, ERBB3, ERBB4, FGF3, FGF4, FGF19, FGFR2, FGFR3, FLT3, HRAS, JAK1, JAK2, KDR, KIT, KRAS, MAP2K1, MTOR, MYC, NRAS, NRG1, NTRK1, NTRK2, NTRK3, PDGFRA, PIK3CA, RTCH1, RAF1, SMO, ATM, BIM, BRCA1, BRCA2, CDKN2A, PTEN, RB1, STK11, TP53, TSC1, TSC2, CYP2D6, DPYD, and UGT1A1.
The 168 genes include APC, B2M, BRCA1, BRCA2, CCND1, CD274, CDK4, CDK6, CDKN2A, FGF19, FGF3, FGF4, KEAP1, KRAS, MLH1, MSH2, MSH6, MYC, NFE2L2, NRAS, PIK3CG, PMS2, POLE, POM121L12, PTEN, RB1, SMAD4, STK11, TP53, VEGFA, YES1, AKT1, ALK, AR, ARID1A, ATM, ATR, BARD1, BCL2L11, BCOR, BLM, BRAF, BRINP3, BRIP1, CARD11, CASP8, CBL, CCNE1, CD74, CDH18, CDKN1A, CDKN1B, CHEK1, CHEK2, CREBBP, CSMD3, CTNNB1, CYP2D2, DIS3, DNMT3A, DPYD, EGFR, EMSY, EP300, EPHA3, EPHA5, EPHA7, EPHB1, ERBB2, ERBB3, ERBB4, ESR1, FANCA, FANCI, FAT3, FBXW7, FGFR1, FGFR2, FGFR3, FLT1, FLT3, FLT4, GATA2, GATA3, GRIN2A, H3F3C, HGF, HIST1H1C, HIST1H3B, HIST1H3G, HRAS, IDH1, IDH2, IGF2, IKZF1, IL7R, INHBA, JAK1, JAK2, KDM5A, KDM6A, KDR, KIT, KMT2D, LRP1B, MAP2K1, MAP3K13, MAX, MCL1, MEN1, MET, MRE11A, MTOR, MUTYH, MYCH, NAV3, NBN, NF1, NOTCH1, NTRK1, NTRK2, NTRK3, PAK7, PALB2, PARK2, PARP1, PDGFRA, PDGFRB, PIK3C2G, PIK3C3, PIK3CA, PIK3R1, POLD1, PPP2R1A, PRKDC, PTPRD, PTPRT, RAD50, RAD51B, RAD51C, RAD51D, RAD54L, RAF1, RARA, RBM10, RET, RNF43, ROS1, RUNX1, SETD2, SMARCA4, SOX2, SOX9, SPOP, SPTA1, SRC, STAG2, TBX3, TERT, TGFBR, TP63, TRIM58, TRPC5, U2AF1, UGT1A1, VEGFB, VEGFC, and VHL.
Sequence data analysis
Sequence data were mapped to the human genome (hg19) using BWA aligner 0.7.10. Local alignment optimization, variant calling, and annotation were performed using GATK 3.2, MuTect, and VarScan. DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3.
Statistical analysis
Estimation of PFS was calculated using the Kaplan–Meier method. Statistical analysis was performed using Statistical Product and Service Solutions (version 5.01). The Cox proportional hazards model was used for multivariable survival analysis. Variables with a P-value <0.1 in the univariate analysis were included in the multivariate analysis. Schoenfeld residuals were used to check the proportional hazards assumption. All tests were two-sided, and P<0.05 was considered to be statistically significant. All statistical analyses were performed with R (version 3.3.3, the R Foundation for Statistical Computing, Vienna, Austria) and RStudio (version 1.1.383).
Results
Patients and clinical characteristics
A total of 1,466 NSCLC patients who received targeted NGS detection from September 2015 to January 2018 were included in this study. Twenty-two patients (1.5%) were identified with ROS1 rearrangement, and 19 patients received crizotinib treatment (first-line treatment, n=14; second-line treatment, n=2; ≥third-line treatment, n=3) (Tables 2 and 3). Among the 22 NSCLC patients with ROS1 rearrangement identified by targeted NGS, 21 patients had a histologic diagnosis of adenocarcinoma, except for 1 patient who was found to have adenocarcinoma mixed with squamous cell carcinoma. Other baseline clinical characteristics including age, gender, smoking status, and PS are outlined in Table 1. The specimens, targeted NGS panels, and genetic results of 19 crizotinib-treated ROS1-rearranged patients are listed in Tables 2 and 3.
Genetic characteristics
Twenty-two patients with NSCLC were detected with ROS1 rearrangement by NGS (Figure 1A). We found that five ROS1-rearranged patients carried dual ROS1 fusion partners: CD74&PUM1, CD74&MRAS, CD74&ADGRV1, EZR&BTBD9, and EZR&XPNPEP1, respectively (Figure 1B). In addition, 10 ROS1 fusion partners were detected, and the most common one was CD74, accounting for 50% (11 of 22); other ROS1 fusion partners included SDC4, TPM3, CCDC6, EZR, MRAS, ADGRV1, PUM1, BTBD9, and XPNPEP1 (Figure 1, Table 3). In addition, we found 23% (5 of 22) of ROS1-positive NSCLC patients with other concomitant mutations, including TP53&PIK3CA&mTOR mutation (n=1), TP53&CDKN2A mutation (n=1), TP53&BRCA2 mutation (n=1), ALK missense mutation (p.R311H) (n=1), and MET amplification (n=1).
  | Figure 1(A) Frequency of ROS1 variants (N=22). (B) ROS1 fusion variants. |
Efficacy of crizotinib in NSCLC patients with ROS1 rearrangement
A total of 19 ROS1-rearranged NSCLC patients were treated with crizotinib. Among them, no one had a complete response, 17 patients had a partial response, 1 patient had stable disease, and 1 patient had progressive disease (Tables 2 and 3 and Figure 2B). The overall response rate (ORR) was 89%, and the median PFS (mPFS) was 13.6 months evaluated by the Kaplan–Meier method (Figure 2C). By the Kaplan–Meier method, we found that NSCLC patients with exclusive ROS1 rearrangement had a longer PFS than those carrying concomitant mutations (mPFS 15.5 vs 8.5 months; P=0.0213) (Figure 2D). There was no significant impact on PFS considering brain metastasis, fusion partner subtypes (CD74 vs non-CD74 group), mutation abundance, and single or dual fusion partners in this small sample size study (Tables 2 and 4).
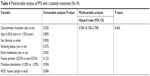  | Table 4 Multivariable analysis of PFS with crizotinib treatment (N=19) |
Safety of crizotinib in NSCLC patients with ROS1 rearrangement
There were no newly occurring or intolerant adverse events compared with other studies. Generally, crizotinib was tolerated and safe in NSCLC patients with ROS1 rearrangement.
Discussion
According to our literature search of April 5, 2018, this was the first real-world study to assess the efficacy of crizotinib and search the correlation between the response duration time of tyrosine kinase inhibitor (TKI) and the presentation of additional mutations in NGS-identified ROS1-rearranged NSCLC patients. Using RT-PCR to identify ROS1 rearrangement, the phase II study conducted in East Asian patients showed an mPFS of 15.9 months and an ORR of 71.7%.20 Another phase I study on which the FDA gave crizotinib approval exhibited an mPFS of 19.2 months and an ORR of 72%.25 As reported by Davies et al, ROS1 rearrangement detection in the clinical setting with break-apart fluorescence in situ hybridization, DNA-based hybrid capture library preparation, followed by NGS and RNA-based anchored multiplex PCR library preparation followed by NGS is complex, and all methodologies have inherent limitations. However, to identify high-degree genotype, our study chose NGS to access ROS1 rearrangement.26 Our results showed that the mPFS was 13.6 months and the ORR was 89%, in spite of the small simple size; these findings represent the robust efficacy of crizotinib in NGS-identified ROS1-rearranged NSCLC patients.
In addition, we found that co-occurring mutations were not rare in patients with ROS1 rearrangement and may negatively affect the PFS of crizotinib. A previous study showed that ROS1 rearrangements were not mutually exclusive with other transformation-associated genetic aberrations, as the majority of the patients presented with additional mutations.27 It was reported in a recent letter to JAMA Oncology that the common presence of co-occurring genetic alterations may be associated with resistance to EGFR TKIs in patients with EGFR-mutant NSCLC.28
We identified that 5 of 22 patients with ROS1 rearrangements had additional mutations, and by Kaplan–Meier curve, we found that these patients had a significantly inferior PFS with crizotinib. We suppose that bypass activation of survival signaling pathways or tumor heterogeneity may contribute to the negative PFS with crizotinib in these patients. With the prevalence of NGS, a genome-identified subgroup showed the important role of precisely targeted treatment. A larger sample size study focusing on NGS identified multiple mutations that are needed for the development of a genome-identified subgroup.
Unlike another study by Li et al that evaluates the ROS1 fusion partners by Sanger sequence, our study used NGS to perform the genotype of ROS1.29 The higher dual fusion partner detection rate may be correlated with the high sensitivity of NGS compared with Sanger sequence.30 Of interest, a recent study has reported that different types of ROS1 fusion partners may be associated with the PFS of crizotinib in patients with ROS1 rearrangements, as the CD74 fusion subgroup showed inferior PFS compared with the non-CD74 group (17.63 vs 12.63 months; P=0.048).29 A tendency of longer PFS was observed in the non-CD74 subgroup, although it had no significance in our study. However, we observed that concomitant mutations were more likely to occur in the CD74 group (four of five), which may provide a rationale to the mechanism underlying the negative impact in PFS of crizotinib for NSCLC patients with CD74-ROS1 rearrangement.
Whereas in the past tyrosine kinase gene mutations were recognized as independent and mutually exclusive in lung cancers, the coexistence of kinase gene mutations such as EGFR and ALK was reported in recent years.31–34 In our study, one patient was identified with ROS1 fusion plus MET amplification and received crizotinib as the first-line treatment; however, this patient eventually proved nonresponsive to crizotinib and died shortly after with an overall survival of 1.5 months. The mechanism underlying this negative response was unknown.
Five patients in our study were detected with dual fusion partners of ROS1. All these patients were found as a common fusion partner (CD74=3, EZR=2) with another unreported fusion partner (MRAS=1, ADGRV1=1, PUM1=1, BTBD9=1, XPNPEP1=1). We found that both the partners were fused to the same site of the ROS1 gene in each patient; however, which one was the driver fusion was unknown. Besides, only one of these five patients with dual ROS1 fusion had disease progression, and whether dual fusion partners might play a role in response to crizotinib was not assessed. To analyze influential genetic factors of crizotinib in patients with ROS1 rearrangement, larger scale clinical studies and basic experiments are required. There are many detailed influential factors of the first-generation ROS1 TKI treatment that remain unknown and require further studies to elucidate.
Considering brain metastases, 5 of 19 crizotinib-treated patients were found to have brain metastases at first diagnosis and, these patients exhibited inferior PFS compared with those without brain metastases (8.5 vs 13.6 months), although with no significance. Whether brain metastases negatively affect the PFS with crizotinib needs more study and larger samples to elucidate.
There are several limitations in our study. First, this study was a retrospective study with a small sample size of ROS1-positive patients conducted in only one center. Second, the results of targeted NGS were not verified by another testing method, and we used different testing panels (56 genes and 168 genes, although all contained ROS1) in the real-world practice, mainly due to patient choice and consideration of disease complexity.
Despite these limitations, this retrospective study demonstrated that NGS-identified patients with ROS1-rearranged NSCLC responded well to crizotinib in real-world practice, whereas concomitant mutations may reduce the length of PFS with crizotinib. In addition, our study highlights the importance of multiplex molecular profiling for lung cancers, as the discovery of multiple mutations may have potential clinical impact on these patients. We believed that “dual-targeted” profiling, a combination of targeted NGS and targeted TKI, will result in a better real-world practice for the management of patients with NSCLC. Further, a similar analysis on EGFR mutations and ALK arrangements will be conducted in our center in the near future.
Acknowledgment
This work was partially supported by the National Natural Science Foundation (NO 81401902 and NO 81501992), Hunan Natural Science Foundation (2017SK2134 and 2018JJ2238).
Disclosure
The authors report no conflicts of interest in this work.
References
Nanavaty P, Alvarez MS, Alberts WM. Lung cancer screening: advantages, controversies, and applications. Cancer Control. 2014;21(1):9–14. | ||
Gkolfinopoulos S, Mountzios G, Beyond E. Beyond EGFR and ALK: targeting rare mutations in advanced non-small cell lung cancer. Ann Transl Med. 2018;6(8):142. | ||
Economopoulou P, Mountzios G. The emerging treatment landscape of advanced non-small cell lung cancer. Ann Transl Med. 2018;6(8):138. | ||
Teicher BA. Targets in small cell lung cancer. Biochem Pharmacol. 2014;87(2):211–219. | ||
Tanaka K, Kumano K, Ueno H. Intracellular signals of lung cancer cells as possible therapeutic targets. Cancer Sci. 2015;106(5):489–496. | ||
Wood SL, Pernemalm M, Crosbie PA, Whetton AD. Molecular histology of lung cancer: from targets to treatments. Cancer Treat Rev. 2015;41(4):361–375. | ||
Riccardo F, Arigoni M, Buson G, et al. Characterization of a genetic mouse model of lung cancer: a promise to identify non-small cell lung cancer therapeutic targets and biomarkers. BMC Genomics. 2014;15(Suppl 3):S1. | ||
Politi K, Herbst RS. Lung cancer in the era of precision medicine. Clin Cancer Res. 2015;21(10):2213–2220. | ||
Sadiq AA, Salgia R. MET as a possible target for non-small-cell lung cancer. J Clin Oncol. 2013;31(8):1089–1096. | ||
Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371(21):1963–1971. | ||
Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–1203. | ||
Rimkunas VM, Crosby KE, Li D, et al. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res. 2012;18(16):4449–4457. | ||
Cai W, Li X, Su C, et al. ROS1 fusions in Chinese patients with non-small-cell lung cancer. Ann Oncol. 2013;24(7):1822–1827. | ||
Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30(8):863–870. | ||
Rogers TM, Russell PA, Wright G, et al. Comparison of methods in the detection of ALK and ROS1 rearrangements in lung cancer. J Thorac Oncol. 2015;10(4):611–618. | ||
Yoon HJ, Sohn I, Cho JH, et al. Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung adenocarcinoma using a radiomics approach. Medicine. 2015;94(41):e1753. | ||
Uguen A, de Braekeleer M. ROS1 fusions in cancer: a review. Future Oncol. 2016;12(16):1911–1928. | ||
Chin LP, Soo RA, Soong R, Ou SH. Targeting ROS1 with anaplastic lymphoma kinase inhibitors: a promising therapeutic strategy for a newly defined molecular subset of non-small-cell lung cancer. J Thorac Oncol. 2012;7(11):1625–1630. | ||
Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res. 2012;18(17):4570–4579. | ||
Wu YL, Yang JC, Kim DW, et al. Phase II study of crizotinib in East Asian patients with ROS1-positive advanced non-small-cell lung cancer. J Clin Oncol. 2018;36(14):1405–1411. | ||
Niu X, Chuang JC, Berry GJ, Wakelee HA. Anaplastic lymphoma kinase testing: IHC vs. FISH vs. NGS. Curr Treat Options Oncol. 2017;18(12):71. | ||
Cao B, Wei P, Liu Z, et al. Detection of lung adenocarcinoma with ROS1 rearrangement by IHC, FISH, and RT-PCR and analysis of its clinicopathologic features. Onco Targets Ther. 2016;9:131–138. | ||
Bubendorf L, Büttner R, Al-Dayel F, et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Arch. 2016;469(5):489–503. | ||
Kalemkerian GP, Narula N, Kennedy EB. Molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement summary of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J Oncol Pract. 2018;14(5):323–327. | ||
Arnaoutakis K. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2015;372(7):683. | ||
Davies KD, Le AT, Sheren J, et al. Comparison of molecular testing modalities for detection of ROS1 rearrangements in a cohort of positive patient samples. J Thorac Oncol. 2018;13(10):1474–1482. | ||
Scheffler M, Schultheis A, Teixido C, et al. ROS1 rearrangements in lung adenocarcinoma: prognostic impact, therapeutic options and genetic variability. Oncotarget. 2015;6(12):10577–10585. | ||
Hong S, Gao F, Fu S, et al. Concomitant genetic alterations with response to treatment and epidermal growth factor receptor tyrosine kinase inhibitors in patients with EGFR-mutant advanced non-small cell lung cancer. JAMA Oncol. 2018;4(5):739–742. | ||
Li Z, Shen L, Ding D, et al. Efficacy of crizotinib among different types of ROS1 fusion partners in patients with ROS1-rearranged non-small cell lung cancer. J Thorac Oncol. 2018;13(7):987–995. | ||
Xu T, Kang X, You X, et al. Cross-platform comparison of four leading technologies for detecting EGFR mutations in circulating tumor DNA from non-small cell lung carcinoma patient plasma. Theranostics. 2017;7(6):1437–1446. | ||
Yang JJ, Zhang XC, Su J, et al. Lung cancers with concomitant EGFR mutations and ALK rearrangements: diverse responses to EGFR-TKI and crizotinib in relation to diverse receptors phosphorylation. Clin Cancer Res. 2014;20(5):1383–1392. | ||
Won JK, Keam B, Koh J, et al. Concomitant ALK translocation and EGFR mutation in lung cancer: a comparison of direct sequencing and sensitive assays and the impact on responsiveness to tyrosine kinase inhibitor. Ann Oncol. 2015;26(2):348–354. | ||
Sweis RF, Thomas S, Bank B, Fishkin P, Mooney C, Salgia R. Concurrent EGFR mutation and ALK translocation in non-small cell lung cancer. Cureus. 2016;8(2):e513. | ||
Lou NN, Zhang XC, Chen HJ, et al. Clinical outcomes of advanced non-small-cell lung cancer patients with EGFR mutation, ALK rearrangement and EGFR/ALK co-alterations. Oncotarget. 2016;7(40):65185–65195. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



