Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 11
Comprehensive comparison of MetAP2 tissue and cellular expression pattern in lean and obese rodents
Authors Han J, Tang Y, Lu M, Hua H
Received 14 April 2018
Accepted for publication 19 June 2018
Published 28 September 2018 Volume 2018:11 Pages 565—577
DOI https://doi.org/10.2147/DMSO.S171109
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Ming-Hui Zou
Jing Han, Yang Tang, Mingjian Lu, Haiqing Hua
Lilly China Research and Development Center, Shanghai, People’s Republic of China
Background: Methionine aminopeptidase 2 (MetAP2) cleaves the initiator methionine from nascent peptides during translation. In both preclinical and clinical studies, the pharmacological inhibition of MetAP2 in obese subjects results in the suppression of food intake and body weight loss. However, the mechanism of action of body weight loss caused by MetAP2 inhibition remains to be elucidated, and the sites of action by pharmacological MetAP2 inhibition remain unknown.
Methods: In the present study, a comprehensive analysis of the MetAP2 expression pattern in mice was performed.
Results: Except for the relatively low expression in adipose tissues, MetAP2 protein was well-expressed in tissues important for metabolism, including liver, whole brain, skeletal muscle and intestine tissues. In comparison to lean mice, MetAP2 mRNA level was elevated in the intestines of diet-induced obese (DIO) mice. At the cellular level, MetAP2 exhibited a distinct high expression in central and peripheral neurons, as well as in epithelial cells lining both the small intestine and colon. In the liver of lean mice, MetAP2 protein exhibited punctate staining, which was enriched in zone three hepatocytes surrounding the central veins. In contrast, MetAP2 expression was diffuse in the liver of DIO mice. Furthermore, MetAP2 was highly expressed in immune cells that infiltrated DIO livers.
Conclusion: Overall, these results delineate the MetAP2 expression at both tissue and cellular levels and highlight the altered MetAP2 expression under pathological conditions.
Keywords: MetAP2, obesity, diabetes
Introduction
In both prokaryotes and eukaryotes, mRNA translation starts with the initiator methionine, which is often cleaved during translation when nascent peptides exit the ribosome. The functional consequences of the removal of the initiator methionine remain not well understood. For myristoylated proteins that start with methionine and glycine, the cleavage of the initiator methionine and exposure of the amine group of glycine at the penultimate position is a prerequisite to co-translational myristoylation.1–3 For proteins with cysteine at the second position, the cleavage of the initiator methionine and exposure of cysteine may modulate the protein turnover rate through the N-degron pathway.4–6 For the majority of proteins, the functional relevance of the co-translational removal of the initiator methionine has not been characterized.7
Two methionine aminopeptidases (MetAP1 and MetAP2) have been identified in eukaryotes.8 Both enzymes favor substrates with a small amino acid at the penultimate position, including glycine, alanine, cysteine, serine and valine.9,10 Among these, proteins with valine at the second position are the preferred substrates for MetAP2 over MetAP19. Although there is still a lack of molecular and cellular understanding on the functional consequences of the MetAP2-dependent removal of the initiator methionine, the physiological importance of MetAP2 during development is highlighted by the embryonic lethality in the whole body of MetAP2 knockout mice.11
Fumagillin is a natural product originally isolated from Aspergillus fumigatus.12 It demonstrates anti-angiogenesis and cytostasis effects in cultured cells or in a model organism.13–20 Fumagillin exerts these effects by covalently modifying and irreversibly inhibiting MetAP2.10,21–24 Several fumagillin analogs have been developed and tested in clinical trials for various cancers.21,25–28 It has been originally observed that cancer patients who are overweight lost a significant amount of body weight after MetAP2 inhibitor treatment.29 Subsequently, one of the fumagillin analogs – beloranib – was tested clinically in obese subjects.30,31 In addition, structurally distinct non-covalent MetAP2 inhibitors were developed for the potential treatment of cancer or obesity in human subjects.28,32–35 In both clinical and preclinical studies, pharmacological MetAP2 inhibition markedly reduced body weight mainly due to the loss of fat mass.30,36 This can be largely attributable to the suppression of food intake.29 However, the exact mechanism underlying the suppression of food intake remains unknown. Despite the remarkable weight loss, the therapeutic window of MetAP2 inhibition remains limited. At higher doses, beloranib treatment was associated with sleep disorders and a drop in blood cell count (www.zafgen.com). In order to develop better MetAP2 inhibitors with greater safety margins, it is important to understand which cell types or tissues MetAP2 inhibition mediates the suppression of food intake and body weight loss. The logical first step is to delineate the MetAP2 expression pattern in tissues and cells relevant to the regulation of satiety and body weight. In the present study, we report the comprehensive mapping of MetAP2 protein expression in the liver, brain, skeletal muscle, adipose tissues and intestines. Immunofluorescence imaging demonstrated a high level of MetAP2 protein in central and peripheral neurons, as well as in intestine epithelial cells. The data also indicated altered MetAP2 expression patterns in hepatocytes obtained from diet-induced obese (DIO) mice. Overall, these results provide directions to generate tissue-specific MetAP2 knockout mice and design the next generation of MetAP2 inhibitors.
Materials and methods
Animal studies
All C57BL/6 mice were housed in the animal services facility of Covance Shanghai or ChemPartner Shanghai under a 12/12 h light-dark cycle. Mice were given free access to food and water ad libitum. Standard laboratory rodent chow diet was provided to the animals. In order to prepare the DIO (diet-induced obesity) mice, 10-week old mice were provided with high-fat diet (60% kcal from fat, Research Diet) for more than 16 weeks. The lead mice used to compare with the DIO mice were age-matched. For tissue dissection, mice were anesthetized and perfused from the blood vessel using PBS for 5 minutes and 4% paraformaldehyde (PFA), and this was allowed to flow in from the left ventricle and flow out through the damaged right atrium. After 15–20 minutes, the target tissues were cut and fixed in 4% PFA for another 4 hours on ice. All studies were performed at the vivarium facility of Covance (Shanghai, People’s Republic of China) or Chem Partner (Shanghai, People’s Republic of China) and in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal studies were approved by the Institutional Animal Care and Use Committee of Covance Shanghai or ChemPartner Shanghai.
Immunohistochemistry
The tissues were placed in 15% sucrose and incubated overnight at 4°C. On the next day, the tissues were switched into 30% sucrose solution, and incubated for 2 days at 4°C. Then, when all tissues sunk to the bottom, the solution was replaced with a fresh 30% sucrose solution for the second time for another 2 days at 4°C. Subsequently, the tissues were embedded in optimal cutting temperature compound (OCT compound), solidified by liquid nitrogen and cut into 10-μm thin slices using a hard tissue slice machine. Next, the slices were washed twice with PBS for 5 minutes each time, and blocked with a blocking solution (P0102, Beyotime) for 30 minutes at room temperature. Then, the slices were incubated with 200 µL of primary antibody (CST, #12547) diluted in Dako diluent (#S0809) for 2 hours at room temperature. Subsequently, the slices were washed three times with PBS for 5 minutes each time and incubated with the secondary antibody (#711-175-152, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 1 hour at room temperature. After that, 100 µL of Hochest (1:100) was added to each sliced section, incubated for 10 minutes, and the slices were washed three times before imaging. The imaging was performed using a confocal microscope (SP8 #8100000338, Leica Microsystems, Wetzlar, Germany).
Western blot
For the cultured cells, the cells were collected using a scraper and mixed with 500 µL RIPA buffer containing a 1% phosphatase inhibitor cocktail. Then, cells were sonicated using a Scientz-IID sonicator, centrifuged at 14,000 rpm for 10 minutes at 4°C, the supernatant was transferred, and the protein concentration was determined by BCA (bicinchoninic acid) assay, according to the manual. For the tissue samples, the samples were sonicated using a Scientz-IID sonicator and centrifuged at 14,000 rpm for 10 minutes at 4°C, the supernatant was transferred, and the protein concentration was determined by BCA assay, according to the manual. Western blot was performed using precast SDS-PAGE gels, and the membrane transfer was performed using the iBlot system. Equal amounts of protein (50 microgram) were loaded for Western blot. Then, the membranes were blocked for 1 hour at room temperature, and the primary antibody (CST, #12547) was added at a dilution of 1:1,000 in blocking buffer and incubated at 4°C. The secondary antibody (#SA5-10034, Thermo Fisher Scientific, Waltham, MA, USA) was diluted in TBST at 1:5,000 and was used for membrane incubation at room temperature for 1 hour. The membrane blot detection was performed with the ChemiDoc™ Imaging system using the Dylight™ 650 protocol.
mRNA analysis
Mouse tissue RNA extraction was performed by homogenize 35 mg tissue in 700 μL lysis buffer from MagMAX™–96 Total RNA Isolation Kit (AM1830, Thermo Fisher Scientific, Waltham, MA, USA). RNA samples were extracted from the homogenized tissue by using the kit and followed the instruction. cDNA samples were obtained by using the iScript® Advanced cDNA Synthesis Kit for RT-qPCR (Bio-Rad Laboratories Inc., Hercules, CA, USA, 1708843). qPCR performed by using TaqMan® Universal Master Mix II and the primers were purchased from Thermo Fisher Scientific (MetAP2, Mm00721762; Tbp, Mm00446973).
Results
MetAP2 protein expression in liver, brain, skeletal muscle and adipose tissues
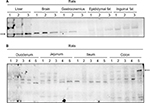
In order to systematically characterize the protein expression of MetAP2, tissue lysates obtained from mice liver, whole brain, gastrocnemius, epididymal and inguinal fat pads were probed by Western blot. MetAP2 protein was readily detected in the liver and brain (Figure 1A). The expression of MetAP2 was lower in the gastrocnemius than in the liver and brain. Furthermore, the expression of MetAP2 in adipose tissues was weak. The investigators also attempted to detect the protein expression of MetAP2 in different regions of the intestine. However, the antibody only detected small protein bands (Figure 1B), suggesting that MetAP2 was degraded by various proteases in the intestine during the tissue collection and lysate preparation, despite the presence of protease inhibitors.
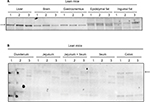
In lean mice, the protein expression pattern of MetAP2 was similar to that in DIO mice. However, the protein level of MetAP2 was slightly higher in the gastrocnemius than in the brain, and no MetAP2 protein was detected in inguinal fat (Figure 2A). Furthermore, the expression in adipose tissues was weakly relative to the other tissues examined (Figure S1). Hence, MetAP2 protein was extensively degraded in the lysates prepared from the various regions of the intestine (Figure 2B).
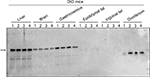
The MetAP2 protein expression pattern in DIO mice was similar to that in lean mice, in which MetAP2 level in the gastrocnemius was lower than that in the liver but was slightly higher than that in the brain. However, no MetAP2 protein was detected in the epididymal and inguinal adipose tissues. Furthermore, a degraded MetAP2 fragment was merely detected in the intestine (Figure 3).
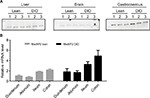
In order to understand the relationship between MetAP2 expression and metabolic disease condition (Figures S2, S3), the protein expression of MetAP2 was compared side by side between lean mice and DIO mice (Figure 4A). It was found that there was significant difference in the liver, brain and gastrocnemius between these two mouse models. In order to further investigate whether the difference in protein levels between lean and obese mice was due to transcriptional regulation or post-transcriptional regulation, mRNA was extracted from different regions of the intestine, and the MetAP2 mRNA level was quantified by real-time qPCR (Figure 4B). It was found that MetAP2 mRNA levels were higher in the ileum and colon, when compared with the duodenum and jejunum, in both lean and obese mice. Compared to lean mice, the MetAP2 mRNA levels in the DIO mice were higher in all the regions along the intestine.
High MetAP2 protein expression in central and peripheral neurons
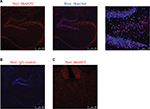
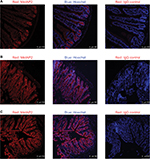
There were different cell types in each of the tissues investigated above. Although Western blot analysis was able to determine the total MetAP2 protein levels in the tissues of interest, it does not provide special information within the tissue and among different cell types. In order to gain deeper insight into the protein expression of MetAP2 at the cellular level, MetAP2 immunofluorescence (IF) imaging was performed. It is noteworthy that an IgG antibody was used as the negative control in the immunostaining experiments to ensure signal specificity (Figure 5B). As shown in Figure 5A, MetAP2 was highly expressed in hippocampal neurons, but was not expressed in non-neuronal cells in the same region. The MetAP2 IF signal was also intense in neurons in other regions of the brain, such as in the cortex of the front lobe (Figure 5C). Furthermore, similar MetAP2 IF patterns were observed in the brain regions of DIO mice (Figure 6).
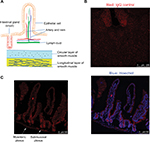
High MetAP2 immunofluorescence signals were detected in enteric neurons in the myenteric plexus and submucosal plexus (Figure 7). MetAP2 was also highly and specifically expressed in neurons within the nodose ganglia dissected from mice (Figure 8).
High MetAP2 protein expression in intestinal epithelial cells
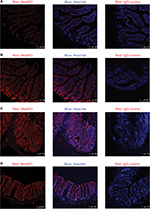
In all the regions along the intestine in lean mice, MetAP2 protein was highly expressed in epithelial cells, including those in the villi and crypts (Figures 7 and 9). In contrast, its expression was very weak in smooth muscles and endothelial cells lining the blood capillaries and lymphatic venules. Similar patterns were observed in intestine tissues obtained from DIO mice (Figure 10).
Diffuse MetAP2 protein expression in the gastrocnemius
In both lean mice and DIO mice, MetAP2 protein exhibited a diffuse expression in the gastrocnemius (Figure 11). The signals were relatively weak when compared to those in neurons and intestinal epithelial cells.
Altered MetAP2 protein expression pattern in livers of DIO mice
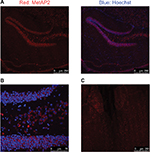
In livers obtained from lean mice, MetAP2 IF signals revealed punctate staining in the cytosol of hepatocytes (Figure 12A). Furthermore, a zoning effect appeared, with the MetAP2 expression level being higher in zone 3 (the region surrounding the central vein), when compared to zone 1.
In livers obtained from DIO mice, there was no apparent difference in MetAP2 expression between zone 1 and zone 3 (Figure 12B). Furthermore, MetAP2 exhibited a diffuse, rather than punctuate, IF staining in the cytosol of hepatocytes. Another feature unique to these DIO livers was the high IF staining in non-parenchymal cells, and it was likely that immune cells infiltrated these DIO livers.
Discussion
At present, MetAP2 protein expression patterns in mice are characterized at both the tissue and cellular level. By Western blot with a MetAP2-specific antibody, it was revealed that MetAP2 is expressed in the liver, brain, skeletal muscle, and to a lesser extent, in epididymal and inguinal adipose tissues. Due to uncontrollable protein degradation, the investigators were unable to definitively determine the level of MetAP2 protein in the intestine. However, MetAP2 mRNA was readily detected in the intestine. The high expression of MetAP2 protein along the intestine was also apparent via immunostaining.
In the present study, the immunofluorescence imaging provided important information to illustrate the unique expression patterns of the MetAP2 protein. Compared to other cell types in the same tissues, MetAP2 protein was highly enriched in the neurons of the central nervous system and peripheral nerve system, including the enteric nerve system and vagal nerves. Given the strong effect of MetAP2 inhibition on the suppression of food intake, and the established roles of central and peripheral nervous systems on the regulation of satiety, it is conceivable that MetAP2 inhibition in the neurons may play an important role in marked weight loss after pharmacological intervention. In addition to neurons, MetAP2 protein is also highly expressed in epithelial cells in the intestine. Intestinal epithelial cells are specialized for nutrients absorption. Furthermore, these cells are short lived, and are rapidly replenished with progenitor cells that proliferate from stem cells residing at the bottom of the crypts. However, it remains to be determined whether MetAP2 functions in nutrients absorption or the replenishment of intestinal epithelial cells.
MetAP2 expression in mice on chow diet or high-fat diet was also compared. MetAP2 mRNA level increased in all regions of the intestine after feeding with high-fat diet, when compared to chow diet. In livers obtained from lean mice, MetAP2 protein exhibited punctate staining in the cytosol of hepatocytes. Furthermore, the expression of MetAP2 also exhibited a zoning effect, with a higher level in zone 3, when compared to zone 1. In contrast, in livers obtained from DIO mice, MetAP2 exhibited diffuse staining in the cytosol, and no apparent zoning effect was observed. Hence, the physiological or pathological relevance of the altered expression pattern in DIO livers remains to be investigated. Another distinct feature in livers obtained from DIO mice was the high MetAP2 expression in non-parenchymal cells, which was most likely from the immune cells that infiltrated the DIO livers.
In summary, the present study reports the highly enriched protein expression of MetAP2 in central and peripheral neurons, as well as in intestinal epithelial cells and also provides evidence for the differential expression of MetAP2 in the intestine and liver between DIO mice and lean mice. The characterization of MetAP2 expression in normal and disease animal models provides guidance to generate tissue-specific MetAP2 knockout mice lines in the future, in order to further investigate the mechanism of action of pharmacological MetAP2 inhibition.
Acknowledgments
The authors would like to thank the following colleagues for their technical assistance: Shiqiang Li, Xudong Mao, Xiankang Fang, Xilin Liu and Wai Mai Shek. The authors are also grateful to Haixia Zou, Jiangwei Zhang, Bei Shan and Bei Zhang for their valuable suggestions.
Disclosure
The authors report no conflicts of interest in this work.
References
Thinon E, Serwa RA, Broncel M, et al. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat Commun. 2014;5:4919. | ||
Broncel M, Serwa RA, Ciepla P, et al. Myristoylation profiling in human cells and zebrafish. Data Brief. 2015;4:379–383. | ||
Martin DD, Beauchamp E, Berthiaume LG. Post-translational myristoylation: Fat matters in cellular life and death. Biochimie. 2011;93(1):18–31. | ||
Eldeeb M, Fahlman R. The-N-end rule: the beginning determines the end. Protein Pept Lett. 2016;23(4):343–348. | ||
Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20(8):1298–1345. | ||
Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci. 2007;32(11):520–528. | ||
van Damme P, Hole K, Gevaert K, Arnesen T. N-terminal acetylome analysis reveals the specificity of Naa50 (Nat5) and suggests a kinetic competition between N-terminal acetyltransferases and methionine aminopeptidases. Proteomics. 2015;15(14):2436–2446. | ||
Dummitt B, Micka WS, Chang YH. N-terminal methionine removal and methionine metabolism in Saccharomyces cerevisiae. J Cell Biochem. 2003;89(5):964–974. | ||
Xiao Q, Zhang F, Nacev BA, Liu JO, Pei D. Protein N-terminal processing: substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry. 2010;49(26):5588–5599. | ||
Addlagatta A, Hu X, Liu JO, Matthews BW. Structural basis for the functional differences between type I and type II human methionine aminopeptidases. Biochemistry. 2005;44(45):14741–14749. | ||
Yeh JJ, Ju R, Brdlik CM, et al. Targeted gene disruption of methionine aminopeptidase 2 results in an embryonic gastrulation defect and endothelial cell growth arrest. Proc Natl Acad Sci U S A. 2006;103(27):10379–10384. | ||
Lefkove B, Govindarajan B, Arbiser JL. Fumagillin: an anti-infective as a parent molecule for novel angiogenesis inhibitors. Expert Rev Anti Infect Ther. 2007;5(4):573–579. | ||
Boxem M, Tsai CW, Zhang Y, Saito RM, Liu JO. The C. elegans methionine aminopeptidase 2 analog map-2 is required for germ cell proliferation. FEBS Lett. 2004;576(1-2):245–250. | ||
Bradshaw RA, Yi E. Methionine aminopeptidases and angiogenesis. Essays Biochem. 2002;38:65–78. | ||
Hannig G, Lazarus DD, Bernier SG, et al. Inhibition of melanoma tumor growth by a pharmacological inhibitor of MetAP-2, PPI-2458. Int J Oncol. 2006;28(4):955–963. | ||
Ma AC, Fung TK, Lin RH, et al. Methionine aminopeptidase 2 is required for HSC initiation and proliferation. Blood. 2011;118(20):5448–5457. | ||
Selvakumar P, Lakshmikuttyamma A, Das U, Pati HN, Dimmock JR, Sharma RK. NC2213: a novel methionine aminopeptidase 2 inhibitor in human colon cancer HT29 cells. Mol Cancer. 2009;8:65. | ||
Turk BE, Griffith EC, Wolf S, Biemann K, Chang YH, Liu JO. Selective inhibition of amino-terminal methionine processing by TNP-470 and ovalicin in endothelial cells. Chem Biol. 1999;6(11):823–833. | ||
Wang J, Sheppard GS, Lou P, et al. Tumor suppression by a rationally designed reversible inhibitor of methionine aminopeptidase-2. Cancer Res. 2003;63(22):7861–7869. | ||
Zhang Y, Griffith EC, Sage J, Jacks T, Liu JO. Cell cycle inhibition by the anti-angiogenic agent TNP-470 is mediated by p53 and p21WAF1/CIP1. Proc Natl Acad Sci U S A. 2000;97(12):6427–6432. | ||
Addlagatta A, Matthews BW. Structure of the angiogenesis inhibitor ovalicin bound to its noncognate target, human Type 1 methionine aminopeptidase. Protein Sci. 2006;15(8):1842–1848. | ||
Chun E, Han CK, Yoon JH, Sim TB, Kim YK, Lee KY. Novel inhibitors targeted to methionine aminopeptidase 2 (MetAP2) strongly inhibit the growth of cancers in xenografted nude model. Int J Cancer. 2005;114(1):124–130. | ||
Griffith EC, Su Z, Niwayama S, Ramsay CA, Chang YH, Liu JO. Molecular recognition of angiogenesis inhibitors fumagillin and ovalicin by methionine aminopeptidase 2. Proc Natl Acad Sci U S A. 1998;95(26):15183–15188. | ||
Griffith EC, Su Z, Turk BE, et al. Methionine aminopeptidase (type 2) is the common target for angiogenesis inhibitors AGM-1470 and ovalicin. Chem Biol. 1997;4(6):461–471. | ||
Arico-Muendel CC, Belanger B, Benjamin D, et al. Metabolites of PPI-2458, a selective, irreversible inhibitor of methionine aminopeptidase-2: structure determination and in vivo activity. Drug Metab Dispos. 2013;41(4):814–826. | ||
Arico-Muendel CC, Benjamin DR, Caiazzo TM, et al. Carbamate analogues of fumagillin as potent, targeted inhibitors of methionine aminopeptidase-2. J Med Chem. 2009;52(24):8047–8056. | ||
Fardis M, Pyun HJ, Tario J, et al. Design, synthesis and evaluation of a series of novel fumagillin analogues. Bioorg Med Chem. 2003;11(23):5051–5058. | ||
Morgen M, Jöst C, Malz M, et al. Spiroepoxytriazoles are fumagillin-like irreversible inhibitors of metAP2 with potent cellular activity. ACS Chem Biol. 2016;11(4):1001–1011. | ||
Joharapurkar AA, Dhanesha NA, Jain MR. Inhibition of the methionine aminopeptidase 2 enzyme for the treatment of obesity. Diabetes Metab Syndr Obes. 2014;7:73–84. | ||
Hughes TE, Kim DD, Marjason J, Proietto J, Whitehead JP, Vath JE. Ascending dose-controlled trial of beloranib, a novel obesity treatment for safety, tolerability, and weight loss in obese women. Obesity. 2013;21(9):1782–1788. | ||
Rohn J. Newsmaker: Zafgen. Nat Biotechnol. 2011;29(12):1068. | ||
Cheruvallath Z, Tang M, Mcbride C, et al. Discovery of potent, reversible MetAP2 inhibitors via fragment-based drug discovery and structure based drug design-Part 1. Bioorg Med Chem Lett. 2016;26(12):2774–2778. | ||
Garrabrant T, Tuman RW, Ludovici D, et al. Small molecule inhibitors of methionine aminopeptidase type 2 (MetAP-2). Angiogenesis. 2004;7(2):91–96. | ||
Marino JP, Fisher PW, Hofmann GA, et al. Highly potent inhibitors of methionine aminopeptidase-2 based on a 1,2,4-triazole pharmacophore. J Med Chem. 2007;50(16):3777–3785. | ||
Morowitz MJ, Barr R, Wang Q, et al. Methionine aminopeptidase 2 inhibition is an effective treatment strategy for neuroblastoma in preclinical models. Clin Cancer Res. 2005;11(7):2680–2685. | ||
Lijnen HR, Frederix L, van Hoef B. Fumagillin reduces adipose tissue formation in murine models of nutritionally induced obesity. Obesity. 2010;18(12):2241–2246. |
Supplementary materials
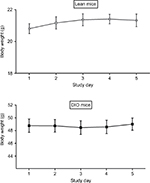  | Figure S2 Body weight measurement for 5 consecutive days. Abbreviations: MetAP2, methionine aminopeptidase 2; DIO, diet-induced obese. |
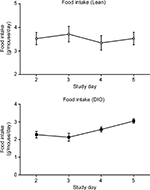  | Figure S3 Food intake measurement for 5 consecutive days. Abbreviations: DIO, diet-induced obese. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.