Back to Journals » Infection and Drug Resistance » Volume 12
Comparative Study Of Genetic Diversity, Virulence Genotype, Biofilm Formation And Antimicrobial Resistance Of Uropathogenic Escherichia coli (UPEC) Isolated From Nosocomial And Community Acquired Urinary Tract Infections
Authors Souza GM, Neto ERDS, da Silva AM, Iacia MVMS, Rodrigues MVP, Pereira VC , Winkelstroter LK
Received 25 August 2019
Accepted for publication 10 October 2019
Published 22 November 2019 Volume 2019:12 Pages 3595—3606
DOI https://doi.org/10.2147/IDR.S228612
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Gabrielle Messias De Souza, Estevan Rodrigues Dos Santos Neto, Alaor Martins da Silva, Maria Vitoria Minzoni de Souza Iacia, Marcus Vinícius Pimenta Rodrigues, Valéria Cataneli Pereira, Lizziane Kretli Winkelstroter
Health Sciences Faculty, University of Western Sao Paulo, Sao Paulo, Brazil
Correspondence: Lizziane Kretli Winkelstroter
Health Sciences Faculty, University of Western Sao Paulo, 700, Jose Bongiovani St, Presidente Prudente, Sao Paulo 19050-920, Brazil
Tel +55 18 3229-1289
Email [email protected]
Introduction: Escherichia coli is a Gram-negative opportunistic human pathogen, which has aroused considerable medical interest for being involved in cases of urinary tract infection.
Aim: Characterize the E. coli isolated both in the hospital and in the community.
Methodology: A total of 200 E. coli isolated in urine samples from hospital and community were evaluated in biofilm formation assay and hydrophobicity MATS method. Antimicrobial susceptibility was performed through agar-diffusion technique. Virulence and ESBL production genes were observed through the polymerase chain reaction amplification of papC, fimH, fliC, kpsMTII, blaTEM, blaCTX-M, blaSHV, and blaOXA.The phylogenetic classification was based on the pattern chuA and yjaA and the region TspE4.C2 by PCR Multiplex.
Results: A higher frequency of non-adherent or poorly adherent isolates was observed in the community group. Approximately 85% of the community isolates were distributed in the highest hydrophilicity group (p<0.05). The level of resistant microorganisms was present at the same level in both source (p>0.05). About 14% of the hospital isolates were positive in the ESBL phenotypic detection test (p>0.05). Among the samples, 95% presented ESBL-encoding genes. The predominant phylogenetic group was B2 (78%). Community isolates showed a higher prevalence of virulence genes fimH, papC, and kpsMTII when compared to hospital samples.
Conclusion: These data confirm the worldwide trend that isolates in the community present sometimes higher levels of virulence and antimicrobial resistance.
Keywords: adhesion, control, ESBL, UTI
Introduction
Urinary tract infections (UTIs) are one of the most common diseases worldwide and it is estimated that approximately 150 million cases occur every year.1–3 Urinary tract infections occur due to invasion and multiplication of microorganisms in the urinary tract. UTIs can be characterized by several aspects such as location, complication, origin, and chronicity.1,2,4,5
The etiology of UTIs is related to a great diversity of microorganisms, such as bacteria, viruses, and fungi.2 Escherichia coli is one of the principal etiological agents and is involved in approximately 70% to 85% of UTI.4,6
Uropathogenic Escherichia coli (UPEC) harbor genes encoding various virulence factors, which contribute to increased pathogenicity. The molecular characteristics and functions of these virulence genes have been well established in some studies.7,8 The presence of some of these genes has been related to virulence factors such as the fliC gene (flagellin), papC gene (fimbria P) and kpsMTII gene (capsule).8 In this way, the severity of the UTI reflects the virulence profile or the phenotype of the infecting strain.9,10
Biofilm formation, an important virulence factor, plays an important role in urinary tract infections (UTIs), accounting for persistent infections and acute prostatitis. The presence of virulence factors may also be related to a greater or lesser ability to form biofilms.8,10,11 Factors such as cellular hydrophobicity, surface electric charge, the presence of proteins in the outer membrane, and properties of the adhesion material also play an important role in the formation of biofilms on abiotic surfaces. Hydrophobicity is generally associated with bacterial adhesiveness and varies according to the microorganism, growth phase, and bacterial surface structures.12
Antimicrobials are the medicines of choice in the treatment of UTIs. However, the delay in laboratory microbiological diagnosis leads to the adoption of empirical treatment. Studies have shown that the empirical, indiscriminate, prolonged, or incorrect use of antimicrobials contributes significantly to the selection of resistant strains in urinary infections. Thus, knowledge of the resistance profile of the isolates found in the community and the hospital is important for better orientation of the initial drug treatment approach for the patient.3,13–15
Bacterial resistance to β-lactam antibiotics has increased significantly in recent years. One of the main mechanisms is the production of β-lactamases, enzymes capable of cleaving the β-lactam ring from its chemical structure.16–18 Extended-spectrum β-lactamases (ESBL) are a type of enzymes, mainly produced by Gram-negative bacilli, which have aroused great concern worldwide, as they cause resistance to all penicillins, cephalosporins (including third and fourth generations), and aztreonam.19,20 In addition, these bacteria are often resistant to trimethoprim/sulfamethoxazole and quinolones.21 ESBLs are classified into several groups according to their amino acid sequences, which are encoded by genes often located on mobile elements like plasmids consequently they are easily transferred between the bacteria.20 Currently, the most common genetic variants are CTX-M, TEM, SHV, and OXA. The presence of these resistance genes can significantly affect the course and outcome of infections, both in the community and in the hospital environment.16,19,21
The spread of resistant E. coli is a major challenge, especially in developing countries, due to poor hygiene, poor access to health care, and low financial investment in health systems.22–26
In this context, the objective of this work was to evaluate the formation of biofilms, hydrophobicity, the presence of virulence genes, and genes for the production of extended spectrum β-lactamase. Also, it will be evaluated the susceptibility to antimicrobials, identification of extended spectrum β-lactamase, and phylogenetic classification of microorganisms isolated from both the hospital and the community. Knowledge of these data is essential for more in-depth knowledge of the epidemiological profile of E. coli urinary tract infections as they provide an epidemiological tool that could help to plan and implement measures of infection prevention and control. In addition, the results will provide information on the health status of a population, contribute to disease management in a region, and be crucial in guiding the empirical treatment of UTIs.
Materials And Methods
Samples Of Escherichia coli
In the present study, a total of 100 Escherichia coli samples isolated from urine in the Hospitalar Clinical Analysis Laboratory and 100 Escherichia coli samples isolated from urine and provided by the Community Clinical Analysis Laboratory from period of august 2017 to September 2018 were used. The collected samples were inoculated into Brain Heart Infusion (BHI) broth 37°C/24h, then supplemented with glycerol and stored at −80°C until the moment of use.
Evaluation Of Biofilm Formation
The isolates were cultured in BHI broth at 37 ° C for 24 hrs. The cultures were adjusted by spectrophotometry at 600 nm to the value 0.1. Aliquots of 20 μL of the cell suspension from each isolate were added to 200 μL of BHI broth present in the wells of 96-well microtiter plates, and then incubated at 37°C/24h. The plates were subsequently washed three times with 0.9% (w/v) saline to remove the unadhered cells. The adhered cells were stained with 200 μl of 0.1% (w/v) crystal violet for 5 mins. The dye was removed, the microplate washed again three times and, after drying for 30 mins in an oven, the dye solubilization was performed with alcohol/acetone solution (80:20). The optical densities of the solution were read at a wavelength of 600 nm. The value found is representative for the bacterial cell adhesion.8,27 Negative controls were performed with BHI alone, omitting bacteria. The mean optical density (OD) of the negative control (DOc) was used as the cut-off point. The isolates were classified as: DO ≤ DOc Non-adherent; DOc< DO ≤ 2x DOc Weak adherent; 2x DOc< DO ≤ 4x DOc Moderate adherent; 4x DOc< DO Strong adherent.
Determination Of Cellular Hydrophobicity
The hydrophobicity of the bacterial cell surface was evaluated by the measurement of MATS (microbial adhesion to solvents) as described.28,29 E. coli cultures were adjusted to OD 600nm of 1.0 in 0.9% saline (A1). A 0.4 mL volume of xylene was added to 3.6 mL of bacterial suspension. Stirring was performed for 1 min in a vortex and then the tubes were conditioned at room temperature for 20min. The hydrocarbon and aqueous phases of the mixtures were allowed to separate and the optical density of aqueous phase was measured spectrophotometrically at 600 nm (A2). The percentage of cell adhesion to hydrocarbons was calculated upon the following formula: A (%) = [(A1–A2)/A1] × 100%. The degree of a strain’s hydrophobicity was assigned as strongly hydrophobic, moderately hydrophobic and hydrophilic within percentage adhesion values equal >50%, 20–50% and <20% respectively. The results obtained were based on the average of three experiments.
Antimicrobial Disc-Diffusion Technique In Agar
The antimicrobial susceptibility test was performed using the agar diffusion technique, as recommended by the Clinical Laboratory Standards Institute-CLSI.30 For inoculation, bacterial cultures in BHI broth were used, previously incubated for 4 to 6 hrs and adjusted to the turbidity of a 0.5 McFarland standard.
After adjustment for the inoculum density, the sowing was performed using a sterile swab on the surface of Mueller Hinton Agar, and then the disks impregnated with the antimicrobials were applied. The plates were incubated at 35°C for 24 hrs. The activity of the antimicrobial was evaluated by the diameter of the inhibition halo and then interpreted based on the norms established by the CLSI.30
The antimicrobials evaluated for E. coli isolates were: ampicillin 10 μg (AMP), cephalothin 30 μg (CFL), cefotaxime 30 μg (CTX), gentamicin 10 μg (GEN), sulfamethoxazole/trimethoprim 1.25/23.75 μg (SUT), ciprofloxacin 5 μg (CIP), and ceftazidime 30 μg (CAZ).30,31
The index of multiple antimicrobial resistance (MAR index) was calculated from the number of antimicrobials to which the isolate was resistant over the total number tested, multiplying the final value by 100 to obtain the results in percentages.32
Detection Of Extended-Spectrum Beta-Lactamase Production (ESBL)
The isolates which presented reduced inhibition halos for extended-spectrum β-lactamase (ceftazidime [CAZ] ≤ 22 mm, aztreonam ≤ 27 mm, cefotaxime [CTX] ≤ 27 mm; ceftriaxone ≤ 25 mm, cefpodoxime ≤ 17 mm) were selected to confirm the presence of ESBL. Confirmation was performed with discs of ceftazidime (CAZ) 30 μg, ceftazidime + clavulanic acid (CCAZ) 30 μg/10 μg, cefotaxime CTX 30 μg, and cefotaxime + clavulanic acid (CCTX) 30 μg/10 μg. The formation of an irregular area of inhibition (ghost zone) is indicative of a positive result for ESBL production.30,33 E. coli ATCC 25922 was used as a control.
Phylogenetic Classification Of Bacterial Samples
The DNA of the isolates was extracted using the phenol-chloroform technique. Subsequently, the DNA was quantified, evaluated for purity and quality, and maintained at - 20°C.34
The phylogenetic origin of the samples was determined following the methodology described by Clermont et al.35 This classification is based on the PCR amplification pattern (Polymerase Chain Reaction) of two genes (chuA and yjaA) and a region of the DNA (TspE4.C2). The primers used in the PCRs are described in Table 1.
 |
Table 1 Oligonucleotide primers used in this study |
PCR was performed using 25 μl reactions containing 100 ng DNA, 30 pmol of each primer, 1X Taq buffer (10 mM Tris, 50 mM KCl, 2.5 mM MgCl2), 200 μM dNTP, and 1 U Taq polymerase. Gene amplification was obtained with the following cycling: 5 mins denaturation at 94°C, followed by 30 cycles of 30 sec at 94°C, 30 sec at 55°C, and 30 sec at 72°C, with a final extension of 7 mins at 72°C.
Aliquots of 10 μl of amplification products were electrophoresed for 45 mins at 100 V on 1.5% agarose gel stained with ethidium bromide. The DNA fragments were compared to a 1000 bp DNA marker. The gel was visualized and photographed under ultraviolet light. The presence (+) and absence (-) of the genes were used in combination for the classification of the samples in phylogenetic groups B2, D, B1, and A.35–37
Evaluation Of The Presence And Absence Of Virulence Genes
The extracted DNA was diluted to a concentration of 100ng/ul for the polymerase chain reaction (PCR). The polymerase chain reaction was performed using specific primers for the genes: papC (outer membrane protein of fimbriae P), fimH (fimbrial adhesin type 1), kpsMTII (capsule), and fliC (flagellum).
The sequence of the initiators and molecular weight of the products are described in Table 1 and the cycling protocol was performed according to the work of Tiba et al.8
The presence and absence of virulence factors were evaluated through the visualization of PCR products. The samples were submitted to 1.5% −2% agarose gel electrophoresis and were identified by incubation in ethidium bromide solution and visualized in a UV light transilluminator. For registration, the gels were photographed in a gel documentation system.
Detection Of The Genes blaTEM, blaCTX-M, blaSHV, And blaOXA By Multiplex PCR
The isolated bacteria were genotyped by multiplex PCR. The primers used for gene detection, blaTEM, blaCTX-M, blaSHV, and blaOXA are listed in Table 1. All reactions were performed with 100 ng of DNA from each microorganism, 0.65 μM of specific primers, 1.25 U Taq DNA polymerase, 5x buffer, 2 mM MgCl2, 0.25 mM dNTPs, and miliQ water to a final volume of 25 μL. The conditions for amplification were as follows: initial denaturation at 95°C for 15 mins, 30 cycles of denaturation at 94°C for 30 seconds, annealing at 60° for 30 seconds, extension at 72°C for 2 mins, followed by final extension at 72°C for 10 mins. The amplified product was electrophoresed, and then the agarose gel was stained with ethidium bromide and visualized in ultraviolet light with the aid of a transilluminator. The presence of bands on the agarose gel is indicative of the presence of the genes studied. Klebsiella pneumoniae ATCC 700603 and E. coli ATCC 35218 were used as positive controls and the strain E. coli ATCC 25922, as a negative control.38–40
Data Analysis
The experiments were carried out in triplicate and the final results were submitted to statistical analysis. For statistical analysis, the chi-square test was used to compare the results found in the community and hospital isolates. To evaluate the correlation between biofilm formation and hydrophobicity, the Pearson Correlation Coefficient was used. The data were analyzed with Prism software, considering a level of significance of 0.05. Similarities between patterns were determined based on the Dice similarity coefficient. The resulting similarities in the matrix were further processed by employing the unweighted-pair group method using average linkages to create a dendrogram that depicted the genetic relatedness between Escherichia coli isolates. Interactive Tree Of Life (http://itol.embl.de) is a web-based tool for the display, manipulation and annotation of phylogenetic trees.
Results
The present work evaluated the biofilm formation of E. coli isolated from urine samples from a hospital and community clinical laboratory. As shown in Table 2, the data demonstrated a higher frequency of non-adherent or poorly adherent isolates in the community group compared to the hospital group (p<0.05). Also, cellular hydrophobicity of the E. coli isolates was evaluated (Table 2). It was observed that E. coli obtained from community samples presented approximately 85% of the isolates distributed in the two groups with the highest hydrophilicity (p<0.5). The correlation between biofilm formation and cellular hydrophobicity was evaluated. A strong correlation of R2 of 0.8153 was observed between these two parameters in the community group, suggesting that those with a low hydrophobicity present a low biofilm formation capacity.
 |
Table 2 Distribution of the isolates according to the level of cellular hydrophobicity and formation of biofilms |
For the antimicrobial resistance profile, ampicillin presented the highest resistance rates among community and hospital isolates, presenting very similar percentages of 50 and 52% (p> 0.05), respectively. Gentamicin demonstrated the lowest resistance rates in the two groups analyzed (Table 3) (p>0.05). No significant differences were found between the two groups, demonstrating that resistant microorganisms are present at the same level in both the hospital environment and the community.
 |
Table 3 Percentage of antimicrobial resistance by E. coli isolated from the nosocomial and community samples |
The MAR index (multiple antibiotic resistance) is an important parameter, used to determine the level of multiresistance among isolates. In the samples obtained from the community, approximately 39% of the isolates obtained a MAR index equal to or greater than 28.5%, representing combined resistance to two or more antimicrobials. In the hospital setting, the percentage for the same index was significantly higher, with a percentage of 55% (p <0.05). In addition, 3% of hospital isolates can be characterized as PDR (Pandrug-resistant), since they showed resistance to all classes of antimicrobials tested.
Isolates producers of extended-spectrum beta-lactamases (ESBL) are generally associated with multidrug resistance. Community isolates showed 6% of positive samples for ESBL production in the disc approach test. Isolates from the hospital environment presented 14% of the positive samples in the ESBL phenotypic detection test (p>0.05). All ESBL-producing positive strains also demonstrated resistance to the antimicrobials: ampicillin, ceftazidime, and ceftriaxone, and 83.3% also presented resistance to ciprofloxacin.
In the present work the presence of the genes blaTEM, blaCTX-M, blaoxa, and blaSHV was identified (Table 4). Of all the samples analyzed, 95% presented genes encoding ESBL. The gene bla TEM was more prevalent, being present in 89% of the isolates (p <0.05). Some isolates had more than one resistance gene related to ESBL production. Approximately 7% presented more than one gene encoding the resistance and 2% of the isolates demonstrated the genes blaoxa, blaTEM, and blaCTX-M concomitantly (Figures 1 and 2). We highlight in this work that 88.8% of non-ESBL isolates in the phenotypic tests presented at least one resistance gene related to ESBL production (Figures 1 and 2).
 |
Table 4 Percentage of the presence of virulence genes and antimicrobial resistance related to esbl production in E. coli isolated from nosocomial and community samples |
Phylogenetic classification showed a high percentage of the B2 group both in the hospital environment (78%) and community isolates (78%). At lower frequencies, percentages of 16, 5, and 1% for the community group and 18, 4, and 0% for the hospital group were observed for the phylogenetic classifications D, A, and B1, respectively, with no statistical difference between the groups (hospital/community) (p>0.05).
The presence of the virulence genes: papC (outer membrane protein of fimbriae P), fimH (fimbrial adhesin type 1), kpsMTII (capsule), and fliC (flagellum) was evaluated. As shown in Table 4, in general, E. coli isolated from the community presented a higher prevalence of virulence genes fimH, papC, and kpsMTII (p<0.05).
The phenotypic and genotypic characteristics evaluated in this study were organized in dendrograms as shown in Figures 1 and 2. Cophenetic correlation coefficients of 0.46 and 0.52 were obtained for hospital and community samples respectively. It was possible to observe that there is no predominance of a profile among the isolates since the characteristics evaluated were variable. However, it was noted that all isolates from the community that presented an ESBL-positive phenotype had the blaTEM gene and the virulence genes papC, fimH, kpsMTII and fliC. In addition, they belonged to phylogenetic group B2, had a MAR index> 57.1%, were non-forming or weak biofilm forming agents, and presented moderate to strong hydrophilicity.
Of the 14 ESBL positive isolates from the hospital, only one did not present the blaTEM gene, eight presented three of the virulence genes, and in one isolate, two virulence genes were presented. It was also observed that only one was from phylogenetic group D and the other 13 from B2. Nine isolates presented a MAR index <57.1%. Regarding the formation of biofilms, seven presented moderate-strong adhesion and nine presented weak to moderate hydrophobicity.
The radial dendogram containing the results of hospital and community samples is shown in Figure 3. Little variability was observed in the profile of the isolates. These data confirm the worldwide trend that isolates with high levels of virulence and antimicrobial resistance are widespread in both the community and hospital environment.
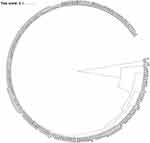 |
Figure 3 Radial phylogenetic tree for comparing the E. coli isolated from nosocomial and community acquired urinary tract infections. |
Discussion
Escherichia coli is a Gram-negative opportunistic human pathogen, which has aroused considerable medical interest for being involved in cases of urinary tract infection.41 There is no fixed pattern in the formation of uropathogenic E. coli biofilms since the adhesion process depends on a variety of factors such as adhesion proteins, formation of exopolysaccharides, type of material, electrostatic charge, and hydrophobicity. The hydrophobic properties of microbial surfaces favor adhesion to abiotic and biotic surfaces and penetration of host tissues. In the present study, a correlation was found between biofilm formation and cell hydrophobicity, suggesting that those with a low hydrophobicity present a low biofilm formation capacity. Considering that medical implants, such as catheters, mechanical heart valves, or pacemakers, are constructed from hydrophobic materials (silicon, stainless steel, Teflon), we highlight the relative ease that hydrophobic microorganisms present in adhering to them.42,43
World phylogenetic analyzes have shown that virulent extraintestinal E. coli strains belong mainly to group B2 and, to a lesser extent, to group D. In contrast, the majority of commensal strains are associated with group A or group B1.44 The data obtained in the present study corroborate with recently published studies. Ghauor and Salehzadeh45 evaluated the phylogenetic profile of E. coli isolated from urine and found a distribution of groups B2, D, A, and B1 with respective percentages 64%, 24%, 12%, and 0%. Iranpour et al44 evaluated the phylogenetic distribution of E coli isolated from patients with urinary tract infection and also observed a predominance of isolates from group B2 (39.3%).
Uropathogenic (UPEC) isolates are a genetically heterogeneous group that exhibits various virulence factors associated with colonization and persistence of the bacterium in the urinary tract.8 However, an unexpected high prevalence of virulence genes was observed in the E. coli isolated from the community. These results demonstrated the new trend in the microorganism profile involved in UTI. However, it must be taken into account that the evolution of the infectious process does not depend only on the characteristics evaluated.
Based on this study, it was possible to compare the profile of E. coli isolated from urine infections from hospital and community samples. It was notable that the hospital environment favors multiresistance to antimicrobials; however, the susceptibility profile and presence of ESBL were similar in both environments. Of all the samples analyzed here, 95% presented genes encoding ESBL. Carmo et al,39 in agreement to our results, found the prevalent gene was blaTEM and in sequence, the gene blaCTX-M. The blaSHV gene is involved with plasmid transmission, where this facility in gene transmission is possibly directly involved with the highest percentage found in hospital isolates, since this environment is very susceptible to the most diverse types of microorganisms and genetic recombination among them.39
Elsayed et al46 determined the occurrence of multidrug resistant strains in urinary tract infections caused by E. coli. In that study, of the 100 clinical isolates, 68 presented one or more plasmids responsible for ESBL resistance. Thus, the high frequency of the presence of these genes in the isolates investigated in this study was highlighted, especially those obtained from the community.
The high prevalence of ESBL resistance genes in the negative samples for the phenotypic detection test is also noted. This fact is of great concern, since under some stimulus these isolates may begin to express these genes, initiating the enzyme synthesis process, and acting as potential sources of resistance genes.47 Thus, it demonstrates the importance of molecular tests and the need for control measures to avoid the dissemination of these microorganisms both in the community and in the hospital environment.
Generally, hospital isolates are associated with higher antimicrobial resistance than community isolates. Wollheim et al48 evaluated the presence of ESBL in microorganisms isolated from the community and hospital. The authors demonstrated that the presence of E. coli producing ESBL is especially a hospital problem and its dissemination to the community is relatively limited. Matta et al49 characterized the resistance of species of different pathogens between infections acquired in the community and acquired in hospitals. These authors demonstrated that hospital-acquired infections/bacteria demonstrate higher resistance rates when compared to community acquired. The data presented here highlight the new trend and perspective in the prevalence of antimicrobial-resistant, since community-acquired bacteria can be till more virulent and resistant than hospital-acquired bacteria.
There is no doubt that ESBL-producing infections are a major concern for the medical world. These are usually associated with increased morbidity/mortality and may present a difficult diagnosis. The fact that prevalence rates are increasing globally, even in non-hospital contexts, and the terrible lack of effective antimicrobial therapy demonstrates the tremendous concern in this regard.
Discussion with patients during every prescribing encounter and education should be routine in the clinical environment. Also, collaboration with local health departments and local laboratory technicians to learn resistance patterns improves patients’ recovery and initial treatment to cure. Thus, the data here suggests that in the clinical sphere decision-making strategies based on antibiogram data and clinical practice guideline recommendations will favor the clinical practice management of urinary tract infections with consequent improvements in patient health.
Author contributions
All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the verison to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Amin M, Mehdinejad M, Pourdangchi Z. Study of bacteria isolated from urinary tract infections and determination of their susceptibility to antibiotics. Jundishapur J Microbiol. 2009;3:118–123.
2. Barros ICAR, Ribeiro AU, Costa ACN, et al. Microorganisms prevalent in urinary tract infections and antimicrobial sensitivity profile: analysis of patients attended at the Military Police Hospital of the State of Goiás, Brazil, in the period from 1998 to 2008. J Health Sci Inst. 2011;29:243–247.
3. Cunha MA, Assunção GLM, Medeiros IM, Freitas MR. Antibiotic resistance patterns of urinary tract infections in a north eastern brazilian capital. Rev Inst Med Trop. 2016;58:1–4. doi:10.1590/S1678-9946201658002
4. Lopes HV, Tavares W. Diagnóstico das infecções do trato urinário. Ver Assoc Med Bras. 2005;51:300–312.
5. Tan CT, Chlebicki MP. Urinary tract infections in adults. Singapore Med J. 2006;57:485–490. doi:10.11622/smedj.2016153
6. Lüthje P, Brauner A. Novel strategies in the prevention and treatment of urinary tract infections. Pathogens. 2016;5:1–14. doi:10.3390/pathogens5010013
7. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13:269–284. doi:10.1038/nrmicro3432
8. Tiba MR, Yano T, Leite DS. Genotypic characterization of virulence factors in Escherichia coli strains from patients with cystitis.. Rev Inst MedTrop S Paulo. 2008;50:255–260. doi:10.1590/S0036-46652008000500001
9. Yun KW, Kim HY, Park HK, Kim W, Lima IS. Virulence factors of uropathogenic Escherichia coli of urinary tract infections and asymptomatic bacteriuria in children. J Microbiol Immunol Infect. 2014;47:455–461. doi:10.1016/j.jmii.2013.07.010
10. Pompilio A, Crocetta V, Savini V, et al. Phylogenetic relationships, biofilm formation, motility, antibiotic resistance and extended virulence genotypes among Escherichia coli strains from women with community-onset primitive acute pyelonephritis. PLoS ONE. 2018;13:1–26. doi:10.1371/journal.pone.0196260
11. Eberly AR, Floyd KA, Beebout CJ, et al. Biofilm formation by uropathogenic Escherichia coli is favored under oxygen conditions that mimic the bladder environment. Int J Mol Sci. 2017;18:1–12. doi:10.3390/ijms18102077
12. Tyfa A, Kunicka-Styczyńska A, Zabielska J. Evaluation of hydrophobicity and quantitative analysis of biofilm formation by Alicyclobacillus sp. Acta Biochim Pol. 2015;62:789–790. doi:10.18388/abp.2015_1133
13. Grigoryan L, Trautner BW, Gupta K. Diagnosis and management of urinary tract infections in the out patient setting a review. JAMA. 2014;312:1677–1684. doi:10.1001/jama.2014.12842
14. Rodrigues WF, Miguel CB, Nogueira APO, et al. Antibiotic resistance of bacteria involved in urinary infections in Brazil: a cross-sectional and retrospective study. Int J Environ Res Public Health. 2016;6:1–10.
15. Ventola CV. The antibiotic resistance crisis part 1: causes andthreats. P&T. 2015;40:277–283.
16. Aboumarzouk OM. Extended spectrum beta-lactamase urinary tract infections. Urol Ann. 2015;6:114–115.
17. Holten KB, Onusko EWM. Appropriate prescribing of oral beta-lactam antibiotics. Am Fam Physician. 2000;62:611–620.
18. Öztürk H, Ozkirimli E, Özgür A. Classification of beta-lactamases and penicillin binding proteins using ligand. PLoS ONE. 2015;10:1–23. doi:10.1371/journal.pone.0117874
19. Fernando MMPSC, Luke WANV, Miththinda JKNDR, et al. Extended spectrum beta lactamase producing organisms causing urinary tract infections in Sri Lanka and their antibiotic susceptibility pattern –a hospital based cross sectional study. BMC Infect Dis. 2017;17:1–10. doi:10.1186/s12879-017-2250-y
20. Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology and treatment. Saudi J Biol Sci. 2015;22:90–101. doi:10.1016/j.sjbs.2014.08.002
21. Picozzi SCM, Casellato S, Rossini M, et al. Extended-spectrum beta-lactamase-positive Escherichia coli causing complicated upper urinary tract infection: urologist should act in time. Urol Ann. 2014;6:107–112. doi:10.4103/0974-7796.130536
22. Adenipekun EO, Jackson CR, Ramadan H, et al. Prevalence and multidrug resistance of Escherichia coli from community acquired infections in Lagos, Nigeria. J Infect Dev Ctries. 2016;30:920–931. doi:10.3855/jidc.7997
23. Blaettler L, Mertz D, Frei R, Elzi L, Widmer A, Battegay M. Secular trend and risk factors for antimicrobial resistance in Escherichia coli isolates in Switzerland1997–2007. Infection. 2009;37:534–539. doi:10.1007/s15010-009-8457-0
24. Fasugba O, Mitchell BG, Mnatzaganian G, Das A, Collignon P, Gardner A. Five-Year antimicrobial resistance patterns of urinary Escherichia coli at an Australian tertiary hospital: time series analyses of prevalence data. PLoS ONE. 2016;3:1–14.
25. Moremi N, Claus H, Mshana SE. Antimicrobial resistance pattern: a report of microbiological cultures at a tertiary hospital in Tanzania. BMC Infect Dis. 2016;16:1–7. doi:10.1186/s12879-016-2082-1
26. Nzalie RN, Gonsu KH, Koulla-Shiro S. Bacterial etiology and antibiotic resistance profile of community-acquired urinary tract infections in a Cameroonian City. Int J Microbiol. 2016;2016:1–6. doi:10.1155/2016/3240268
27. Møretrø T, Langsrud S. Listeria monocytogenes: biofilm formation and persistence in food-processing environments. Biofilms. 2004;1:107–121. doi:10.1017/S1479050504001322
28. Rodrigues LB, Santos LR, Rizzo NN, et al. Hydrophobicity and biofilm formation on polystyrene by Salmonella Heidelberg isolated from a poultry slaughterhous. Acta Sci Vet. 2009;37:225–230. doi:10.22456/1679-9216.16333
29. Locatelli CI, Englert GE, Kwitko S, Simonetti AB. In vitro bacterial adherence to silicone and polymetylmethacrylate intraocular lenses. Arq Bras Oftalmol. 2004;67:241–248. doi:10.1590/S0004-27492004000200011
30. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Approved Standard M100-S19 20016 29 (3).
31. Guimarães AG, Cardoso RCV, Azevêdo PF, Meneses RB. Profile of susceptibility of bacteria, isolated from curdle cheese. Rev Inst Adolfo Lutz. 2012;71:259–265.
32. Vasconcelos FR, Rebouças RH, Evangelista-Barreto NS, De Sousa OV, Vieira RHSF. Resistance profile of Escherichia coli isolated from the environment. Arq Inst Biol. 2010;77:405–410.
33. Martins AC, Picoli SU. Métodos alternativos para detecção de betalactamase de espectro estendido em Escherichia coli e Klebsiella pneumoniae. J Bras Patol Med. 2011;47:421–426. doi:10.1590/S1676-24442011000400005
34. Sambrook J, Russel WD. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory; 2001.
35. Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi:10.1128/AEM.66.10.4555-4558.2000
36. Girardini LK, Siqueira FM, Krewer CC, Krewer CC, Costa MM, Vargas AC. Phylogenetic and pathotype analysis of Escherichia coli swine isolates from Southern Brazil. PesqVet Bras. 2012;32:374–378. doi:10.1590/S0100-736X2012000500002
37. Santos ACM, Pignatari ACC, Silva RM, Zidko ACM, Gales AC. The virulence of extra-intestinal pathogenic Escherichia coli (ExPEC)regarding host age and gender . O Mundo Da Saúde. 2009;33:392–400.
38. Amaya E, Reyes D, Paniagua M, et al. Antibiotic resistance patterns of Escherichia coli isolates from diferente aquatic environmental sources in Leon, Nicaragua. Clin Microbiol Infect. 2012;18:E347- E354. doi:10.1111/j.1469-0691.2012.03930.x
39. Carmo MS, Marques AM, Gonçalves LHB, et al. Detection of extended spectrum β-lactamases (ESBLS) in isolates of uropathogenic Escherichia coli (UPECs) from patients in the communityRev Patol Trop. 2012;41:419–426. doi:10.5216/rpt.v41i4.21708
40. Fang H, Ataker F, Hedin G, Dornbusch K. Molecular epidemiology of extended-spectrum β-Lactamases among Escherichia coli isolates collected in a Swedish Hospital and its associated health care facilities from 2001 to 2006. J Clin Microbiol. 2008;46:707–712. doi:10.1128/JCM.01943-07
41. Poursina F, Sepehrpour S, Mobasherizadeh S. Biofilm formation in nonmultidrug-resistant Escherichia coli isolated from patients with urinary tract infection in Isfahan, Iran. Adv Biomed Rev. 2018;7:1–9.
42. Krasowska A. Sigler K How microorganisms use hydrophobicity and what does this mean for human needs? Front Cell Infect Microbiol. 2014;4:112. doi:10.3389/fcimb.2014.00112
43. Mirani ZA, Fatima A, Urooj S, Aziz M, Khan M, Abbas T. Relationship of cell surface hydrophobicity with biofilm formation and growth rate: a study on Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli. Iran J Basic MedSci. 2018;21:760–769.
44. Iranpour D, Hassanpour M, Ansari H, et al. Phylogenetic groups of Escherichia coli strains from patients with urinary tract infection in Iran based on the new clermont phylotyping method. Bio Med Res Int. 2015;846219.
45. Ghauor M, Salenhzadeh A. Phylogenetic classification of Escherichia coli isolated from urinary tract infections in the Central Regions of Guilan Province Maryam Ghauor. J Med Microbiol Infec Dis. 2017;5:17–20.
46. Elsayed TI, Ismail HA, Elgamal SA, Gad AHA. The occurrence of multidrug resistant E. Coli which produce ESBL and cause urinary tract infections. J Appl Microbiol Biochem. 2017;1:2–8. doi:10.21767/2576-1412.100008
47. Alberts B, Johnson A, Walter P. Biologia Molecular da Célula.
48. Wolheim C, Guerra FMI, Conte DV, et al. Nosocomial and community infections due to class A extended-spectrum β-lactamase (ESBLA)-producing Escherichia coli and Klebsiella spp. in Southern Brazil. Braz J Infect Dis. 2001;15:138–143.
49. Matta R, Souheil H, Hallit R, Bawab W, Rogues MA, Salameh P. Epidemiology and microbiological profile comparison between community and hospital acquired infections: a multicenter retrospective study in Lebanon. J Infect Public Health. 2018;11:405–411. doi:10.1016/j.jiph.2017.09.005
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


