Back to Journals » OncoTargets and Therapy » Volume 9
Circulating interleukin-10 levels and human papilloma virus and Epstein–Barr virus-associated cancers: evidence from a Mendelian randomization meta-analysis based on 11,170 subjects
Authors Qu K, Pang Q, Lin T, Zhang L , Gu M, Niu W , Liu C, Zhang M
Received 21 September 2015
Accepted for publication 22 November 2015
Published 7 March 2016 Volume 2016:9 Pages 1251—1267
DOI https://doi.org/10.2147/OTT.S96772
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Kai Qu,1,* Qing Pang,1,* Ting Lin,1 Li Zhang,2 Mingliang Gu,3 Wenquan Niu,4 Chang Liu,1,* Ming Zhang5
1Department of Hepatobiliary Surgery, The First Affiliated Hospital of Xi’an Jiaotong University College of Medicine, Xi’an, Shaanxi Province, People’s Republic of China; 2Department of Ultrasound Diagnostics, Tangdu Hospital, Fourth Military Medical University, Xi’an, Shaanxi Province, People’s Republic of China; 3Chinese Academy of Sciences Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing, People’s Republic of China; 4State Key Laboratory of Medical Genomics, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, People’s Republic of China; 5Department of Otolaryngology, Eye, Ear, Nose and Throat Hospital, Fudan University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Abstract: Recent studies have showed interleukin 10 (IL-10) is a critical cytokine that determines antiviral immune response and is related to virus-associated cancers. However, whether genetically elevated circulating IL-10 levels are associated with the risk of human papilloma virus and Epstein–Barr virus-associated cancers (HEACs) is still unclear. Mendelian randomization method was implemented to meta-analyze available observational studies by employing IL-10 three variants (-592C>A, -819C>T, and -1082A>G) as instruments. A total of 24 articles encompassing 11,170 subjects were ultimately eligible for the meta-analysis. Overall, there was a significant association between IL-10 promoter variant -1082A>G and HEACs under allelic and dominant models (both P<0.01). Subgroup analysis by cancer type indicated that the risk estimate of -1082A>G was significant for nasopharyngeal cancer under allelic, homozygous genotypic and dominant models (all P<0.001). Moreover by ethnicity, carriers of -1082G allele had a 74% increased risk for nasopharyngeal cancer in Asians under dominant model (odds ratio [OR] =1.737; 95% confidence interval [CI]: 1.280–2.358; P<0.001). In further Mendelian randomization analysis, the predicted OR for 10 pg/mL increment in IL-10 levels was 1.14 (95% CI: 1.01–16.99) in HEACs. Our findings provided strong evidence for a critical role of genetically elevated circulating IL-10 levels in the development of HEACs, especially in Asian population and for nasopharyngeal cancer.
Keywords: interleukin-10, human papilloma virus, Epstein–Barr virus, meta-analysis, Mendelian randomization
Introduction
There is growing recognition that viruses are capable of causing cancer in humans, and approximately 15% of all human malignancies are estimated to be attributable to viruses, creating a major global health burden.1 Among various cancer viruses, human papilloma virus (HPV) and Epstein–Barr virus (EBV) are exhaustively investigated and are considered to account for 38% of all virus-associated cancers.2 HPV infection is associated with more than 90%3 and 60%4,5 cases of cervical and oropharyngeal cancers, respectively, and in contrast, EBV is associated with nearly 90% of nasopharyngeal cancer,6,7 Burkitt’s,8 and Hodgkin’s lymphoma.5,7,9 Moreover, the two viruses could also exhibit synergistic or cooperative effects on carcinogenesis. For example, EBV is deemed as a “helper virus” for HPV-induced carcinogenesis.10 Coinfection of HPV and EBV was observed in 30%–50% of patients with oral cancer11,12 and cervical cancer.13,14 Although the obvious association between HPV/EBV and human papilloma virus and Epstein–Barr virus-associated cancers (HEACs) has been universally accepted, the inherited procancer mechanisms so far remain unclear. It should be pointed out that the infection of HPV and EBV affects more than 40% of general population; however, only a small proportion of infected cases develop cancer.15 Interindividual differences of innate antiviral immunity that is affected by hereditary factors might be involved in the underlying pathological process of HEACs.16
Among the antiviral-relevant immune factors, interleukin-10 (IL-10) is a key cytokine that determines viral clearance or persistence17 and is involved in carcinogenesis.18,19 Recent studies have observed a marked high level of circulating IL-10 in patients with HEACs and its association with poor prognosis.20,21 Emerging evidence suggested that interindividual differences in circulating IL-10 levels might be due to the polymorphic defects of IL-10. Recently, three variants located within IL-10 promoter region, viz, −592C>A (rs1800872), −819C>T (rs1800871), and −1082A>G (rs1800896), have been well defined in association with the changes of IL-10 production.22,23 It is, therefore, reasonable to hypothesize that if IL-10 is involved in the carcinogenesis process of HEACs, the inherited genetic determinants that alter IL-10 production should affect cancer susceptibility in the direction and magnitude predicted by its circulating levels.
Mendelian randomization approach, which is based on Mendel’s second law, uses measured variation in genes of known function to examine the effect of a modifiable exposure on disease in observational studies.24,25 This method could partially provide evidence for the causal nature of the target phenotype influenced by genetic defects. To test the hypothesis that genetically elevated levels of IL-10 due to IL-10 genetic defects cause an increased risk of cancer, in this study, we first decided to perform a meta-analysis to evaluate the association of IL-10 three variants with both circulating IL-10 levels and the risk for HEACs. If the variant under study is found to be predictive of both cancer and circulating IL-10 levels, Mendelian randomization approach will be further implemented to test the possible association of circulating IL-10 levels with HEACs.
Materials and methods
This meta-analysis was undertaken according to the guidelines set forth by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (Table S1).
Search strategy
A literature search for observational studies investigating the association between IL-10 three variants (−592C>A [rs1800872], −819C>T [rs1800871], and −1082A>G [rs1800896]) and all types of HEACs was conducted of PubMed and Google Scholar databases covering the period from the earliest possible year to August 1, 2015. Subject terms used for the search were: “interleukin 10”, “interleukin-10”, “IL 10”, “IL-10”, “oral or mouth cancer”, “nasopharyngeal cancer”, “oropharyngeal cancer”, “Hodgkin or Burkitt lymphoma”, “cervical or vaginal or vulvar cancer”, “anus or anal cancer”, combined with “polymorphism”, “genetic”, “variant”, “mutation”, “allele”, or “genotype”. The reference lists of all the retrieved articles as well as those of reviews on the same topic were also searched to supplement the additional missing articles. Searching results was limited to studies with a case–control design and articles published in the English language.
Trial selection
Two investigators (Kai Qu and Ming Zhang) independently obtained the full texts of potentially eligible articles based on the titles and abstracts. If necessary, we emailed the corresponding authors to avoid double counting of participants recruited in more than one publication. In case of more than one publication from the same study population, we abstracted data from the most recent or most complete publication.
Inclusion/exclusion criteria
For inclusion, the studies should strictly fulfill the following inclusion criteria (all points must be satisfied for inclusion): 1) clinical endpoint (dependent variable): HEACs including oropharyngeal cancer, nasopharyngeal cancer, cervical cancer, Hodgkin’s lymphoma, and Burkitt’s lymphoma; 2) study design: either retrospective or prospective case–control design; and 3) independent variables: the genotype and/or allele counts of at least one of IL-10 three variants (−592C>A, −819C>T, and −1082A>G). Studies were excluded (one point was sufficient for exclusion) if they investigated the gene function, disease progression, severity, and the response to treatment or survival. Additionally, conference abstracts, case reports or series, editorials, narrative reviews, meta-analysis, and the non-English articles were also excluded.
Data extraction
Two investigators (Kai Qu and Ming Zhang) independently extracted data using a standardized Excel template. Disagreements were resolved by consensus or by a third investigator (Wenquan Niu). Data were collected on the first author, year of publication, ethnicity and country of the study population, cancer type, case–control status, study design, sample size, the genotypes/alleles of IL-10 three variants (−592C>A, −819C>T, and −1082A>G) between cases and controls, and characteristics of participants, if available, including age, sex, body mass index, smoking, drinking, family history of cancer, and virus infection status.
Statistical analysis
In this meta-analysis, three genetic models of inheritance were performed for IL-10 variants, including allelic model (the A allele versus the C allele for −592C>A SNP [single nucleotide polymorphism]; the T allele versus the C allele for −819C>T SNP; and the G allele versus the A allele for −1082A>G SNP), homozygous (the AA genotype versus the CC genotype for −592C>A SNP; the TT genotype versus the CC genotype for −819C>T SNP; and the GG genotype versus the AA genotype for −1082A>G) and dominant model (the AA genotype plus the AC genotype versus the AA genotype for −592C>A SNP; the TT genotype plus the TC genotype versus the CC genotype for −819C>T SNP; and the GG genotype plus the GA genotype versus the AA genotype for −1082A>G).
Weighted odds ratios (ORs) and the corresponding 95% confidence intervals (CIs) were evaluated by a random-effects model using the DerSimonian and Laird method. Heterogeneity between studies was computed by the χ2 test, and was quantified by the inconsistency index (I2) statistic, which ranges from 0% to 100% and is defined as the percentage of the observed between-study variability that is due to heterogeneity rather than chance.
Predefined subgroup analyses were performed a priori according to the cancer types (oral cancer, nasopharyngeal cancer, cervical cancer or lymphoma [including Hodgkin and Burkitt lymphoma]), ethnicity of the study populations (Caucasian, Asian, Latinos, or African), study design (population-based or hospital-based), and the total sample size (<300 subjects or ≥300 subjects). The data were presented and summarized if there were three or more independent studies that provided the genotype or allele counts of the IL-10 three variants between cases and controls.
Genetic association studies have been considered more closely relevant to randomized trials than other types of epidemiological study due to independent assortment of alleles that theoretically should not be confounded by environmental or behavioral factors.25 Therefore, we employed Mendelian randomization model to test the hypothesis that genetically elevated level of IL-10 because of variants in IL-10 cause an increased risk of HEACs. In Mendelian randomization analysis, risk estimate was computed from the ratio of the coefficient of the association between a variant and a disease to that of the association between the variant and biomarker as a reflection of the potential effect of circulating IL-10 levels on cancer risk.
Publication bias was assessed by visual inspection of Begg’s and Egger’s funnel plots, accompanied by the corresponding Begg’s and Egger’s tests. The trim and fill method was implemented to estimate the number and outcomes of potentially missing trials resulting from publication bias. Data management and statistical analyses were conducted using STATA software (StataCorp, College Station, TX, USA, version 11.2 for Windows). P<0.05 was considered statistically significant. For Begg’s and Egger’s statistics, a significance level was defined as P<0.10.
Results
Eligible articles
A flow diagram schematizing the process of article selection with specific reasons is presented in Figure 1. In total, 103 potentially relevant articles were identified after the initial search, and 24 of them that satisfied inclusion/exclusion criteria were deemed as eligible.26–49 All 24 qualified articles written in English were published between 2001 and 2014.
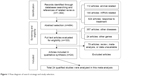  | Figure 1 Flow diagram of search strategy and study selection. |
Study characteristics
The basic characteristics of all 24 qualified articles are listed in Table 1, and the genotype distributions and allele frequencies of IL-10 three variants (−592C>A, −819C>T, and −1082A>G) between cases and controls are listed in Table S2. In this meta-analysis, 12 articles were conducted for cervical cancer, 7 for lymphoma (including Hodgkin’s lymphoma and Burkitt’s lymphoma), 4 for nasopharyngeal cancer, and 1 for oral cancer. Additionally, there were 8 articles involving Asians, 9 involving Caucasians, 3 involving Latinos, 2 involving Africans, and 1 involving the mixed populations. There were 11 articles conducted on a population-based design and 13 on a hospital-based design. Of 24 qualified articles, 14 (58.33%) had the total sample size (the sum of patients and controls) of at least 300 subjects.
Overall and subgroup analysis of IL-10 variants and HEACs
Pooling all 24 qualified articles together indicated a significant association between IL-10–1082A>G variant and HEACs under allelic (OR=1.283; 95% CI: 1.071–1.537; P=0.007) and dominant models (OR=1.382; 95% CI: 1.128–1.694; P=0.002; Table 2). Conversely, we failed to find any significance for the other two variants (−592C>A and −819C>T) in association with HEACs (Tables S3 and S4).
To account for the potential sources of between-study heterogeneity, a set of predefined subgroup analyses were conducted (Table 2). By ethnicity, an extremely significant association between IL-10–1082G allele and HEACs in Asians was observed under allelic (OR=2.009; 95% CI: 1.566–2.578; P<0.001), homozygous genotypic (OR=2.832; 95% CI: 1.831–4.379; P<0.001), and dominant (OR=2.101; 95% CI: 1.694–2.607; P<0.001) models (Figure 2A and B). In Caucasians, Latinos, Africans, or mixed populations, there was no significant association observed in this meta-analysis. By cancer type, the magnitude of risk estimates was significant for nasopharyngeal cancer under allelic (OR=1.530; 95% CI: 1.063–2.200; P=0.022) and dominant models (OR=1.737; 95% CI: 1.280–2.358; P<0.001; Figure 2C and D), whereas no significance was reached for the other cancer types under investigation. By study design, there was a significant association between IL-10–1082G allele and HEACs in population-based studies under allelic (OR=1.365; 95% CI: 1.047–1.780; P=0.022) and dominant (OR=1.492; 95% CI: 1.157–1.924; P=0.002) models. Additionally, IL-10–1082G allele was more significantly associated with HEACs in those case–control-matched studies under allelic (OR=1.407; 95% CI: 1.091–1.814; P=0.009), homozygous genotypic (OR=1.612; 95% CI: 1.024–2.536; P=0.039), and dominant (OR=1.557; 95% CI: 1.214–1.999; P<0.001) models (Table 2).
There was no publication bias of three IL-10 promoter variants either in overall or subgroup analysis as reflected by the funnel plots, Egger’s tests, and Begg–Mazemdar tests (Figure 3).
Association of IL-10 variants with circulating IL-10 levels
Genotype–phenotype association was based on four articles with circulating IL-10 levels measured in HEAC cancer patients (Table S5).37,41,50,51 We compared averaged circulating IL-10 levels under homozygous genotypic and dominant models. Circulating IL-10 level was significantly elevated in −1082G allele carriers under homozygous genotypic model (standard mean difference [SMD] =25.692; 95% CI: 1.303–50.081; P=0.039) and dominant model (SMD =13.313; 95% CI: 0.901–25.725; P=0.036; Figure 4). There were low probabilities of publication bias for both models as reflected by the Begg’s funnel plots (both P=0.296) and the Egger’s tests (P=0.308 and P=0.442, respectively). As expected, there were no significant differences in the changes of circulating IL-10 level for −592C>A and −819C>T under both models.
Predicted association of circulating IL-10 levels with HEACs from Mendelian randomization
We assumed a linear–logistic relationship between difference of circulating IL-10 level and odds of HEACs when implementing Mendelian randomization method. The predicted OR for 5 and 10 pg/mL IL-10 increment were 1.13 (95% CI: 1.02–18.64) and 1.28 (95% CI: 1.05–347.26), respectively. Because the 95% CIs of both estimated OR did not include the null hypothesis value of 1.00, it was safe to reject the null hypothesis at a 5% significance level, and reveal a potential role of elevated circulating IL-10 level in development of HEACs.
Discussion
On the basis of a meta-analysis of the data from 24 studies involving 5,390 cases and 5,780 controls, we investigated IL-10 three promoter variants (−592C>A, −819C>T, and −1082A>G) and circulating IL-10 levels in relation to the risk for HEACs. One principal finding of this study was the significant association of IL-10–1082A>G variant with HEACs, especially in Asians and for nasopharyngeal cancer. On the basis of aforementioned results, we further employed −1082A>G variant as an instrument to surrogate circulating IL-10 levels, and revealed the association of IL-10 levels with risk for HEACs. To our knowledge, this is the first meta-analysis demonstrating the association between circulating IL-10 levels and HEACs by implementing Mendelian randomization approach.
Previous studies have revealed that HPV and EBV infection is the main etiologic risk factors for many epithelial malignancies such as oropharyngeal cancer,4 nasopharyngeal cancer,6 cervical cancer,3 and some subtypes of lymphoma such as Hodgkin’s9 and Burkitt’s lymphoma.8 HPV and EBV are ubiquitous, double-stranded DNA viruses, which can be found in the upper aerodigestive tract. Epidemiologic data showed HPV and EBV infection affected over 10% and 90% of the general population, respectively, but only a small percentage of those infected developed cancer, probably because of lowered immune response and virus clearance due to interindividual genetic variations that result in persistent virus infection. IL-10, a cytokine with multiple effects in immunoregulation and inflammation, has a central role in infection by limiting the immune response to pathogens. Given the essential role of IL-10 in the antiviral response in vitro52 and in vivo,17 it is reasonable to expect that IL-10 is implicated in the tumorigenesis of HEACs. Indeed, elevation of circulating IL-10 levels was detected in patients with cervical cancer,53 oropharyngeal cancer,54 and Hodgkin’s lymphoma,55 and was associated with poor prognoses of these patients. Consistent with above evidence, in this study, our pooled results using Mendelian randomization approach indicated that 5 and 10 pg/mL increments in circulating IL-10 levels were 1.13 and 1.28 times more likely to develop HEACs in a significant manner, respectively. However, considering the unstable status of circulating IL-10 levels in time as previously described (plasma half-life ranged from 2.7 to 4.5 hours),56 which may cause a weak association between IL-10 level and HEAC risk, well-designed studies with precise IL-10 measurement are required to quantify this effect size reliably.
Nasopharyngeal cancer is a quite common malignancy in Eastern Asians, especially in Chinese, as well as in migrants from those areas,57 with its incidence rates peaking at 30 cases/100,000 in males and at 10 cases/100,000 in females.58 Conversely, it is rare in Europe and North America, accounting for less than 1% of all cancer cases, with incidence rates generally below 2 cases/100,000 in males and 1 case/100,000 in females.58 The obvious various incidence of nasopharyngeal cancer among different ethnic groups suggests this cancer is influenced by heredity factors. In this study, our results robustly confirmed the association between −1082A>G allele and nasopharyngeal cancer, especially in Asian population with relative low between-study heterogeneity. Our findings added a potential explanation for varying incidence of nasopharyngeal cancer worldwide. Further studies are necessary to confirm our findings, and IL-10–1082A>G allele, once validated, might be a specific biomarker for patients with nasopharyngeal cancer.
Despite the clear strengths including the large sample sizes and implementation of Mendelian randomization approach, several possible limitations in the present meta-analysis should also be noted. First, to avoid the impact of low-quality studies, we only included articles written in English, which might cause publication bias, even though our funnel plots and statistical tests did not tell. Second, we only examined three promoter variants in IL-10 gene, and investigation on other variants in or flanking IL-10 gene, especially some low-penetrance genes will be encouraged. Third, the single-locus-based nature of meta-analysis precluded the possibility of gene–gene59 and gene–environment interactions, but whether this variant integrated with other genetic or environmental risk factors will enhance prediction requires additional research. For instance, it is found that different types of HPV proteins exhibited varying abilities in inducing promoter activity of IL-10 gene.60 Therefore, it is also necessary to perform a HPV type-stratified analysis in further study. Fourth, nearly all involved studies in this meta-analysis had circulating IL-10 measured only once and did not reflect its long-term level in the development of HEACs. Therefore, because of the above limitations, the jury must refrain from drawing a firm conclusion until a large-scale and well-designed study confirms or refutes our findings.
In summary, our findings provided evidence for a critical role of genetically elevated circulating IL-10 in the development of HEACs by employing IL-10 gene −1082A>G as an instrument, and the risk association of this variant with HEACs was more evident in Asian patients with nasopharyngeal cancer. Additional studies examining biological function of elevated circulating IL-10 level in HEACs, as well as studies seeking to provide clinical validations of our findings, are warranted.
Acknowledgments
The authors are thankful to Dr Qichao Huang in the Fourth Military Medical University for his technical assistance in statistics. This work was supported by the grants of National Natural Science Foundation of China (Grant numbers: 81201549, 81272644, and 81472247), the Project of Innovative Research Team for Key Science and Technology in Xi’an Jiaotong University (number: 2013KCJ-23), and the Scientific Research Project of Shanghai Municipal Bureau of Health (number: 20114y069).
Disclosure
The authors report no conflicts of interest in this work.
References
McLaughlin-Drubin ME, Munger K. Viruses associated with human cancer. Biochim Biophys Acta. 2008;1782(3):127–150. | ||
Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009;392(1):1–10. | ||
Bosch FX, Manos MM, Munoz N, et al. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87(11):796–802. | ||
Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–4301. | ||
de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13(6):607–615. | ||
Gu AD, Zeng MS, Qian CN. The criteria to confirm the role of Epstein–Barr virus in nasopharyngeal carcinoma initiation. Int J Mol Sci. 2012;13(10):13737–13747. | ||
Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118(12):3030–3044. | ||
Brady G, MacArthur GJ, Farrell PJ. Epstein–Barr virus and Burkitt lymphoma. J Clin Pathol. 2007;60(12):1397–1402. | ||
Glaser SL, Lin RJ, Stewart SL, et al. Epstein–Barr virus-associated Hodgkin’s disease: epidemiologic characteristics in international data. Int J Cancer. 1997;70(4):375–382. | ||
Al Moustafa AE, Chen D, Ghabreau L, Akil N. Association between human papillomavirus and Epstein–Barr virus infections in human oral carcinogenesis. Med Hypotheses. 2009;73(2):184–186. | ||
Jalouli J, Jalouli MM, Sapkota D, Ibrahim SO, Larsson PA, Sand L. Human papilloma virus, herpes simplex virus and epstein barr virus in oral squamous cell carcinoma from eight different countries. Anticancer Res. 2012;32(2):571–580. | ||
Tyan YS, Liu ST, Ong WR, Chen ML, Shu CH, Chang YS. Detection of Epstein–Barr virus and human papillomavirus in head and neck tumors. J Clin Microbiol. 1993;31(1):53–56. | ||
Khenchouche A, Sadouki N, Boudriche A, et al. Human papillomavirus and Epstein–Barr virus co-infection in cervical carcinoma in Algerian women. Virol J. 2013;10:340. | ||
Prayitno A. Cervical cancer with human papilloma virus and Epstein Barr virus positive. J Carcinog. 2006;5:13. | ||
Hariri S, Unger ER, Sternberg M, et al. Prevalence of genital human papillomavirus among females in the United States, the National Health And Nutrition Examination Survey, 2003–2006. J Infect Dis. 2011;204(4):566–573. | ||
Beskow AH, Gyllensten UB. Host genetic control of HPV 16 titer in carcinoma in situ of the cervix uteri. Int J Cancer. 2002;101(6):526–531. | ||
Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12(11):1301–1309. | ||
Dennis KL, Blatner NR, Gounari F, Khazaie K. Current status of interleukin-10 and regulatory T-cells in cancer. Curr Opin Oncol. 2013;25(6):637–645. | ||
Bermudez-Morales VH, Gutierrez LX, Alcocer-Gonzalez JM, Burguete A, Madrid-Marina V. Correlation between IL-10 gene expression and HPV infection in cervical cancer: a mechanism for immune response escape. Cancer Invest. 2008;26(10):1037–1043. | ||
El-Sherif AM, Seth R, Tighe PJ, Jenkins D. Quantitative analysis of IL-10 and IFN-gamma mRNA levels in normal cervix and human papillomavirus type 16 associated cervical precancer. J Pathol. 2001;195(2):179–185. | ||
Viviani S, Notti P, Bonfante V, Verderio P, Valagussa P, Bonadonna G. Elevated pretreatment serum levels of Il-10 are associated with a poor prognosis in Hodgkin’s disease, the Milan Cancer Institute Experience. Med Oncol. 2000;17(1):59–63. | ||
Suarez A, Castro P, Alonso R, Mozo L, Gutierrez C. Interindividual variations in constitutive interleukin-10 messenger RNA and protein levels and their association with genetic polymorphisms. Transplantation. 2003;75(5):711–717. | ||
Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24(1):1–8. | ||
Proitsi P, Lupton MK, Velayudhan L, et al. Genetic predisposition to increased blood cholesterol and triglyceride lipid levels and risk of Alzheimer disease: a Mendelian randomization analysis. PLoS Med. 2014;11(9):e1001713. | ||
Niu W, Liu Y, Qi Y, Wu Z, Zhu D, Jin W. Association of interleukin-6 circulating levels with coronary artery disease: a meta-analysis implementing mendelian randomization approach. Int J Cardiol. 2012;157(2):243–252. | ||
Andrie E, Michos A, Kalampoki V, et al. Genetic variants in immunoregulatory genes and risk for childhood lymphomas. Eur J Haematol. 2009;83(4):334–342. | ||
Barbisan G, Perez LO, Contreras A, Golijow CD. TNF-alpha and IL-10 promoter polymorphisms, HPV infection, and cervical cancer risk. Tumour Biol. 2012;33(5):1549–1556. | ||
Chagas BS, Gurgel AP, da Cruz HL, et al. An interleukin-10 gene polymorphism associated with the development of cervical lesions in women infected with Human Papillomavirus and using oral contraceptives. Infect Genet Evol. 2013;19:32–37. | ||
Cunningham LM, Chapman C, Dunstan R, Bell MC, Joske DJ. Polymorphisms in the interleukin 10 gene promoter are associated with susceptibility to aggressive non-Hodgkin’s lymphoma. Leuk Lymphoma. 2003;44(2):251–255. | ||
da Silva GN, Bacchi MM, Rainho CA, de Oliveira DE. Epstein–Barr virus infection and single nucleotide polymorphisms in the promoter region of interleukin 10 gene in patients with Hodgkin lymphoma. Arch Pathol Lab Med. 2007;131(11):1691–1696. | ||
Farhat K, Hassen E, Gabbouj S, Bouaouina N, Chouchane L. Interleukin-10 and interferon-gamma gene polymorphisms in patients with nasopharyngeal carcinoma. Int J Immunogenet. 2008;35(3):197–205. | ||
Fernandes AP, Goncalves MA, Simoes RT, Mendes-Junior CT, Duarte G, Donadi EA. A pilot case–control association study of cytokine polymorphisms in Brazilian women presenting with HPV-related cervical lesions. Eur J Obstet Gynecol Reprod Biol. 2008;140(2):241–244. | ||
Govan VA, Carrara HR, Sachs JA, Hoffman M, Stanczuk GA, Williamson AL. Ethnic differences in allelic distribution of IFN-g in South African women but no link with cervical cancer. J Carcinog. 2003;2(1):3. | ||
Ivansson EL, Gustavsson IM, Magnusson JJ, et al. Variants of chemokine receptor 2 and interleukin 4 receptor, but not interleukin 10 or Fas ligand, increase risk of cervical cancer. Int J Cancer. 2007;121(11):2451–2457. | ||
Matsumoto K, Oki A, Satoh T, et al. Interleukin-10 −1082 gene polymorphism and susceptibility to cervical cancer among Japanese women. Jpn J Clin Oncol. 2010;40(11):1113–1116. | ||
Minnicelli C, Barros MH, Klumb CE, Romano SO, Zalcberg IR, Hassan R. Relationship of Epstein–Barr virus and interleukin 10 promoter polymorphisms with the risk and clinical outcome of childhood Burkitt lymphoma. PLoS One. 2012;7(9):e46005. | ||
Munro LR, Johnston PW, Marshall NA, et al. Polymorphisms in the interleukin-10 and interferon gamma genes in Hodgkin lymphoma. Leuk Lymphoma. 2003;44(12):2083–2088. | ||
Nieters A, Beckmann L, Deeg E, Becker N. Gene polymorphisms in Toll-like receptors, interleukin-10, and interleukin-10 receptor alpha and lymphoma risk. Genes Immun. 2006;7(8):615–624. | ||
Oduor CI, Chelimo K, Ouma C, et al. Interleukin-6 and interleukin-10 gene promoter polymorphisms and risk of endemic Burkitt lymphoma. Am J Trop Med Hyg. 2014;91(3):649–654. | ||
Pratesi C, Bortolin MT, Bidoli E, et al. Interleukin-10 and interleukin-18 promoter polymorphisms in an Italian cohort of patients with undifferentiated carcinoma of nasopharyngeal type. Cancer Immunol Immunother. 2006;55(1):23–30. | ||
Roh JW, Kim MH, Seo SS, et al. Interleukin-10 promoter polymorphisms and cervical cancer risk in Korean women. Cancer Lett. 2002;184(1):57–63. | ||
Shekari M, Kordi-Tamandani DM, MalekZadeh K, Sobti RC, Karimi S, Suri V. Effect of anti-inflammatory (IL-4, IL-10) cytokine genes in relation to risk of cervical carcinoma. Am J Clin Oncol. 2012;35(6):514–519. | ||
Singh H, Jain M, Sachan R, Mittal B. Association of TNFA (−308G>A) and IL-10 (−819C>T) promoter polymorphisms with risk of cervical cancer. Int J Gynecol Cancer. 2009;19(7):1190–1194. | ||
Stanczuk GA, Sibanda EN, Perrey C, et al. Cancer of the uterine cervix may be significantly associated with a gene polymorphism coding for increased IL-10 production. Int J Cancer. 2001;94(6):792–794. | ||
Tsai CW, Chang WS, Lin KC, et al. Significant association of Interleukin-10 genotypes and oral cancer susceptibility in Taiwan. Anticancer Res. 2014;34(7):3731–3737. | ||
Tsai CW, Tsai MH, Shih LC, Chang WS, Lin CC, Bau DT. Association of interleukin-10 (IL10) promoter genotypes with nasopharyngeal carcinoma risk in Taiwan. Anticancer Res. 2013;33(8):3391–3396. | ||
Wang Q, Zhang C, Walayat S, Chen HW, Wang Y. Association between cytokine gene polymorphisms and cervical cancer in a Chinese population. Eur J Obstet Gynecol Reprod Biol. 2011;158(2):330–333. | ||
Wei YS, Kuang XH, Zhu YH, et al. Interleukin-10 gene promoter polymorphisms and the risk of nasopharyngeal carcinoma. Tissue Antigens. 2007;70(1):12–17. | ||
Zoodsma M, Nolte IM, Schipper M, et al. Interleukin-10 and Fas polymorphisms and susceptibility for (pre)neoplastic cervical disease. Int J Gynecol Cancer. 2005;15(Suppl 3):282–290. | ||
Hohaus S, Giachelia M, Massini G, et al. Clinical significance of interleukin-10 gene polymorphisms and plasma levels in Hodgkin lymphoma. Leuk Res. 2009;33(10):1352–1356. | ||
Jin L, Sturgis EM, Cao X, et al. Interleukin-10 promoter variants predict HPV-positive tumors and survival of squamous cell carcinoma of the oropharynx. FASEB J. 2013;27(6):2496–2503. | ||
Santin AD, Hermonat PL, Ravaggi A, et al. Interleukin-10 increases Th1 cytokine production and cytotoxic potential in human papillomavirus-specific CD8(+) cytotoxic T lymphocytes. J Virol. 2000;74(10):4729–4737. | ||
Chopra V, Dinh TV, Hannigan EV. Circulating serum levels of cytokines and angiogenic factors in patients with cervical cancer. Cancer Invest. 1998;16(3):152–159. | ||
Fujieda S, Sunaga H, Tsuzuki H, Fan GK, Saito H. IL-10 expression is associated with the expression of platelet-derived endothelial cell growth factor and prognosis in oral and oropharyngeal carcinoma. Cancer Lett. 1999;136(1):1–9. | ||
Bohlen H, Kessler M, Sextro M, Diehl V, Tesch H. Poor clinical outcome of patients with Hodgkin’s disease and elevated interleukin-10 serum levels. Clinical significance of interleukin-10 serum levels for Hodgkin’s disease. Ann Hematol. 2000;79(3):110–113. | ||
Huhn RD, Radwanski E, Gallo J, et al. Pharmacodynamics of subcutaneous recombinant human interleukin-10 in healthy volunteers. Clin Pharmacol Ther. 1997;62(2):171–180. | ||
Mousavi SM, Sundquist J, Hemminki K. Nasopharyngeal and hypopharyngeal carcinoma risk among immigrants in Sweden. Int J Cancer. 2010;127(12):2888–2892. | ||
Polesel J, Serraino D, Negri E, et al. Consumption of fruit, vegetables, and other food groups and the risk of nasopharyngeal carcinoma. Cancer Causes Control. 2013;24(6):1157–1165. | ||
Hayashi T, Ito R, Cologne J, et al. Effects of IL-10 haplotype and atomic bomb radiation exposure on gastric cancer risk. Radiat Res. 2013;180(1):60–69. | ||
Bermudez-Morales VH, Peralta-Zaragoza O, Alcocer-Gonzalez JM, Moreno J, Madrid-Marina V. IL-10 expression is regulated by HPV E2 protein in cervical cancer cells. Mol Med Rep. 2011;4(2):369–375. |
Supplementary materials
  | Table S1 PRISMA 2009 Checklist |
  | Table S5 Changes of circulating IL-10 level across genotypes of three examined variants in IL-10 |
References
Moher D, Liberati A, Tetzlaff J, Altman DG; The PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(6):e1000097. | ||
Andrie E, Michos A, Kalampoki V, et al. Genetic variants in immunoregulatory genes and risk for childhood lymphomas. Eur J Haematol. 2009;83(4):334–342. | ||
Barbisan G, Perez LO, Contreras A, Golijow CD. TNF-alpha and IL-10 promoter polymorphisms, HPV infection, and cervical cancer risk. Tumour Biol. 2012;33(5):1549–1556. | ||
Chagas BS, Gurgel AP, da Cruz HL, et al. An interleukin-10 gene polymorphism associated with the development of cervical lesions in women infected with Human Papillomavirus and using oral contraceptives. Infect Genet Evol. 2013;19:32–37. | ||
Cunningham LM, Chapman C, Dunstan R, Bell MC, Joske DJ. Polymorphisms in the interleukin 10 gene promoter are associated with susceptibility to aggressive non-Hodgkin’s lymphoma. Leuk Lymphoma. 2003;44(2):251–255. | ||
da Silva GN, Bacchi MM, Rainho CA, de Oliveira DE. Epstein–Barr virus infection and single nucleotide polymorphisms in the promoter region of interleukin 10 gene in patients with Hodgkin lymphoma. Arch Pathol Lab Med. 2007;131(11):1691–1696. | ||
Farhat K, Hassen E, Gabbouj S, Bouaouina N, Chouchane L. Interleukin-10 and interferon-gamma gene polymorphisms in patients with nasopharyngeal carcinoma. Int J Immunogenet. 2008;35(3): 197–205. | ||
Fernandes AP, Goncalves MA, Simoes RT, Mendes-Junior CT, Duarte G, Donadi EA. A pilot case–control association study of cytokine polymorphisms in Brazilian women presenting with HPV-related cervical lesions. Eur J Obstet Gynecol Reprod Biol. 2008;140(2): 241–244. | ||
Govan VA, Carrara HR, Sachs JA, Hoffman M, Stanczuk GA, Williamson AL. Ethnic differences in allelic distribution of IFN-g in South African women but no link with cervical cancer. J Carcinog. 2003;2(1):3. | ||
Ivansson EL, Gustavsson IM, Magnusson JJ, et al. Variants of chemokine receptor 2 and interleukin 4 receptor, but not interleukin 10 or Fas ligand, increase risk of cervical cancer. Int J Cancer. 2007;121(11):2451–2457. | ||
Matsumoto K, Oki A, Satoh T, et al. Interleukin-10 -1082 gene polymorphism and susceptibility to cervical cancer among Japanese women. Jpn J Clin Oncol. 2010;40(11):1113–1116. | ||
Minnicelli C, Barros MH, Klumb CE, Romano SO, Zalcberg IR, Hassan R. Relationship of Epstein–Barr virus and interleukin 10 promoter polymorphisms with the risk and clinical outcome of childhood Burkitt lymphoma. PLoS One. 2012;7(9):e46005. | ||
Munro LR, Johnston PW, Marshall NA, et al. Polymorphisms in the interleukin-10 and interferon gamma genes in Hodgkin lymphoma. Leuk Lymphoma. 2003;44(12):2083–2088. | ||
Nieters A, Beckmann L, Deeg E, Becker N. Gene polymorphisms in Toll-like receptors, interleukin-10, and interleukin-10 receptor alpha and lymphoma risk. Genes Immun. 2006;7(8):615–624. | ||
Oduor CI, Chelimo K, Ouma C, et al. Interleukin-6 and interleukin-10 gene promoter polymorphisms and risk of endemic Burkitt lymphoma. Am J Trop Med Hyg. 2014;91(3):649–654. | ||
Pratesi C, Bortolin MT, Bidoli E, et al. Interleukin-10 and interleukin-18 promoter polymorphisms in an Italian cohort of patients with undifferentiated carcinoma of nasopharyngeal type. Cancer Immunol Immunother. 2006;55(1):23–30. | ||
Roh JW, Kim MH, Seo SS, et al. Interleukin-10 promoter polymorphisms and cervical cancer risk in Korean women. Cancer Lett. 2002; 184(1):57–63. | ||
Shekari M, Kordi-Tamandani DM, MalekZadeh K, Sobti RC, Karimi S, Suri V. Effect of anti-inflammatory (IL-4, IL-10) cytokine genes in relation to risk of cervical carcinoma. Am J Clin Oncol. 2012;35(6):514–519. | ||
Singh H, Jain M, Sachan R, Mittal B. Association of TNFA (-308G>A) and IL-10 (-819C>T) promoter polymorphisms with risk of cervical cancer. Int J Gynecol Cancer. 2009;19(7):1190–1194. | ||
Stanczuk GA, Sibanda EN, Perrey C, et al. Cancer of the uterine cervix may be significantly associated with a gene polymorphism coding for increased IL-10 production. Int J Cancer. 2001;94(6):792–794. | ||
Tsai CW, Chang WS, Lin KC, et al. Significant association of Interleukin-10 genotypes and oral cancer susceptibility in Taiwan. Anticancer Res. 2014;34(7):3731–3737. | ||
Tsai CW, Tsai MH, Shih LC, Chang WS, Lin CC, Bau DT. Association of interleukin-10 (IL10) promoter genotypes with nasopharyngeal carcinoma risk in Taiwan. Anticancer Res. 2013;33(8):3391–3396. | ||
Wang Q, Zhang C, Walayat S, Chen HW, Wang Y. Association between cytokine gene polymorphisms and cervical cancer in a Chinese population. Eur J Obstet Gynecol Reprod Biol. 2011;158(2):330–333. | ||
Wei YS, Kuang XH, Zhu YH, et al. Interleukin-10 gene promoter polymorphisms and the risk of nasopharyngeal carcinoma. Tissue Antigens. 2007;70(1):12–17. | ||
Zoodsma M, Nolte IM, Schipper M, et al. Interleukin-10 and Fas polymorphisms and susceptibility for (pre)neoplastic cervical disease. Int J Gynecol Cancer. 2005;15(Suppl 3):282–290. | ||
Hohaus S, Giachelia M, Massini G, et al. Clinical significance of interleukin-10 gene polymorphisms and plasma levels in Hodgkin lymphoma. Leuk Res. 2009;33(10):1352–1356. | ||
Jin L, Sturgis EM, Cao X, et al. Interleukin-10 promoter variants predict HPV-positive tumors and survival of squamous cell carcinoma of the oropharynx. FASEB J. 2013;27(6):2496–2503. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.








