Back to Journals » Cancer Management and Research » Volume 12
CircRNA_102272 Promotes Cisplatin-Resistance in Hepatocellular Carcinoma by Decreasing MiR-326 Targeting of RUNX2
Authors Guan Y, Zhang Y, Hao L, Nie Z
Received 7 May 2020
Accepted for publication 30 July 2020
Published 7 December 2020 Volume 2020:12 Pages 12527—12534
DOI https://doi.org/10.2147/CMAR.S258230
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Yonghai Guan,1,* Ying Zhang,2,* Lina Hao,3 Zhenwang Nie1
1Department of Infectious Diseases, The Second Hospital of Dalian Medical University, Dalian 116023, Liaoning, People’s Republic of China; 2Sixth Department of Liver Diseases, The Sixth People’s Hospital of Dalian, Dalian Medical University, Dalian 116031, Liaoning, People’s Republic of China; 3Department of Nephrology, The Third People’s Hospital of Dalian, Dalian 116000, Liaoning, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhenwang Nie
Department of Infectious Diseases, The Second Hospital of Dalian Medical University, No. 467 Zhongshan Road, Shahekou District, Dalian 116023, Liaoning, People’s Republic of China
Tel +86 15541117868
Email [email protected]
Background: Hepatocellular carcinoma (HCC) is the leading cause of tumor-associated death in males and females worldwide. HCC is mostly diagnosed at advanced stages and the chemotherapeutic cisplatin is one of the major therapeutic options in the treatment of patients with treating advanced HCC. Despite several reports on HCC multidrug resistance, the underlying regulatory mechanisms are still unclear.
Methods: RT-PCR was performed to detect circRNA_102272, miR-326 and RUNX2 expression. The CCK8 assay was used to examine cell proliferation and cisplatin IC50 values. The luciferase reporter assay was performed to verify complementary combinations between circRNA_102272 and miR-326 and between miR-326 and RUNX2.
Results: CircRNA_102272 expression was upregulated in HCC tissues and cells. CircRNA_102272 knockdown suppressed HCC cell proliferation and decreased cisplatin-resistance. In addition, circRNA_102272 facilitated HCC cisplatin-resistance by regulating the miR-326/RUNX2 axis.
Conclusion: CircRNA_102272 is significantly increased in HCC tissues and cells and promotes HCC cell proliferation and cisplatin-resistance. More importantly, circRNA acts as a ceRNA to suppress the expression and activity of miR-326, leading to the increase in RUNX2 expression. By elucidating circRNA_102272 role and mechanism of action in HCC, our study provides insights and an opportunity to overcome cisplatin-resistance in HCC.
Keywords: circRNA_102272, cisplatin-resistance, miR-326, RUNX2, HCC
Introduction
Hepatocellular carcinoma (HCC) is the leading cause of tumor-associated death in males and females worldwide.1 HCC is mostly diagnosed at advanced stages of disease, and the chemotherapeutic drug cisplatin is one of the major therapeutic options for the treatment of patients with advanced HCC. Although several studies reported on HCC multidrug resistance, the regulatory mechanisms are still unclear. Thus, it is necessary to further study these mechanisms of resistance.
Circular RNAs (circRNAs) are a new type of non-coding RNAs, characterized by covalently closed-loop structures of endogenous RNAs. Their specificity and stability have been shown to be cell-type dependent.2–4 Recently, several studies pointed out to circRNA association with tumorigenesis and metastasis. For example, circNT5E acts as a sponge of microRNA-422a to promote glioblastoma tumorigenesis.5 CircRNA hsa_circ_0001564 facilitates osteosarcoma tumorigenesis via sponging miR-29c-3p.6 CircRNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through the miR-4641/PCK1 pathway7 and CircRNA hsa_circ_0000064 promotes lung cancer cell proliferation and metastasis.8 Although some studies indicated that circRNAs play a significant role in multidrug resistance, the regulatory mechanisms are still unclear.
Mechanistically, circRNA functions mainly include miRNA binding, proteins interactions and splicing machine regulation.9 Notably, circRNAs affect tumors by sponging miRNA. For example, circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer.10 CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630.11 Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via the miR-1275/FOXK1 axis.12 In addition, miRNA is playing significant regulatory roles in cell growth, proliferation, differentiation and cell death. For example, anti-miR-203 suppresses breast cancer growth and stemness by targeting SOCS3.13 MiR-331-3p inhibits the Hepatocellular Carcinoma (HCC) bel-7402 cell line by downregulation of E2F1.14 Thus, circRNAs regulate the expression of target genes via a circRNA-miRNA-mRNA function network.
Chemotherapy drugs have always been one of the strategies for cancer treatment. For example, bitter melon extract inhibits breast cancer growth in preclinical model by inducing autophagic cell death.15 Bitter melon enhances natural killer-mediated toxicity against head and neck cancer cells.16 Methyl-β-cyclodextrin enhance the efficacy of doxorubicin in solid tumor cells.17 Thus, it is very meaningful to study the multidrug resistance of chemotherapy.
Although there are several studies on multidrug resistance,18,19 the relationship between circRNA and HCC multidrug resistance is still unclear. Thus, in this study, we investigated the relationship between circRNA_102,272 and cisplatin-resistance. Moreover, we report for the first time that circRNA_102272 is upregulated in HCC tissues and cells. In addition, we also found that circRNA_102272 promotes cisplatin-resistance in HCC through the miR-326/RUNX2 axis.
Materials and Methods
HCC Tissues Collection
Human HCC and adjacent normal tissues were obtained from the Second Hospital of Dalian Medical University. This study was approved by the Human Research Ethics Committee of the Second Hospital of Dalian Medical University. Signed written informed consents were obtained from all patients.
Cell Culture
The human normal liver cell line 7701 and the HCC cell lines HepG2 and Huh7 were obtained from the Cell Bank of Type Culture Collection (Shanghai City, China). All cell lines were cultured in DMEM with 10% fetal calf serum in a 37°C incubator with 5% CO2 and 100% humidity. STR profiling of cells was identified by the Cell Bank of Type Culture Collection (Shanghai City, China), and the mycoplasmas were tested in culture once a month.
Cells (2 x 105 cells/well) were seeded onto 6-well dishes at 24 h and then transfected with control or experimental siRNA for circRNA_102272 or si-RUNX2 per dish at 80% confluence using the Lipofectamine 3000 (Life Technologies, Shanghai, China) according to the manufacturer’s instructions. The primers of si-circRNA_102272 were 5ʹ-CCGTCGAGCATCATTGGTT-3ʹ; the primers of si-RUNX2 were 5ʹ-GGACTGTGGTTACTGTCAT-3ʹ.
CCK8 Assay
Cell proliferation and cisplatin IC50 were determined using the Cell Counting Kit-8 (CCK-8, Roche, Basel, Switzerland) and following the manufacturer’s instructions. The absorbance was detected spectrophotometrically at 450 nm.
RT-PCR Analysis
Total RNA was collected using the mirVana™ kit (Ambion, TX), according to the manufacturer’s recommendations. After purification, the RNA was transcribed into cDNA using a Reverse Transcription Kit (Takara, Tokyo, Japan). RT-PCR analysis was performed using the SYBR Green (Takara). The primers used for RT-PCR were: circRNA_102272 primers were 5ʹ-GAGTGAGGAGAGATCCCCCTAAC-3ʹ (sense) and 5ʹ-GGCATCATGACGTCATCCGAATC-3ʹ (anti-sense); RUNX2 primers were: 5ʹ-CCGCCTCAGTGATTTAGGGC-3ʹ (sense) and 5ʹ-GGGTCTGTAATCTGACTCTGTCC-3ʹ (anti-sense); GAPDH primers were 5ʹ-ACAACTTTGGTATCGTGGAAGG-3ʹ (sense) and 5ʹ-GCCATCACGCCACAGTTTC-3ʹ (anti-sense).
Mice Xenografts
The in vivo experiments were approved by the Human Research Ethics Committee of the Second Hospital of Dalian Medical University and followed the guidelines on the welfare and use of animals (Laboratory animal—Guideline for ethical review of animal welfare, GB/T 35,892–2018, China) that was issued by China National Standardization Management Committee. BALB/c nude mice were purchased from the Animal Experimental Center, Shanghai University of Traditional Chinese Medicine, and randomly divided into two groups (n=5). The Huh cells (5 x 106) with sh-circRNA_102272 or sh-NC were injected into the right flanks of nude mice and tumors’ volume was measured every 5 days.
Luciferase Reporter Assay
For the luciferase assay, cells (50,000 cells/well) were co-transfected with luciferase reporter plasmids containing wild-type or mutated 3ʹUTR and miR-326 mimics using Lipofectamine 2000 (Invitrogen) and according to manufacturer’s recommendations. The luciferase activity was detected by the Dual-Luciferase Assay kit (Promega, UK) and according to the manufacturer’s protocol.
Statistical Analysis
Each experiment was performed at least 3 times. All values were presented as mean ± SD. Student’s t-test and one-way analysis of variance (ANOVA) were performed to compare differences between the two groups. P < 0.05 was considered statistically significant.
Results
CircRNA_102272 is Upregulated in HCC Tissues and Cells
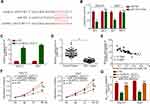
The microarray data in the GEO dataset (GSE97332) were used to initially detect circRNAs expression, and normalized microarray data were measured using GEO2R after applying log2 transformation. The results showed that circRNA_10227 expression was increased in HCC samples compared to normal samples (Figure 1A). Subsequently, circRNA_102272 expression in 60 pairs of HCC tissues was investigated. The result showed that circRNA_102272 expression was significantly upregulated in HCC tissues compared to normal tissues (Figure 1B), and Kaplan–Meier analysis showed that HCC patients with high circRNA_102272 expression had lower survival rates (Figure 1C). In addition, RT-PCR indicated that circRNA_1022272 expression was significantly increased in Hep G2/DDP and Huh 7/DDP cells compared with Hep G2 and Huh 7 cells (Figure 1D). Thus, these results revealed that circRNA_102272 overexpression may be involved in HCC tumorigenesis and multidrug resistance.
CircRNA_102272 Knockdown Suppresses HCC Cell Proliferation and Cisplatin-Resistance
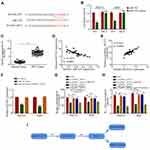
Since circRNA-102272 is upregulated in HCC, RNA interference was used to downregulate circRNA_102272 expression and to assess its biological functions in HCC. RT-PCR analysis showed that circRNA_102272 expression in si-circRNA_102272-transfected HCC cells was decreased (Figure 2A). CCK8 and colony formation assays showed that circRNA_102272 knockdown significantly suppressed cell proliferation of Hep G2 and Huh7 cells (Figure 2B and C). In addition, we further studied the relationship between circRNA_102272 expression and cisplatin-resistance. The result showed that circRNA_102272 knockdown decreases cisplatin IC50 values in Hep G2 and Huh7 cells (Figure 2D). Meanwhile, in vivo experiments indicated that tumors’ volume and weight in the circRNA_102272 knockdown group were significantly suppressed and that circRNA_102272 knockdown, combined with an intraperitoneal injection of cisplatin into the mice, further inhibited tumors’ volume and weight (Figure 2E and F). Thus, these data demonstrate that circRNA_102272 acts as an oncogene in HCC.
CircRNA_102272 Promotes Cisplatin-Resistance Through MiR-326 Sponging
To explore the regulatory mechanisms of circRNA_102272 in cisplatin-resistance, the online software miRBase and circinteractome were used to predict potential miRNAs involvements in these mechanisms. The result showed that circRNA_102272 may bind to miR-326 (Figure 3A), and the luciferase reporter assay confirmed their complementary combinations (Figure 3B). The result also suggested that miR-326 binds and suppresses circRNA_102272 activity. The downregulation of circRNA_102272 in Hep G2 and Huh7 cells also increased miR-326 expression (Figure 3C). In addition, the expression of miR-326 in HCC tissues was decreased compared to normal tissues (Figure 3D), and the Pearson correlation analysis revealed that circRNA_102272 expression inversely correlates with miR-326 expression (Figure 3E). Subsequently, cell proliferation and cisplatin IC50 values in the Hep G2/si-circRNA_102272 and Huh7/si-circRNA_102272 cells, transfected with miR-326 inhibitor, were significantly rescued (Figure 3F and G). Thus, circRNA_102272 promotes cisplatin-resistance through miR-326 sponging.
CircRNA_102272 Facilitates HCC Cisplatin-Resistance by Regulating the MiR-326/RUNX2 Axis
To explore the potential existence of a circRNA-miRNA-mRNA axis in cisplatin-resistance, the online software TargetScan, miRanda and PicTar were used to predict potentially involved mRNAs. The result showed that miR-326 may bind to the 3ʹ-UTR region of RUNX2 (Figure 4A), and the luciferase reporter assay confirmed their complementary combinations (Figure 4B) and revealed that miR-326 binds and suppresses RUNX2 activity. In addition, the expression of RUNX2 in HCC tissues was increased compared to normal tissues (Figure 4C). Notably, Pearson correlation analysis showed that miR-326 expression inversely correlates with RUNX2 expression (Figure 4D), and that circRNA_102272 expression positively correlates with RUNX2 expression (Figure 4E). It also revealed that circRNA_102272 may regulate RUNX2 expression via miR-326. Moreover, miR-326 upregulation in Hep G2 and Huh7 cells significantly suppressed RUNX2 expression (Figure 4F), suggesting that miR-326 suppresses RUNX2 expression. Furthermore, miR-326 overexpression rescued RUNX2 expression in transfected miR-326 mimics Hep G2 and Huh7 cells. Subsequently, the CCK8 assay revealed that RUNX2 knockdown of rescues the inhibitory effects on cell proliferation and cisplatin IC50 values in Hep G2 and Huh 7 cells (Figure 4G and H), suggesting that circRNA_102272 facilitates HCC cisplatin-resistance by regulating the miR-326/RUNX2 axis (Figure 4I).
Discussion
Circular RNAs (circRNAs) are a new type of non-coding RNAs, characterized by covalently closed-loop structures of endogenous RNAs. CircRNAs specificity and stability are cell-type dependent.2–4 Several studies indicated that circRNAs play significant roles in tumor progression, metastasis and multidrug resistance. For example, the circRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulating the miR-1324/FZD5/Wnt/β-catenin axis.20 Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6.21 CircFADS2 regulates lung cancer cells proliferation and invasion via miR-498 sponging.22 Thus, circRNAs are biomarkers for multiple types of tumors.23–25 In addition, several miRNAs and lncRNAs have been reported to regulate HCC development.26,27 However, there are currently few studies that reported on the function of circRNAs in HCC, especially in terms of multidrug resistance.
In this study, we first found that circRNA_102272 was significantly upregulated in HCC tissues and cells, especially in Hep G2/DDP and Huh7/DDP cells. We also found by Kaplan–Meier analysis that HCC patients with high circRNA_102272 expression have lower survival rates. Furthermore, we studied the function of circRNA_102272 in HCC progression and the in vitro results indicated that circRNA_102272 promotes HCC cell proliferation and reduces HCC’s cisplatin-resistance. Notably, circRNA_102272 knockdown inhibited HCC cell proliferation and enhanced HCC cells’ sensitivity to cisplatin in vivo. These data revealed that circRNA_102272 acts as an oncogene in HCC.
Accumulating evidence indicated that circRNA affects tumorigenesis by sponging miRNAs. The online software miRBase and circinteractome were used to predict the potential target miRNAs. The results showed that circRNA_102272 may bind to miR-326, which was confirmed using the luciferase reporter assay. Previous studies revealed that miR-326 acts as a tumor suppressor in human tumors. For example, miR-326 reverses chemoresistance in human lung adenocarcinoma cells by specifically targeting protein 1.28 MiR-326 regulates endometrial cancer EMT and metastasis through TWIST1 targeting.29 In this study, the miR-326 expression in HCC tissues was decreased compared to normal tissues, and Pearson correlation analysis revealed that circRNA_102272 expression inversely correlates with miR-326. Thus, CircRNA_102272 modulates HCC cisplatin-resistance by sponging miR-326.
RUNX2, a member of runt-related transcription factors (RUNX), has been reported to be involved in tumor occurrence, development, and metastasis in several types of cancers, such as breast,30 lung,31 prostate32 and thyroid cancer.33 However, the roles of RUNX2 in HCC cisplatin-resistance remain unclear. In this study, RUNX2 was identified as a direct target of miR-326 and its expression was increased in HCC. Moreover, rescue experiments further indicated the antagonistic roles of si-circRNA_102272 and si-RUNX2 in the modulation of HCC cells’ cisplatin-resistance. Therefore, circRNA_102272 facilitates HCC cisplatin-resistance by regulating the miR-326/RUNX2 axis.
Taken together, our results demonstrated that circRNA_102272 is significantly increased in HCC tissues and cells, and that it promotes HCC cell proliferation and cisplatin-resistance. More importantly, circRNA_102272 acted as a ceRNA that suppresses miR-326 expression and activity, leading to RUNX2 increased expression. By elucidating circRNA_102272 role and mechanism of action in HCC, our study provides insights and an opportunity to overcome multidrug resistance in HCC.
Acknowledgments
This work was supported by Liaoning Province Natural Science Foundation Project (20180551027).
Disclosure
The authors declare no conflicts of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
3. Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi:10.1038/nsmb.2959
4. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO, Preiss T. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi:10.1371/journal.pone.0030733
5. Wang R, Zhang S, Chen X, et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 2018;78(17):4812–4825. doi:10.1158/0008-5472.CAN-18-0532
6. Li J-F, Song Y-Z. Circular RNA hsa_circ_0001564 facilitates tumorigenesis of osteosarcoma via sponging miR-29c-3p. Tumor Biol. 2017;39(8):101042831770998. doi:10.1177/1010428317709989
7. Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499(4):1044–1049. doi:10.1016/j.bbrc.2018.03.221
8. Luo YH, Zhu XZ, Huang KW, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898. doi:10.1016/j.biopha.2017.12.015
9. Huang S, Yang B, Chen BJ, et al. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017;109(5–6):401–407. doi:10.1016/j.ygeno.2017.06.005
10. Sang Y, Chen B, Song X, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther. 2019;27(9):1638–1652. doi:10.1016/j.ymthe.2019.05.011
11. Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging. 2017;9(6):1585–1594. doi:10.18632/aging.101254
12. Ma X, Yang X, Bao W, et al. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem Biophys Res Commun. 2018;498(4):1009–1015. doi:10.1016/j.bbrc.2018.03.105
13. Muhammad N, Bhattacharya S, Steele R, Ray RB. Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget. 2016;7(36):58595–58605. doi:10.18632/oncotarget.11193
14. Jin W, Zhong N, Wang L, Yu J, Yin F, Zhang K. MiR-331-3p inhibition of the hepatocellular carcinoma (HCC) Bel-7402 cell line by down-regulation of E2F1. J Nanosci Nanotechnol. 2019;19(9):5476–5482. doi:10.1166/jnn.2019.16535
15. Muhammad N, Steele R, Isbell TS, Philips N, Ray RB. Bitter melon extract inhibits breast cancer growth in preclinical model by inducing autophagic cell death. Oncotarget. 2017;8(39):66226–66236. doi:10.18632/oncotarget.19887
16. Bhattacharya S, Muhammad N, Steele R, Kornbluth J, Ray RB. Bitter melon enhances natural killer-mediated toxicity against head and neck cancer cells. Cancer Prev Res (Phila). 2017;10(6):337–344. doi:10.1158/1940-6207.CAPR-17-0046
17. Mohammad N, Singh SV, Malvi P, et al. Strategy to enhance efficacy of doxorubicin in solid tumor cells by methyl-beta-cyclodextrin: involvement of p53 and Fas receptor ligand complex. Sci Rep. 2015;5:11853. doi:10.1038/srep11853
18. Xu N, Zhang J, Shen C, et al. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Commun. 2012;423(4):826–831. doi:10.1016/j.bbrc.2012.06.048
19. Song IS, Jun SY, Na HJ, et al. Inhibition of MKK7-JNK by the TOR signaling pathway regulator-like protein contributes to resistance of HCC cells to TRAIL-induced apoptosis. Gastroenterology. 2012;143(5):1341–1351. doi:10.1053/j.gastro.2012.07.103
20. Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun. 2018;497(2):626–632. doi:10.1016/j.bbrc.2018.02.119
21. Gong Y, Mao J, Wu D, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 2018;18:116. doi:10.1186/s12935-018-0602-3
22. Zhao F, Han Y, Liu Z, Zhao Z, Li Z, Jia K. circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci Rep. 2018;38(4). doi:10.1042/BSR20180570
23. Sun H, Tang W, Rong D, et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomarkers. 2018;21(2):299–306. doi:10.3233/CBM-170379
24. Yin WB, Yan MG, Fang X, Guo JJ, Xiong W, Zhang RP. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363–368. doi:10.1016/j.cca.2017.10.011
25. Xuan L, Qu L, Zhou H, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8(2):932–939.
26. Dai W, Wang C, Wang F, et al. Anti-miR-197 inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem Biophys Res Commun. 2014;446(2):541–548. doi:10.1016/j.bbrc.2014.03.006
27. Xiong D, Sheng Y, Ding S, et al. LINC00052 regulates the expression of NTRK3 by miR-128 and miR-485-3p to strengthen HCC cells invasion and migration. Oncotarget. 2016;7(30):47593–47608. doi:10.18632/oncotarget.10250
28. Li J, Li S, Chen Z, et al. miR-326 reverses chemoresistance in human lung adenocarcinoma cells by targeting specificity protein 1. Tumor Biol. 2016;37(10):13287–13294. doi:10.1007/s13277-016-5244-2
29. Liu W, Zhang B, Xu N, Wang MJ, Liu Q. miR-326 regulates EMT and metastasis of endometrial cancer through targeting TWIST1. Eur Rev Med Pharmacol Sci. 2017;21(17):3787–3793.
30. Onodera Y, Miki Y, Suzuki T, et al. Runx2 in human breast carcinoma: its potential roles in cancer progression. Cancer Sci. 2010;101(12):2670–2675. doi:10.1111/j.1349-7006.2010.01742.x
31. Li H, Zhou RJ, Zhang GQ, Xu JP. Clinical significance of RUNX2 expression in patients with nonsmall cell lung cancer: a 5-year follow-up study. Tumor Biol. 2013;34(3):1807–1812. doi:10.1007/s13277-013-0720-4
32. Brubaker KD, Vessella RL, Brown LG, Corey E. Prostate cancer expression of runt-domain transcription factor Runx2, a key regulator of osteoblast differentiation and function. Prostate. 2003;56(1):13–22. doi:10.1002/pros.10233
33. Niu DF, Kondo T, Nakazawa T, et al. Transcription factor Runx2 is a regulator of epithelial-mesenchymal transition and invasion in thyroid carcinomas. Lab Invest. 2012;92(8):1181–1190. doi:10.1038/labinvest.2012.84
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.