Back to Journals » Cancer Management and Research » Volume 12
Circ_0032821 Facilitates Gastric Cancer Cell Proliferation, Migration, Invasion and Glycolysis by Regulating MiR-1236-3p/HMGB1 Axis
Authors Chen L, Chi K, Xiang H, Yang Y
Received 30 June 2020
Accepted for publication 10 September 2020
Published 12 October 2020 Volume 2020:12 Pages 9965—9976
DOI https://doi.org/10.2147/CMAR.S270164
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Lei Chen,1 Kun Chi,2 Huaguo Xiang,3 Yan Yang4
1Department of Emergency Medicine, Xiang’an Hospital of Xiamen University, Xiamen, 361101, People’s Republic of China; 2Department of Nursing, Qingdao Municipal Hospital (Group), Qingdao 266071, People’s Republic of China; 3Medical Laboratory, Shenzhen Baoan District Fuyong People’s Hospital, Shenzhen 518103, People’s Republic of China; 4Department of Gastroenterology, Central People’s Hospital of Tengzhou, Tengzhou 277500, People’s Republic of China
Correspondence: Yan Yang
Department of Gastroenterology, Central People’s Hospital of Tengzhou, Tengzhou, Shandong 277500, People’s Republic of China
Email [email protected]
Background: Circular RNAs (circRNAs) play an essential role in the pathogenesis of malignant tumors, including gastric cancer (GC). However, the effect of circ_0032821 on GC remains largely unknown.
Methods: QRT-PCR assay was employed to examine the levels of circ_0032821, CEP128 mRNA and miR-1236-3p. RNase R digestion assay was utilized to verify the feature of circ_0032821. Cell Counting Kit-8 (CCK-8) assay and transwell assay were adopted to evaluate cell proliferation and metastasis. The level of glycolysis was evaluated through detecting ECAR, OCR, lactate production, glucose uptake and ATP synthesis. Dual-luciferase reporter assay and RIP assay were conducted to analyze the relationship between miR-1236-3p and circ_0032821 or HMGB1. Western blot assay was adopted for high mobility group box 1 (HMGB1) level. Murine xenograft model assay was utilized for the effect of circ_0032821 in vivo.
Results: High level of circ_0032821 was observed in GC tissues and cells. Silencing of circ_0032821 markedly repressed cell proliferation, metastasis and glycolysis in GC cells in vitro and blocked tumorigenesis of GC in vivo. For mechanism analysis, circ_0032821 was identified as the sponge of miR-1236-3p and HMGB1 was the target gene of miR-1236-3p. Moreover, miR-1236-3p suppression restored the influences of circ_0032821 deficiency on GC cell proliferation, metastasis and glycolysis. Overexpression of miR-1236-3p relieved the malignant behaviors of GC cells by targeting HMGB1.
Conclusion: Circ_0032821 accelerated GC development through elevating HMGB1 expression via sponging miR-1236-3p.
Keywords: gastric cancer, circ_0032821, miR-1236-3p, HMGB1
Introduction
Gastric cancer (GC) is one of the most common malignant tumors in the digestive tract, ranking the third cancer-related death worldwide.1 As a global high incidence tumor, the treatment of GC has attracted much attention. In recent years, despite much progression has been made in the diagnosis and therapy methods of GC, it still has high incidence and mortality rates.2,3 It is thus urgent to understand the pathogenesis responsible for GC and develop new treatment methods for this lethal disease.
Circular RNAs (circRNAs) are a newly identified type of non-coding RNAs (ncRNAs) featured by closed-loop structures.4,5 In recent years, the functions of circRNAs have received the researchers’ attention with the progression of bioinformatics and high throughput sequencing technologies.6 Emerging evidence has reported that circRNAs exert essential functions in the carcinogenesis of GC. The abnormal elevation of circ_0009910 was linked to the worse clinical characteristics and facilitated GC cell progression and metastasis.7 Circ-MAT2B accelerated GC cell colony formation, viability and glycolysis via modulation of miR-515-5p/HIF-1α.8 As for circ_0032821, Jiang et al disclosed that circ_0032821 promoted the malignant phenotypes of GC cells by activating MEK1/ERK1/2 signaling pathway.9 Nevertheless, the exact roles and underlying mechanisms of circ_0032821 are barely understood.
MicroRNAs (miRNAs) are small ncRNAs with ~22 nucleotides and predominantly alter gene expression through recognition of 3ʹUTR of target genes.10 The abnormal expression of miRNAs, such as miR-615-3p,11 miR-491-5p12 and miR-181b,13 has been detected and demonstrated to play a crucial role in GC progression. Recently, the tumor-suppressive roles of miR-1236-3p have been reported in human cancers, including GC.14–16 For instance, An et al unraveled that miR-1236-3p restrained the malignant biological behaviors of GC cells via interaction with MTA2.17 High mobility group box 1 (HMGB1) is a nonhistone chromosomal protein and has a vital role in GC development.18,19 For example, HMGB1 contributed to GC cell invasion and growth by modulating NF-κB pathway.20 Li et al claimed that HMGB1 took part in regulating GC cell growth via acting as the target of miR-1179.21 Nonetheless, it is still unclear whether HMGB1 can be targeted by miR-1236-3p to alter GC development.
The purpose of this research was to decipher the expression pattern of circ_0032821 in GC, and then further explore the specific functions and regulatory mechanisms of circ_0032821 in GC carcinogenesis.
Materials and Methods
Specimens Acquisition
Fifty-one pairs of GC tissue specimens and adjacent normal tissue specimens were collected from GC patients at Xiang’an Hospital of Xiamen University. This work was allowed by the Ethics Committee of Xiang’an Hospital of Xiamen University and was conducted in accordance with the Helsinki Declaration. Written informed consents were provided by the enrolled patients. The collected tissues were stored at −80°C prior to use.
Cell Culture
GC cell lines (MKN-45, N87, HGC27 and AGS) and gastric epithelial cell line (GES-1) were bought from Procell (Wuhan, China). The cells were kept in RPMI 1640 medium (Procell) added with 10% FBS (Procell) and 1% penicillin-streptomycin (Procell) at 37°C in a humidified environment consisting of 5% CO2.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The RNA in tissues and cells was extracted utilizing TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) and quantified utilizing a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific). Then M-MLV First Strand Kit (Takara, Dalian, China) or miRNA first strand cDNA Synthesis Kit (GeneCopoeia, Wuhan, China) was employed to reversely transcribe RNA into cDNA. Then qRT-PCR reaction was manipulated utilizing BeyoFast™ SYBR Green qPCR Mix (Beyotime, Shanghai, China) and specific primers (GeneCopoeia) on CFX96 Touch Real-time PCR detection system (Bio-Rad, Hercules, CA, USA). The primers were: circ_0032821: (F: 5ʹ-AGGAATCTGAGTTGCAGTGTCTC-3ʹ and R: 5ʹ-TGATCCTTGAGCTGCAATCTGG-3ʹ); CEP128: (F: 5ʹ-TGATGCCTCTCTGCCTCTTT-3ʹ and R: 5ʹ-GAAAAGCCAACTGCCAGAAG-3ʹ); miR-1236-3p: (F: 5ʹ-CCAATCAGCCTCTTCCCCTT-3ʹ and R: 5ʹ-TATGGTTGTTCACGACTCCTTCAC-3ʹ); HMGB1: (F: 5ʹ-TGCAGATGACAAGCAGCCTT-3ʹ and R: 5ʹ-GCTGCATCAGGCTTTCCTTT-3ʹ); β-actin: (F: 5ʹ-CCCGAGCCGTGTTTCCT-3ʹ and R: 5ʹ-GTCCCAGTTGGTGACGATGC-3ʹ); U6: (F: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and R: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ). The results were computed through the 2-ΔΔCt strategy with β-actin or U6 as an internal reference.
RNase R Digestion Assay
5 μg total RNA isolated from HGC27 and AGS cells was treated with 20 U RNase R (Epicentre, Madison, Wisconsin, USA) for 30 min at 37°C. Thereafter, the abundance of circ_0032821 and CEP128 mRNA was estimated via qRT-PCR analysis.
Cell Transfection
Three circ_0032821 small interfering RNAs (si-circ_0032821#1, si-circ_0032821#2 and si-circ_0032821#3) and scramble siRNA (si-NC), circ_0032821 overexpression vector (circ_0032821) and its control (Vector), miR-1236-3p mimics (miR-1236-3p) and miR-NC, miR-1236-3p inhibitors (in-miR-1236-3p) and in-miR-NC, the overexpression vector of HMGB1 (HMGB1) and its control (pcDNA), short hairpin RNA against circ_0032821 (sh-circ_0032821) and sh-NC were all synthesized by GeneCopoeia. HGC27 and AGS cells were cultured in 6-well plates and transfected with synthetic plasmids or oligonucleotides utilizing Lipofectamine 3000 (Life Technologies, Darmstadt, Germany).
Cell Counting Kit-8 (CCK-8) Assay
CCK-8 assay was executed to determine the proliferation capacity of HGC27 and AGS cells. In short, transfected cells (2×103) were seeded into each well of the 96-well plates and cultivated for 24 h. Then 10 μL CCK-8 (5 mg/mL; Boster, Wuhan, China) was supplemented into the well at 0 h, 24 h, 48 h and 72 h with incubation for an additional 2 h. The absorption was recorded at 450 nm with a microplate reader (Potenov, Beijing, China).
Transwell Assay
The transwell inserts (BD Bioscience, San Jose, CA, USA) precoated with or without 5% Matrigel (BD Biosciences) were adopted to test the invasion and migration of HGC27 and AGS cells, respectively. Briefly, 2×104 transfected cells suspended in serum-free medium were added into the top chambers. 600 μL full culture medium was added into the bottom chambers. After incubation for 24 h, cells that migrated/invaded into the lower membrane surface were fixed in methanol and dyed with 0.1% crystal violet (Sangon, Shanghai, China), and then counted under a light microscope (Olympus, Tokyo, Japan) at the magnification of 100×.
Measurements of Extracellular Acidification Rate (ECAR) and Oxygen Consumption Rate (OCR)
The ECAR and OCR of GC cells were examined with a Seahorse XFe96 Extracellular Flux Analyzer (Seahorse Bioscience, Billerica, MA, USA) according to the instructions of manufacturer. The Seahorse XF Glycolysis Stress Test kit (Seahorse Bioscience) and Seahorse XF Cell Mito Stress Test kit (Seahorse Bioscience) were adopted to measure ECAR and OCR of GC cells. Briefly, 1×105 cells were plated into Seahorse plates, maintained overnight and then washed with Seahorse buffer. Next, Seahorse buffer including glucose (Glc), oligomycin (Oligo) and 2-deoxyglucose (2-DG) were injected into the analyzer in the proper order to measure ECAR. For OCR measurement, Seahorse buffer including oligomycin (Oligo), p-trifluoromethoxy carbonyl cyanide phenylhydrazone (FCCP) and rotenone + antimycin A (Rot + AA) were sequentially injected. The results were analyzed using software XF-96 wave (Seahorse Bioscience).
Detection of Lactate Production, Glucose Uptake and ATP Synthesis
The levels of lactate production, glucose consumption and ATP synthesis in HGC27 and AGS cells were measured using lactate assay kit (Sigma-Aldrich, St. Louis, MO, USA), glucose assay kit (Sigma-Aldrich) and ATP assay kit (Sigma-Aldrich) in line with the manufacturer’s instructions, respectively.
Dual-Luciferase Reporter Assay
The circ_0032821 and HMGB1 3ʹUTR wild-type with potential miR-1236-3p binding sites and mutant without miR-1236-3p binding sites were amplified and cloned into pmirGLO plasmid (Promega, Madison, WI, USA), establishing circ_0032821 WT, circ_0032821 MUT, HMGB1 3ʹUTR WT and HMGB1 3ʹUTR MUT. Next, the indicated luciferase vectors were transfected into HGC27 and AGS cells in combination with miR-1236-3p or miR-NC. After 48 h, the measurement of luciferase activity was done using Dual-Luciferase Reporter Assay Kit (Promega).
RNA Immunoprecipitation (RIP) Assay
RIP assay was implemented with Imprint® RNA Immunoprecipitation Kit (Sigma-Aldrich). In brief, HGC27 and AGS cells transfected with miR-1236-3p or miR-NC were lysed in RIP lysis buffer and then cultivated with magnetic beads conjugated with Ago2 and IgG. Next, the samples were incubated with proteinase K (Solarbio, Beijing, China) for 30 min. Afterward, the immunoprecipitated RNA was extracted and subjected to qRT-PCR analysis for circ_0032821 enrichment.
Western Blot Assay
The isolation of total protein was executed utilizing RIPA buffer (CWBio, Beijing, China) and the concentrations of proteins were measured with a BCA Protein Quantification Kit (Vazyme, Nanjing, China). Then equal amounts of protein were separated through sodium dodecyl sulfonate-polyacrylamide gel (Solarbio) electrophoresis followed by transferring into polyvinylidene difluoride membranes (Amersham Biosciences, Chicago, IL, USA). After blocking in 5% non-fat milk for 1 h at indoor temperature, the membranes were incubated overnight with primary antibodies: β-actin (bs-0061R; Bioss, Beijing, China) and HMGB1 (bs-0664R; Bioss) at 4°C and probed with horseradish peroxidase-conjugated secondary antibody (bs-0295M-HRP; Bioss) for 1 h at indoor temperature. The signals were examined using chemiluminescent substrate kit (Millipore, Bedford, MA, USA).
Murine Xenograft Model
AGS cells stably transfected with sh-circ_0032821 or sh-NC were suspended in 100 μL PBS (Solarbio) and then inoculated subcutaneously into the BALB/c nude mice (4–6 weeks old; Vital River Laboratory, Beijing, China). The volume of xenograft neoplasms was examined every 7 days and estimated via the equation: (length×width2)/2. On day 28, the mice were sacrificed and the xenograft neoplasms were harvested, weighed and then saved at −80°C for the subsequent experiments. The animal studies were permitted by the Ethics Committee of Animal Research of Xiang’an Hospital of Xiamen University and carried out following the guidelines of the national animal protection and ethics institute.
Statistical Analysis
The experiments were manipulated in triplicate. Data analysis was finished using GraphPad Prism 7 and the results were exhibited as mean ± SD. The differences were analyzed by Student’s t-test or one-way analysis of variance (ANOVA). The correlations among circ_0032821, miR-1236-3p and HMGB1 were estimated through Pearson correlation coefficient analysis. P value less than 0.05 denoted statistical significance.
Results
Circ_0032821 Was Upregulated in GC Tissues and Cells
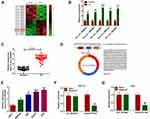
By analyzing public GEO dataset GSE78092, we found that 10 circRNAs were abnormally upregulated in GC samples compared to normal samples (Figure 1A). Next, we selected 5 circRNAs (hsa_circ_0005533, hsa_circ_0004099, hsa_circ_0032821, hsa_circ_0000144 and hsa_circ_0023642) and verified their levels in GC tissues and normal tissues by qRT-PCR assay. The results showed that they were increased in GC tissues compared to normal tissues (Figure 1B). We chose hsa_circ_0032821 as our research target because it had a more significant difference between GC and normal tissues than the other 4 circRNAs. Moreover, circ_0032821 was conspicuously elevated in the clinical GC tissues in comparison with adjacent non-tumor tissues (Figure 1C). As shown in Figure 1D, circ_0032821 originated from the exons (16–18) of CEP128 gene and its mature length after splicing was 430 nts. Furthermore, our results exhibited that circ_0032821 was evidently increased in GC cell lines (MKN-45, N87, HGC27 and AGS) compared to GES-1 cell line (Figure 1E). In addition, we determined the feature of circ_0032821 in HGC-27 and AGS cells using RNase R assay, showing that circ_0032821 was resistant to RNase R digestion, while CEP128 was digested by RNase R (Figure 1F and G). Taken together, the dysregulation of circ_0032821 might be linked to the progression of GC.
Silencing of Circ_0032821 Inhibited GC Cell Proliferation, Migration, Invasion and Glycolysis
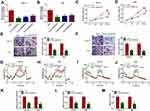
To explore the roles of circ_0032821 in GC development, si-circ_0032821#1, si-circ_0032821#2 or si-circ_0032821#3 was transfected into HGC27 and AGS cells to knockdown the expression of circ_0032821. As exhibited in Figure 2A and B, si-circ_0032821#1, si-circ_0032821#2 or si-circ_0032821#3 transfection remarkably reduced the level of circ_0032821 in HGC27 and AGS cells, and the knockdown efficiency of si-circ_0032821#1 was more significant than si-circ_0032821#2 and si-circ_0032821#3; thus, si-circ_0032821#1 was selected to conduct further functional experiments. CCK-8 assay showed that circ_0032821 knockdown drastically suppressed the proliferation ability of HGC27 and AGS cells (Figure 2C and D). The results of transwell assay indicated the migration and invasion capacities of HGC27 and AGS cells were strikingly repressed by circ_0032821 knockdown in comparison with control groups (Figure 2E and F). Subsequently, we investigated the effect of circ_0032821 on glycolysis in HGC27 and AGS cells. For ECAR analysis, circ_0032821 knockdown distinctly reduced the maximal ECAR in both cell types compared to control groups (Figure 2G and H). For OCR analysis, circ_0032821 deficiency evidently elevated the basal and maximum OCR relative to control groups (Figure 2I and J). Additionally, we determined the levels of lactate production, glucose uptake and ATP synthesis using relevant kits. The results showed that circ_0032821 interference markedly decreased the levels of lactate production, glucose uptake and ATP synthesis in HGC27 and AGS cells in comparison with control groups (Figure 2K–M). Collectively, circ_0032821 knockdown restrained the proliferation, metastasis and glycolysis in GC cells.
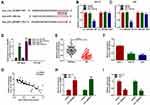
Circ_0032821 Functioned as the Sponge of MiR-1236-3p in GC Cells
To clarify the underlying mechanism of circ_0032821 in GC progression, online tool circinteractome was utilized to analyze the potential targets of circ_0032821. As presented in Figure 3A, miR-1236-3p was found to be a target of circ_0032821 and their complementary sequences were shown. To verify this prediction, we firstly conducted dual-luciferase reporter assay. The results showed that miR-1236-3p transfection apparently inhibited the luciferase activity of circ_0032821 WT, but did not affect the luciferase activity of circ_0032821 MUT in both HGC27 and AGS cells (Figure 3B and C). Then the RIP results showed that miR-1236-3p transfection evidently enriched the level of circ_0032821 in Ago2 in both HGC27 and AGS cells compared to miR-NC groups, whereas circ_0032821 was undetectable in IgG all along, further confirming the interaction between circ_0032821 and miR-1236-3p (Figure 3D). Next, our results exhibited that miR-1236-3p was lowly expressed in GC tissues and cells compared to normal tissues and cells (Figure 3E and F). Moreover, our results presented that there was an inverse correlation between the expression of circ_0032821 and miR-1236-3p in GC tissues (Figure 3G). Afterward, we transfected circ_0032821 or Vector into HGC27 and AGS cells to analyze the effect of circ_0032821 on miR-1236-3p expression. The transfection efficiency was evaluated by qRT-PCR assay, showing that circ_0032821 was successfully transfected into HGC27 and AGS cells (Figure 3H). In addition, we observed that circ_0032821 transfection markedly reduced miR-1236-3p level in HGC27 and AGS cells (Figure 3I). To summarize, circ_0032821 negatively modulated miR-1236-3p expression via acting as the sponge of miR-1236-3p.
Inhibition of MiR-1236-3p Abrogated the Malignant Behaviors of GC Cells Mediated by Circ_0032821 Deficiency
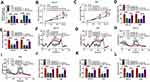
To elucidate whether circ_0032821 could alter GC cell progression by targeting miR-1236-3p, rescue experiments were conducted by administrating si-NC, si-circ_0032821#1, si-circ_0032821#1+in-miR-NC or si-circ_0032821#1+in-miR-1236-3p into HGC27 and AGS cells. As shown in Figure 4A, si-circ_0032821#1 transfection obviously enhanced miR-1236-3p expression in HGC27 and AGS cells, while the administration of in-miR-1236-3p partially restored the impact. As illustrated by CCK-8 experiment and transwell experiment, the suppressive roles of circ_0032821 knockdown in cell proliferation, migration and invasion were ameliorated by reducing miR-1236-3p expression in both HGC27 and AGS cells (Figure 4B–E). Additionally, the association between circ_0032821 and miR-1236-3p in regulating glycolysis level was explored. Our results indicated that circ_0032821 knockdown decreased the maximal ECAR and increased the consumption of oxygen in HGC27 and AGS cells, while miR-1236-3p inhibition effectively abolished the impacts (Figure 4F–I). Additionally, circ_0032821 deficiency reduced the levels of lactate production, glucose consumption and ATP synthesis in HGC27 and AGS cells, while the effects were overturned by reducing miR-1236-3p (Figure 4J–L). These results demonstrated that circ_0032821 knockdown suppressed GC cell malignant phenotypes by binding to miR-1236-3p.
HMGB1 Was a Direct Target Gene of MiR-1236-3p
Through searching online software TargetScan, HMGB1 was found to be the target gene of miR-1236-3p and their potential binding sites are exhibited in Figure 5A. To verify this prediction, dual-luciferase reporter assay was carried out. The results showed that miR-1236-3p transfection resulted in a remarkable suppression in the luciferase activity of HMGB1 3ʹUTR WT in both HGC27 and AGS cells compared to miR-NC groups, while the luciferase activity of HMGB1 3ʹUTR MUT was not changed by miR-1236-3p transfection, further confirming the combination between HMGB1 and miR-1236-3p (Figure 5B and C). As expected, the protein level of HMGB1 was evidently enhanced in GC tissues and cells relative to normal tissues and cells (Figure 5D and E). Furthermore, our results showed that HMGB1 mRNA level was negatively correlated with miR-1236-3p level and positively correlated with circ_0032821 in GC tissues (Figure 5F and G). We also demonstrated that miR-1236-3p transfection led to a distinct downregulation in HMGB1 protein level in HGC27 and AGS cells in comparison with miR-NC groups (Figure 5H). In addition, circ_0032821 silencing drastically decreased the protein level of HMGB1 in HGC27 and AGS cells, while the effect was weakened by the inhibition of miR-1236-3p (Figure 5I). All these findings suggested that circ_0032821 positively regulated HMGB1 expression via targeting miR-1236-3p in GC cells.
MiR-1236-3p Overexpression Relieved the Malignant Phenotypes of GC Cells by Targeting HMGB1
To further investigate the effects of miR-1236-3p and HMGB1 on GC cell progression, HGC27 and AGS cells were assigned to 4 groups: miR-NC, miR-1236-3p, miR-1236-3p+pcDNA and miR-1236-3p+HMGB1. As we observed in Figure 6A, the downregulation of HMGB1 protein level caused by miR-1236-3p transfection in HGC27 and AGS cells were overturned by the transfection of HMGB1. The results of CCK-8 assay and transwell assay indicated that miR-1236-3p overexpression conspicuously restrained the proliferation, migration and invasion capacities of HGC27 and AGS cells, while the effects were relieved by elevating HMGB1 expression (Figure 6B–E). In addition, we found that the maximal ECAR was reduced and the consumption of oxygen was increased in HGC27 and AGS cells overexpressing miR-1236-3p, while HMGB1 upregulation effectively reversed the impacts (Figure 6F–I). Furthermore, the elevation of miR-1236-3p markedly reduced the levels of lactate production, glucose consumption and ATP generation in HGC27 and AGS cells, but HMGB1 overexpression partially overturned the effects (Figure 6J–L). Taken together, miR-1236-3p repressed GC cell proliferation, migration, invasion and glycolysis by targeting HMGB1.
Circ_0032821 Knockdown Blocked Tumor Growth of GC in vivo
At last, the function of circ_0032821 in the tumorigenesis of GC was explored through in vivo murine xenograft models. Circ_0032821-silenced AGS cells were administrated into the nude mice. Tumor volume was monitored every 7 days and tumors were harvested and weighed after 28 days. As a result, reduced tumor volume and weight were observed in mice inoculated with circ_0032821-silenced AGS cells compared to control groups (Figure 7A and B). Moreover, we found that circ_0032821 and HMGB1 protein levels were lower and miR-1236-3p level was higher in the xenograft tumor tissues from sh-circ_0032821 groups than sh-NC control groups (Figure 7C–E). These results suggested that circ_0032821 knockdown hampered tumor growth of GC in vivo.
Discussion
Accumulating studies have pointed out that circRNAs play vital roles in tumorigenesis, including GC. However, the effects of circ_0032821 on GC have not been well defined yet. Herein, we focused on the functional roles and underlying mechanisms of circ_0032821 in GC. As a result, we demonstrated that circ_0032821 overexpression facilitated cell proliferation, migration, invasion and glycolysis in GC cells. Moreover, the regulatory axis circ_0032821/miR-1236-3p/HMGB1 in GC was discovered.
In the previous reports, circRNAs have been claimed to participate in regulating GC development via the competitive endogenous RNAs (ceRNAs) regulatory mechanism of circRNA/miRNA/mRNA axis.22 For instance, circ-SERPINE2 accelerated GC cell growth and restrained apoptosis via modulation of miR-375/YWHAZ axis.23 Circ_0092306 repressed cell apoptosis and promoted viability and motility in GC cells by decreasing miR-197-3p and increasing PRKCB.24 As a member of circRNAs, Jiang et al declared that circ_0032821 was highly expressed in GC and its silencing led to an obvious suppression in cell proliferation and motility and a distinct promotion in autophagy in GC cells.9 In line with the results of Jiang et al, we also observed that circ_0032821 level was raised in GC tissues and cell lines. Circ_0032821 interference predominantly impeded the proliferation and metastasis abilities of GC cells. It has been widely accepted that tumors cells can overcome metabolic stress and promote energy replenishment to meet the rapid growth of tumor cells.25,26 Thus, we therefore investigated whether circ_0032821 could alter the level of glycolysis in GC cells. Our results showed that circ_0032821 knockdown decelerated the glycolysis level in GC cells, as demonstrated by decreased ECAR, lactate production, glucose consumption and ATP synthesis and increased OCR. Moreover, we found that circ_0032821 deficiency blocked the tumorigenicity of GC in vivo. All these results illustrated the oncogenic role of circ_0032821 in GC.
For mechanism analysis, circ_0032821 was demonstrated to enhance HMGB1 expression via functioning as the sponge of miR-1236-3p. In this paper, miR-1236-3p level was found to be reduced in GC tissues and cells. Moreover, circ_0032821 knockdown-mediated malignant GC cell phenotypes were effectively recovered by reducing miR-1236-3p, indicating that circ_0032821 facilitated GC progression via targeting miR-1236-3p. Previous studies indicated that miR-1236-3p level was declined in GC tissues and low expression of miR-1236-3p was related to worse clinical characteristics and poor prognosis of GC patients.16,27 Moreover, miR-1236-3p could interact with MTA2 to repress GC cell proliferation and metastasis.17 Consistently, our results exhibited that miR-1236-3p upregulation drastically hampered GC cell growth, motility and glycolysis. Additionally, HMGB1 was demonstrated to be a target gene of miR-1236-3p for the first time. In GC, HMGB1 has been verified to act as the target of miR-1179,21 miR-129-5p28 and miR-50529 to modulate GC progression. In addition to a nuclear function as a DNA chaperone and cell death regulator, HMGB1 can be released into the extracellular environment to act as the prototypic damage-associated molecular pattern molecule.19 Moreover, a previous study showed that HMGB1 could regulate YAP-mediated HIF1α pathway to activate the transcription of glycolysis genes, which in turn mediated the glycolysis to obtain energy to meet tumor cell proliferation.30 In this research, we demonstrated that HMGB1 overexpression abated the suppressive roles of miR-1236-3p in GC cell proliferation, metastasis and glycolysis, suggesting that miR-1236-3p played an anti-carcinogenic role in GC via binding to HMGB1. However, how HMGB1 modulating glycolysis level still need further exploration. In addition, our results showed that circ_0032821 knockdown led to a remarkable decrease in HMGB1 protein level in GC cells, while miR-1236-3p inhibition reversed the effect. The findings suggested that circ_0032821 could sponge miR-1236-3p to modulate HMGB1 expression in GC cells.
Combining all these results, we drew a conclusion that circ_0032821 accelerated the malignancy of GC via elevating HMGB1 expression through sponging miR-1236-3p. This work facilitated our understanding to the pathogenesis of GC and implicated the therapeutic possibility for GC patients.
Funding
There is no funding to report.
Disclosure
The authors declare no potential conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Shen L, Shan YS, Hu HM, et al. Management of gastric cancer in Asia: resource-stratified guidelines. Lancet Oncol. 2013;14(12):e535–547. doi:10.1016/S1470-2045(13)70436-4
3. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi:10.3322/caac.20006
4. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
5. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
6. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
7. Liu M, Liu KD, Zhang L, et al. Circ_0009910 regulates growth and metastasis and is associated with poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22(23):8248–8256.
8. Liu J, Liu H, Zeng Q, Xu P, Liu M, Yang N. Circular RNA circ-MAT2B facilitates glycolysis and growth of gastric cancer through regulating the miR-515-5p/HIF-1alpha axis. Cancer Cell Int. 2020;20:171. doi:10.1186/s12935-020-01256-1
9. Jiang Y, Zhang Y, Chu F, Xu L, Wu H. Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int. 2020;20:74. doi:10.1186/s12935-020-1151-0
10. van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11(9):644–656.
11. Wang J, Liu L, Sun Y, et al. miR-615-3p promotes proliferation and migration and inhibits apoptosis through its potential target CELF2 in gastric cancer. Biomed Pharmacother. 2018;101:406–413. doi:10.1016/j.biopha.2018.02.104
12. Yu T, Wang LN, Li W, et al. Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 2018;109(5):1393–1403. doi:10.1111/cas.13583
13. Li LQ, Yang Y, Chen H, Zhang L, Pan D, Xie WJ. MicroRNA-181b inhibits glycolysis in gastric cancer cells via targeting hexokinase 2 gene. Cancer Biomark. 2016;17(1):75–81. doi:10.3233/CBM-160619
14. Bian T, Jiang D, Liu J, et al. miR-1236-3p suppresses the migration and invasion by targeting KLF8 in lung adenocarcinoma A549 cells. Biochem Biophys Res Commun. 2017;492(3):461–467. doi:10.1016/j.bbrc.2017.08.074
15. Wang Y, Yan S, Liu X, et al. miR-1236-3p represses the cell migration and invasion abilities by targeting ZEB1 in high-grade serous ovarian carcinoma. Oncol Rep. 2014;31(4):1905–1910. doi:10.3892/or.2014.3046
16. An JX, Ma ZS, Ma MH, Shao S, Cao FL, Dai DQ. MiR-1236-3p serves as a new diagnostic and prognostic biomarker for gastric cancer. Cancer Biomark. 2019;25(2):127–132. doi:10.3233/CBM-171026
17. An JX, Ma MH, Zhang CD, Shao S, Zhou NM, Dai DQ. miR-1236-3p inhibits invasion and metastasis in gastric cancer by targeting MTA2. Cancer Cell Int. 2018;18:66.
18. Muller S, Scaffidi P, Degryse B, et al. New EMBO members’ review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. EMBO J. 2001;20(16):4337–4340. doi:10.1093/emboj/20.16.4337
19. Kang R, Chen R, Zhang Q, et al. HMGB1 in health and disease. Mol Aspects Med. 2014;40:1–116. doi:10.1016/j.mam.2014.05.001
20. Zhang J, Kou YB, Zhu JS, Chen WX, Li S. Knockdown of HMGB1 inhibits growth and invasion of gastric cancer cells through the NF-kappaB pathway in vitro and in vivo. Int J Oncol. 2014;44(4):1268–1276. doi:10.3892/ijo.2014.2285
21. Li Y, Qin C. MiR-1179 inhibits the proliferation of gastric cancer cells by targeting HMGB1. Hum Cell. 2019;32(3):352–359. doi:10.1007/s13577-019-00244-6
22. Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–73281. doi:10.18632/oncotarget.19154
23. Liu J, Song S, Lin S, et al. Circ-SERPINE2 promotes the development of gastric carcinoma by sponging miR-375 and modulating YWHAZ. Cell Prolif. 2019;52(4):e12648.
24. Chen Z, Ju H, Zhao T, et al. hsa_circ_0092306 targeting miR-197-3p promotes gastric cancer development by regulating PRKCB in MKN-45 cells. Mol Ther Nucleic Acids. 2019;18:617–626. doi:10.1016/j.omtn.2019.08.012
25. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi:10.1016/j.cmet.2015.12.006
26. Shamsi M, Saghafian M, Dejam M, Sanati-Nezhad A. Mathematical modeling of the function of warburg effect in tumor microenvironment. Sci Rep. 2018;8(1):8903. doi:10.1038/s41598-018-27303-6
27. Zhu XP, Wang XL, Ma J, et al. Down-regulation of miR-1236-3p is correlated with clinical progression and unfavorable prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22(18):5914–5919.
28. Wang D, Hu Y. Long non-coding RNA PVT1 competitively binds microRNA-424-5p to regulate CARM1 in radiosensitivity of non-small-cell lung cancer. Mol Ther Nucleic Acids. 2019;16:130–140. doi:10.1016/j.omtn.2018.12.006
29. Tian L, Wang ZY, Hao J, Zhang XY. miR-505 acts as a tumor suppressor in gastric cancer progression through targeting HMGB1. J Cell Biochem. 2019;120:8044–8052.
30. Chen R, Zhu S, Fan XG, et al. High mobility group protein B1 controls liver cancer initiation through yes-associated protein -dependent aerobic glycolysis. Hepatology. 2018;67(5):1823–1841. doi:10.1002/hep.29663
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.