Back to Journals » International Journal of Nanomedicine » Volume 13
Cholesteryl oleate-loaded cationic solid lipid nanoparticles as carriers for efficient gene-silencing therapy
Authors Suñé-Pou M, Prieto-Sánchez S, El Yousfi Y, Boyero-Corral S, Nardi-Ricart A , Nofrerias-Roig I, Pérez-Lozano P , García-Montoya E , Miñarro-Carmona M, Ticó JR , Suñé-Negre JM , Hernández-Munain C, Suñé C
Received 6 December 2017
Accepted for publication 17 March 2018
Published 30 May 2018 Volume 2018:13 Pages 3223—3233
DOI https://doi.org/10.2147/IJN.S158884
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Marc Suñé-Pou,1–3 Silvia Prieto-Sánchez,2 Younes El Yousfi,2 Sofía Boyero-Corral,2 Anna Nardi-Ricart,1 Isaac Nofrerias-Roig,1 Pilar Pérez-Lozano,1,3 Encarna García-Montoya,1,3 Montserrat Miñarro-Carmona,1,3 Josep Ramón Ticó,1,3 Josep Mª Suñé-Negre,1,3 Cristina Hernández-Munain,4 Carlos Suñé2
1Service of Development of Medicines (SDM), Faculty of Pharmacy, University of Barcelona, Barcelona, Spain; 2Department of Molecular Biology, Institute of Parasitology and Biomedicine “López-Neyra” (IPBLN-CSIC), Granada, Spain; 3Pharmacotherapy, Pharmacogenetics and Pharmaceutical Technology Research Group, IDIBELL-UB, Duran i Reynals Hospital, Hospitalet de Llobregat, Barcelona, Spain; 4Department of Cell Biology and Immunology, Institute of Parasitology and Biomedicine “López-Neyra” (IPBLN-CSIC), Granada, Spain
Background: Cationic solid lipid nanoparticles (SLNs) have been given considerable attention for therapeutic nucleic acid delivery owing to their advantages over viral and other nanoparticle delivery systems. However, poor delivery efficiency and complex formulations hinder the clinical translation of SLNs.
Aim: The aim of this study was to formulate and characterize SLNs incorporating the cholesterol derivative cholesteryl oleate to produce SLN–nucleic acid complexes with reduced cytotoxicity and more efficient cellular uptake.
Methods: Five cholesteryl oleate-containing formulations were prepared. Laser diffraction and laser Doppler microelectrophoresis were used to evaluate particle size and zeta potential, respectively. Nanoparticle morphology was analyzed using electron microscopy. Cytotoxicity and cellular uptake of lipoplexes were evaluated using flow cytometry and fluorescence microscopy. The gene inhibition capacity of the lipoplexes was assessed using siRNAs to block constitutive luciferase expression.
Results: We obtained nanoparticles with a mean diameter of approximately 150–200 nm in size and zeta potential values of 25–40 mV. SLN formulations with intermediate concentrations of cholesteryl oleate exhibited good stability and spherical structures with no aggregation. No cell toxicity of any reference SLN was observed. Finally, cellular uptake experiments with DNA- and RNA-SLNs were performed to select one reference with superior transient transfection efficiency that significantly decreased gene activity upon siRNA complexation.
Conclusion: The results indicate that cholesteryl oleate-loaded SLNs are a safe and effective platform for nonviral nucleic acid delivery.
Keywords: cationic solid lipid nanoparticles, SLNs, cholesteryl oleate, plasmid DNA, siRNA, transfection, cytotoxicity, uptake
Introduction
The development of efficient delivery systems is a critical factor for successful gene therapy.1 Numerous studies have described the use of vectors, such as retroviruses and adenoviruses, for delivering and expressing genes.2–4 While this approach often results in high expression efficiencies, it has several disadvantages, including marked immunogenicity, insertional mutagenesis, DNA package size limit, nonspecific effects, and potential hazards to laboratory personnel (comprehensive reviews are given in the studies by Collins and Thrasher,5 van der Loo and Wright,6 and Chira et al7). Alternatively, mechanical transfection methods, such as direct microinjection and the “gene gun,” or physical transfection methods, such as electroporation, sonoporation, or laser irradiation, represent direct methods for delivering genetic material into cells.8 Some of these methods have demonstrated their potential to directly transfer nucleic acids into cells and achieve good transgene expression. However, the instability of the nucleic acid and its rapid degradation, together with the associated toxicity, are still present.8 In recent decades, nanoparticle-mediated therapeutic nucleic acid delivery has been gaining considerable attention. Compared with viral systems, nanoparticles are less immunogenic and oncogenic and have no potential for virus recombination, all of which translates to improved safety.9 Nanoparticles are relatively easy to prepare and can incorporate nucleic acids with little toxicity.10 However, the transfection efficiency is low and a major concern in nanoparticle-mediated targeted therapy.11 While considerable effort is needed to study the biological interactions of nanoparticles with cells and their constituent proteins,12 it has been widely observed that transfection efficiency is dependent on the specific properties of the nanoparticle, ie, its formulation.13 Nanoparticle diameter, shape, surface charge, and components influence cellular uptake and toxicity.14–17 Hence, the research and development of novel nanoparticle formulations able to enter cells with high efficiency have been a priority for the clinical success of nanoparticle delivery systems.11,18
Depending on the manufacturing process and the physicochemical properties, nanoparticles fall into different categories, ie, inorganic and organic nanoparticles. Among the first, a good example is gold nanoparticles, which can be used as cancer immunotherapy carriers or for diagnostic applications because they can be functionalized to detect low levels of specific targets.19–22 Organic nanoparticles have shown tremendous potential for broad clinical applications and gene therapy applications.23–25 As an example, several formulations of polymeric nanoparticles containing poly(lactic-co-glycolic acid) (PLGA), cyclodextrin, and chitosan have been developed for cancer therapy,26,27 and pH-sensitive polymers, which exhibit improved drug delivery to tumors, have shown increased antitumor activity with decreased side effects.28 The fact that many nanoparticle-based approaches have been approved for clinical use serves as a proof of concept.29,30 However, manufacturing processes, costs, toxicity, and transfection efficiency continue to hinder the widespread use of nanotechnology for clinical purposes in humans.1
Cationic solid lipid nanoparticles (SLNs) represent one of the most promising nanoparticle-based methods for gene therapy.31–35 SLNs were developed to overcome the limitations of polymeric nanoparticles and other lipid-based nanoparticles.36 They are based on the use of biocompatible lipids to produce a surface charge, which allows DNA and RNA binding and facilitates good biocompatibility and low cytotoxicity.10,37,38 SLNs are capable of transfecting nucleic acids into cells in vitro and in vivo,10,35 thus supporting their research and development for potential effective gene therapy strategies. SLNs can also be formulated for drug delivery and targeting through antibody-mediated, cationic lipid, and pH-sensitive lipid attachments.33 SLNs can be obtained using several methods with the advantage of avoiding organic solvent incorporation.39,40 Although SLNs have a remarkably wide range of properties and applications, they present some limitations and difficulties that need to be overcome before their translation into the clinic. Poor delivery efficiency, uncontrollable nanoparticle transport inside the body, and problems related to realizing some manufacturing processes at a large scale to meet Good Manufacturing Practice (GMP) standards are limitations that explain the small number of these products on the market.41 Enhanced SLN formulations need to be engineered to overcome these obstacles and produce effective nanoparticle systems for the successful intracellular delivery of nucleic acids and the improvement in nanomedicine development and translation.
The use of the cholesterol derivative cholesteryl oleate in SLN formulations has not been reported. In this study, we proposed the use of cholesteryl oleate to obtain an optimized formula that maintains the nanoparticle structure, morphology, and nucleic acid binding efficiency while improving both cytotoxicity and transfection to induce potent biological activity. We used a simple fabrication method with inexpensive reagents, thus incentivizing the eventual scaled-up production of SLNs.
Materials and methods
SLN production
The following materials were used to synthesize the nanoparticles: poloxamer 188 (Sigma-Aldrich Co., St Louis, MO, USA), octadecylamine (Acros Organics, Geel, Belgium), stearic acid (EMD Millipore, Billerica, MA, USA), cholesteryl oleate (Alfa Aesar, Karlsruhe, Germany), and ultrapure water (EMD Millipore). The SLNs were produced using the hot microemulsification method, as previously described.42 Briefly, all components were melted, and the matrix lipid was poured with stirring onto the cationic lipid, water, and surfactant solution to form a hot emulsion. The emulsion was dispersed into cooled water at 2°C–3°C (ratio 1:5) under stirring to form the SLNs. The final microemulsion was centrifuged at 15,000 rpm (approximately 19,000× g) and filtered. These nanoparticles were lyophilized to preserve their properties and to improve the stability. The amount of the matricial lipid (cholesteryl oleate + stearic acid) varied depending on the formulation (Table 1). All assays except for the electronic microscopy studies were performed with lyophilized SLNs (L-3 Telstar; Telstar, Terrassa, Spain) using trehalose (5% w/v) as a cryoprotectant.
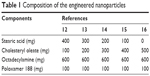  | Table 1 Composition of the engineered nanoparticles |
Plasmid constructs and siRNAs
The major histocompatibility complex (MHC) reporter luciferase plasmid has a minimal fos promoter element and three copies of the MHC class I κΒ element and has been previously described.43 The sequence of the Cy3-labeled siRNA was as follows: 5′-CUACAACAGCCACAACGE-3′ (siEGFP).44 The sequence of the siRNA targeting luciferase (siLUC) was as follows: 5′-UUGUAUUCAGCCCAUAGC-3′.45
Determination of SLN particle size
SLN suspensions were used for particle size determination by laser diffraction according to the Mie theory on a Mastersizer 2000 (Malvern Instruments, Malvern, UK). All samples were measured in triplicate using a sufficient amount of sample to obtain an appropriate obscuration percentage. A Fourier transform was applied to the measured diffracted angles to obtain the particle size data using specialized software, and the mean value was calculated in nanometers.
Determination of SLN surface charge (zeta potential)
A Zetasizer Nano Z (Malvern Instruments) was used to determine the surface charge of the SLNs by laser Doppler microelectrophoresis. Samples were stabilized at 25°C prior to measurement. The electrophoretic mobility between electrodes connected to the cell containing the sample was converted to zeta potential values using specific software. Results are expressed in mV.
Morphological analysis of SLNs
The surface and content homogeneity of the SLNs were analyzed by transmission electron microscopy (TEM) using a Spirit 120 kV microscope (FEI Company, Hillsboro, OR, USA).
Cell culture and transfection assays
HEK293T cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and grown and maintained as previously described.46 For the viability assays, HEK293T cells were grown in 35 mm plates (Fisher Scientific, Madrid, Spain) to approximately 60%–70% confluence. Because the concentration of octadecylamine varied according to the performance of the manufacturing process, we measured its concentration using a Varian 212 liquid chromatography mass spectrometry (LC-MS) system, and the test volumes were selected based on the concentration of octadecylamine present in the samples to ensure that the amount of octadecylamine and the octadecylamine:cholesteryl oleate ratio remained constant. Accordingly, SLNs were added to the cell cultures using 23.4 μL of reference 12, 14.2 μL of reference 13, 5 μL of reference 14, 4.25 μL of reference 15, and 11.6 μL of reference 16 (Table 1). After 24 and 48 h, cells were harvested and processed for cytotoxicity using flow cytometry analysis. For all the biological assays implicating nucleic acids, the complexes were prepared by mixing SLNs with the appropriate amount of plasmid DNA (pDNA) or siRNA as given in the following sections. The mixture was kept at room temperature for 40–45 min to allow the complexes to form before transfection. For the luciferase assays, HEK293T cells were grown in 35 mm plates to approximately 60%–70% confluence, and then the medium was changed to a medium without serum or antibiotics. Lipoplexes were prepared by mixing 1,000 ng of the MHC reporter luciferase plasmid, 100 ng of Renilla plasmid, and the amounts of SLNs derived from the viability assay as mentioned earlier. The mixtures were added to the cells, and after 4–6 h, 200 μL of serum was added to each well. The cells were harvested approximately 48 h after transfection and processed for luciferase activity using the dual-luciferase reporter assay (Promega Corporation, Fitchburg, WI, USA) protocol following the manufacturer’s instructions. For the transfection of Cy3-labeled siRNA, HEK293T cells were grown in 35 mm plates (Falcon) to approximately 60%–70% confluence, and the medium was changed to a medium without serum or antibiotics. Lipoplexes were prepared by mixing 60 or 120 nM of Cy3-labeled siRNA and 13.5 μL of reference 14 SLNs. As a transfection control, 60 nM of Cy3-labeled siRNA was transfected into cells using Lipofectamine 2000 according to the manufacturer’s protocols. Then, the cells were transfected with the lipoplexes. Cells were harvested at different times after transfection and processed for flow cytometry and confocal microscopy analysis. For the transfection of siLUC, we used the TZM-bl cell line. TZM-bl is a HeLa cell line that contains integrated copies of the luciferase and B-galactosidase genes under the control of the HIV-1 promoter (NIH AIDS Reagent Program, Germantown, MD, USA).47 Cells were grown in 35 mm plates to approximately 60%–70% confluence, and the medium was changed to a medium without serum or antibiotics. Lipoplexes were prepared by mixing 120 nM of siLUC and different amounts of reference 14 SLNs. Cells were transfected with the lipoplex mixtures, and after 4–6 h, 200 μL of serum was added to each well.
Cell proliferation
Cell proliferation was detected using the MTT reagent (Hoffman-La Roche Ltd., Basel, Switzerland). Briefly, 10 μL of MTT was added to the plate and incubated for 4 h at 37°C. The purple precipitate of the cells was treated with solubilization buffer, as indicated by the manufacturer, to dissolve the crystals. The absorbance at 570 nm was measured with an enzyme-linked immunosorbent assay (ELISA) reader (Molecular Devices LLC, Sunnyvale, CA, USA) at different time points after treatment.
Flow cytometry
Cytometry was performed on a FACSCalibur (BD Biosciences, San Jose, CA, USA), and the results were analyzed using FlowJo software. The cutoff values were established using unstained HEK293T cells to subtract the autofluorescence signal of the cells. At this setting, only 0.5%–1% of the cells were over the 100 level for the Cy3 fluorescence channel.
Confocal microscopy
Confocal microscopy studies were performed using a Leica SP5 spectral confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany). Images were analyzed and digitally processed for presentation using LAS AF v2.3.6 software (Leica Microsystems) and Adobe Photoshop CS3 extended v10.0 software, respectively.
Statistical analyses
Statistical analyses were performed using Graph Pad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) with one-way analysis of variance (ANOVA) followed by Tukey’s posttest analysis to describe significant differences between groups. The P-values are represented by asterisks (**P = 0.001–0.01 and ***P < 0.001). The absence of an asterisk indicates that the change relative to the control is not statistically significant.
Results
Preparation and characterization of SLNs
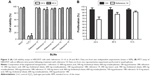
The particle size results from the initial characterization of the different SLN references 12–16 are shown in Figure 1. We obtained an important population of particles approximately 150–200 nm in size, confirming the presence of nanoparticles. References 12–14 contained homogeneous material with almost no aggregates present (Figure 1A–C). We observed an increased amount of aggregation in reference 15, but nanoparticles were also synthesized (Figure 1D). Reference 16, which was synthesized with 100% cholesteryl oleate, contained numerous aggregates (Figure 1E). These results showed that references 12–14 were the best suitable to form lipoplexes for transfection purposes. Concerning zeta potential, there were no differences among the different formulations (Table 2). All the references had zeta potential values from 25 to 40 mV, which indicated a potentially good capacity for nucleic acid binding.
Images of the SLNs were acquired by TEM as shown in Figure 2. We observed the presence of nanostructures in all the formulations. However, the homogeneity and quality of these particles were substantially different in each formulation. For references 12–14, the images showed a high proportion of nanoparticles without any amorphous structures. In contrast, images of references 15 and 16 showed less spherical nanoparticles. In reference 16, we also observed many aggregates using a large field of view (data not shown). For reference 16, it was difficult to find images containing nanoparticles, further suggesting that the presence of 100% cholesteryl oleate in the formula yields nanoparticles that become unstable. These results are consistent with the laser diffraction data and confirm the suitability of references 12–14 for further investigation.
Cytotoxicity
An early and crucial stage in determining the efficacy of nanoparticles is toxicity testing. To determine the effect of the SLNs on cellular viability, a fixed quantity of nanoparticles from each reference was incubated with HEK293T cells for 48 h. We assessed cell viability by counting the number of viable cells using flow cytometry analysis. Variability in the nanoparticle concentration among the samples was minimized by quantifying octadecylamine via LC-MS. No toxic effects of any SLN reference were observed (Figure 3A). Moreover, the viability of SLN-containing cells was similar to that of untreated cells after the incubation time (Figure 3A). These results show that the modified SLNs are not harmful to human cells cultured in vitro. The cytotoxicity against HEK293T cells was also studied using the MTT colorimetric cell proliferation assay. Time-dependent cytotoxic effects of SLNs from reference 14 (selected based on their physicochemical characteristics and good performance as given in the following sections) were observed as shown in Figure 3B. While initial cytotoxicity was observed at 24 h after treatment, the cells recovered quickly and showed almost normal viability at later treatment time points (Figure 3B).
Cellular uptake and biological activity
The cellular uptake of nanoparticles and their capability to generate a biological response can be tested by the luciferase reporter assay, where pDNA driving the expression of the luciferase gene is transfected into cells. Using this assay, we tested the cellular uptake and bioactivity of two different luciferase reporter plasmids (MHC and Renilla) using references 12–16 (Figure 4A and B) in the same quantities employed in the viability assays. While we observed a dose–response effect in the luciferase activity upon transfection with SLNs from references 12 to 14, the enzymatic activity decreased upon transfection with SLNs from references 15 and 16. These results correlate with the physicochemical parameters of the SLNs and suggest that aggregation and poor stability are detrimental to transfection efficiency and bioactivity. Reference 14 resulted in potent luciferase activity (Figure 4A and B), which is consistent with the previously determined size and morphology data of this formulation (our results) and with the binding capacity of these nanoparticles.10,42 We concluded that our cholesteryl oleate-based SLNs can be internalized in cells and generate a biological response. Therefore, these SLNs provided protection against nuclease activity, enabling cellular uptake, intracellular nucleic acid release and RNA transcription in the nucleus, transport to the cytoplasm, and translation into protein, enabling bioactivity. Reference 14 showed good physicochemical parameters and superior transfection efficiency and was therefore suitable for further experimentation.
A more relevant model for RNA interference (RNAi) applications is the siRNA-mediated targeting of endogenous gene products rather than the transient transfection of plasmid-based genes. Therefore, we generated a synthetic siRNA against the luciferase gene to be used in subsequent experiments. First, we used flow cytometry to evaluate the cellular uptake of SLN-Cy3-labeled siRNA complexes using reference 14 in HEK293T cells. SLN complexation was performed with 60 and 120 nM Cy3-labeled siRNA. After the quantification analysis, the transfection efficiency of the SLN-Cy3-labeled siRNA complexes was approximately 40%–45% (Figure 5A). However, the SLNs elicited increased signal intensity compared with Lipofectamine 2000 (Figure 5B). Interestingly, we observed an increase in signal intensity when using larger quantities of SLN-Cy3-labeled siRNA complexes (Figure 5B). Given that both siRNA concentrations exhibited similar transfection efficiency, these data suggest that more fluorescent siRNA is introduced into each individual cell, which may give rise to an increased biological response.
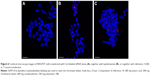
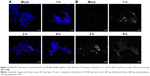
The transfection efficiency data were further supported by the confocal microscopy results. We observed a lack of fluorescence when the Cy3-labeled siRNA alone was added to the cells (Figure 6A). Many large fluorescent dots were observed in the cytoplasm and in the nucleus at 1 h post-transfection of the Cy3-labeled siRNA using Lipofectamine 2000 (Figure 6B). Importantly, the addition of the SLN-Cy3-labeled siRNA complexes resulted in similar fluorescence signals (Figure 6C), reflecting the efficient cell binding and intracellular uptake of the lipoplexes. While strong fluorescence signals were still observed at 2 h post-transfection, weak fluorescence was detected at 6 h post-transfection, as assessed by confocal microscopy (Figure 7A) or flow cytometry (data not shown), thus revealing the rapid intracellular uptake of these lipoplexes. Grayscale images (Figure 7B) showed multiple arrays of fluorescence signals widely spread throughout the cells, which may explain the loss of signal observed with image color information.
To confirm that the SLNs could deliver and release siRNA molecules into cells to result in bioactivity, we performed transfection experiments using TZM-bl cells, which carry integrated copies of the HIV-1 long terminal repeat driving expression of the luciferase gene. Targeting luciferase expression with SLN-siLUC complexes using different quantities of reference 14 resulted in an approximately 35% reduction in luciferase activity after 48 h compared with the control (Figure 8). This downregulation of endogenous luciferase expression validated the use of SLNs incorporating cholesteryl oleate in their formulation as promising tools for gene targeting.
Discussion
Identification of new formulations and cationic compounds for therapeutic use is of fundamental importance for achieving therapeutic nucleic acid transfer. This objective must be attained with consideration for the requirements of cost-optimized large-scale production. The purpose of this study was to formulate and characterize SLNs incorporating the cholesterol derivative cholesteryl oleate to achieve SLN–nucleic acid complexes with more efficient cellular uptake and reduced cytotoxicity. The steroid lipid cholesterol is an essential component of animal cell membranes and has multiple functions. We reasoned that incorporating cholesterol or cholesterol derivatives in the lipoplex formulation might increase the transfection efficiency and biocompatibility of the resulting SLNs. Previous studies have shown that lipid mixtures with cholesterol or cholesterol derivatives, which were generally synthesized using complicated protocols, exhibited enhanced transfection.48–51 Cholesterol may improve transfection efficiency by promoting fusion of the lipoplex with the cell membrane.52 We decided to test cholesteryl oleate because of its low melting point (44°C–47°C versus 148°C for cholesterol), which was compatible with our protocols.42 In this study, we have shown that cholesteryl oleate can be conveniently formulated in SLNs through a simple and uncomplicated protocol. The resulting vectors showed good performance in delivering DNA or siRNA into cells without cytotoxicity and with good transfection efficiency.
We compared in detail the properties of a series of SLNs with increasing quantities of cholesteryl oleate in its formulation. While all SLNs formed suspensions in aqueous media, the stability of the nanoparticles was decreased with high cholesteryl oleate concentrations (references 15 and 16). Accordingly, electron microscopy studies revealed nearly spherical structures in those SLN formulations with low and intermediate concentrations of cholesteryl oleate and less spherical structures with abundant aggregation in those formulations with higher concentrations of cholesteryl oleate. While the toxicity assessment results of the formulations containing any cholesteryl oleate concentration were similar to that observed for the control, the transfection efficiency of each compound differed according to the formulation. In the transient transfection experiments using firefly and Renilla luciferase expression vectors, reference 14 displayed robust, dose–response luciferase activation much higher than that observed for the other references. Therefore, this reference was selected for further studies using siRNA. Cytometry and confocal microscopy characterizations showed the ability of reference 14 to deliver siRNA molecules into cells. We observed massive and rapid uptake, as indicated by many large siRNA-labeled fluorescent dots throughout the cytoplasm at 1 h post-transfection, with a loss of signal at longer times post-transfection. Semiquantitative cytometry analysis showed approximately 45% cell incorporation, which is a remarkable transfection efficiency for a nonfunctionalized nanoparticle-based delivery method.53 In addition to enhanced cellular uptake levels, siRNA delivery requires stabilization and protection against degradation by nucleases, which can impair the biological response. Targeting a chromosome-integrated luciferase gene, we observed an ~35% reduction in enzymatic activity, which demonstrates that upon SLN complexation, siRNA is protected against degradation, internalized by cells, and released intracellularly, ultimately leading to bioactivity. In this study, we used HEK293T and HeLa cells to establish experimental conditions for nucleic acid delivery into cells using SLNs formulated with cholesteryl oleate. Future experiments will test our formulation in hard-to-transfect primary and cell lines to deliver nucleic acids to appropriate targets in order to interfere with relevant gene expression programs.
Conclusion
Nanoparticles can facilitate the intracellular delivery of siRNA, and many studies have shown that siRNA can have a significant therapeutic effect on disease, especially tumors.54 However, the use of these nanoparticle carriers still has disadvantages in terms of transfection efficiency, immunogenicity, and toxicity, all of which limit their therapeutic applications. The experiments reported in this study demonstrate that the use of cholesteryl oleate in SLNs provides an avenue for the development of highly efficient and nontoxic delivery carriers for therapeutic applications.
Acknowledgments
We are grateful to many colleagues for their helpful suggestions, critical discussions, and comments. We thank José Alcamí and Mayte Coiras (National Microbiology Center, Majadahonda, Madrid, Spain) for the TZM-bl cells. This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (grant number BFU2014-54660-R) and the Andalusian Government (Excellence Project BIO-2515/2012) to CS and from the Spanish Ministry of Economy and Competitiveness (grant number BFU2016-79699-P) to CHM. Support from the European Region Development Fund (ERDF [FEDER]) is also acknowledged. MSP was supported by a fellowship from the Spanish Ministry of Education (FPU Program [Training Program for Academic Staff]).
Disclosure
The authors report no conflicts of interest in this work.
References
Wilhelm S, Tavares AJ, Dai Q, et al. Analysis of nanoparticle delivery to tumours. Nat Rev Mater. 2016;1:1–12. | ||
Hacein-Bey-Abina S, Le Deist F, Carlier F, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346(16):1185–1193. | ||
Westphal M, Yla-Herttuala S, Martin J, et al; ASPECT Study Group. Adenovirus-mediated gene therapy with sitimagene ceradenovec followed by intravenous ganciclovir for patients with operable high-grade glioma (ASPECT): a randomised, open-label, phase 3 trial. Lancet Oncol. 2013;14(9):823–833. | ||
Cicalese MP, Ferrua F, Castagnaro L, et al. Update on the safety and efficacy of retroviral gene therapy for immunodeficiency due to adenosine deaminase deficiency. Blood. 2016;128(1):45–54. | ||
Collins M, Thrasher A. Gene therapy: progress and predictions. Proc Biol Sci. 2015;282(1821):20143003. | ||
van der Loo JC, Wright JF. Progress and challenges in viral vector manufacturing. Hum Mol Genet. 2016;25(R1):R42–R52. | ||
Chira S, Jackson CS, Oprea I, et al. Progresses towards safe and efficient gene therapy vectors. Oncotarget. 2015;6(31):30675–30703. | ||
Mehier-Humbert S, Guy RH. Physical methods for gene transfer: improving the kinetics of gene delivery into cells. Adv Drug Deliv Rev. 2005;57(5):733–753. | ||
Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–555. | ||
Fabregas A, Prieto-Sanchez S, Sune-Pou M, et al. Improved formulation of cationic solid lipid nanoparticles displays cellular uptake and biological activity of nucleic acids. Int J Pharm. 2017;516(1–2):39–44. | ||
Al-Dosari MS, Gao X. Nonviral gene delivery: principle, limitations, and recent progress. AAPS J. 2009;11(4):671–681. | ||
Deng J, Gao C. Recent advances in interactions of designed nanoparticles and cells with respect to cellular uptake, intracellular fate, degradation and cytotoxicity. Nanotechnology. 2016;27(41):412002. | ||
Basha G, Novobrantseva TI, Rosin N, et al. Influence of cationic lipid composition on gene silencing properties of lipid nanoparticle formulations of siRNA in antigen-presenting cells. Mol Ther. 2011;19(12):2186–2200. | ||
Chithrani BD, Chan WC. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007;7(6):1542–1550. | ||
Gratton SE, Ropp PA, Pohlhaus PD, et al. The effect of particle design on cellular internalization pathways. Proc Natl Acad Sci U S A. 2008;105(33):11613–11618. | ||
Marano F, Hussain S, Rodrigues-Lima F, Baeza-Squiban A, Boland S. Nanoparticles: molecular targets and cell signalling. Arch Toxicol. 2011;85(7):733–741. | ||
Rauch J, Kolch W, Laurent S, Mahmoudi M. Big signals from small particles: regulation of cell signaling pathways by nanoparticles. Chem Rev. 2013;113(5):3391–3406. | ||
Wang Y, Huang L. Composite nanoparticles for gene delivery. Adv Genet. 2014;88:111–137. | ||
Anselmo AC, Mitragotri S. A review of clinical translation of inorganic nanoparticles. AAPS J. 2015;17(5):1041–1054. | ||
Almeida JP, Figueroa ER, Drezek RA. Gold nanoparticle mediated cancer immunotherapy. Nanomedicine. 2014;10(3):503–514. | ||
Mieszawska AJ, Mulder WJ, Fayad ZA, Cormode DP. Multifunctional gold nanoparticles for diagnosis and therapy of disease. Mol Pharm. 2013;10(3):831–847. | ||
Huang HC, Barua S, Sharma G, Dey SK, Rege K. Inorganic nanoparticles for cancer imaging and therapy. J Control Release. 2011;155(3):344–357. | ||
Gozuacik D, Yagci-Acar HF, Akkoc Y, Kosar A, Dogan-Ekici AI, Ekici S. Anticancer use of nanoparticles as nucleic acid carriers. J Biomed Nanotechnol. 2014;10(9):1751–1783. | ||
Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12(11):967–977. | ||
Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. | ||
Ku SH, Kim K, Choi K, Kim SH, Kwon IC. Tumor-targeting multifunctional nanoparticles for siRNA delivery: recent advances in cancer therapy. Adv Healthc Mater. 2014;3(8):1182–1193. | ||
Wang X, Yang C, Zhang Y, Zhen X, Wu W, Jiang X. Delivery of platinum(IV) drug to subcutaneous tumor and lung metastasis using bradykinin-potentiating peptide-decorated chitosan nanoparticles. Biomaterials. 2014;35(24):6439–6453. | ||
He H, Chen S, Zhou J, et al. Cyclodextrin-derived pH-responsive nanoparticles for delivery of paclitaxel. Biomaterials. 2013;34(21):5344–5358. | ||
Min Y, Caster JM, Eblan MJ, Wang AZ. Clinical translation of nanomedicine. Chem Rev. 2015;115(19):11147–11190. | ||
Bobo D, Robinson KJ, Islam J, Thurecht KJ, Corrie SR. Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm Res. 2016;33(10):2373–2387. | ||
Carrillo C, Sanchez-Hernandez N, Garcia-Montoya E, et al. DNA delivery via cationic solid lipid nanoparticles (SLNs). Eur J Pharm Sci. 2013;49(2):157–165. | ||
Moritz M, Geszke-Moritz M. Recent developments in the application of polymeric nanoparticles as drug carriers. Adv Clin Exp Med. 2015;24(5):749–758. | ||
Rostami E, Kashanian S, Azandaryani AH, Faramarzi H, Dolatabadi JE, Omidfar K. Drug targeting using solid lipid nanoparticles. Chem Phys Lipids. 2014;181:56–61. | ||
Yu YH, Kim E, Park DE, et al. Cationic solid lipid nanoparticles for co-delivery of paclitaxel and siRNA. Eur J Pharm Biopharm. 2012;80(2):268–273. | ||
Apaolaza PS, Del Pozo-Rodriguez A, Solinis MA, et al. Structural recovery of the retina in a retinoschisin-deficient mouse after gene replacement therapy by solid lipid nanoparticles. Biomaterials. 2016;90:40–49. | ||
Madan JR, Khude PA, Dua K. Development and evaluation of solid lipid nanoparticles of mometasone furoate for topical delivery. Int J Pharm Investig. 2014;4(2):60–64. | ||
Ezzati Nazhad Dolatabadi J, Hamishehkar H, Eskandani M, Valizadeh H. Formulation, characterization and cytotoxicity studies of alendronate sodium-loaded solid lipid nanoparticles. Colloids Surf B Biointerfaces. 2014;117:21–28. | ||
Doktorovova S, Kovacevic AB, Garcia ML, Souto EB. Preclinical safety of solid lipid nanoparticles and nanostructured lipid carriers: current evidence from in vitro and in vivo evaluation. Eur J Pharma Biopharm. 2016;108:235–252. | ||
Geszke-Moritz M, Moritz M. Solid lipid nanoparticles as attractive drug vehicles: composition, properties and therapeutic strategies. Mater Sci Eng C Mater Biol Appl. 2016;68:982–994. | ||
Muller RH, Mader K, Gohla S. Solid lipid nanoparticles (SLN) for controlled drug delivery – a review of the state of the art. Eur J Pharm Biopharm. 2000;50(1):161–177. | ||
Ezzati Nazhad Dolatabadi J, Valizadeh H, Hamishehkar H. Solid lipid nanoparticles as efficient drug and gene delivery systems: recent breakthroughs. Adv Pharm Bull. 2015;5(2):151–159. | ||
Fabregas A, Sanchez-Hernandez N, Tico JR, et al. A new optimized formulation of cationic solid lipid nanoparticles intended for gene delivery: development, characterization and DNA binding efficiency of TCERG1 expression plasmid. Int J Pharm. 2014;473(1–2):270–279. | ||
Sánchez-Álvarez M, Montes M, Sánchez-Hernández N, Hernández-Munain C, Suñé C. Differential effects of sumoylation on transcription and alternative splicing by transcription elongation regulator 1 (TCERG1). J Biol Chem. 2010;285(20):15220–15233. | ||
Becerra S, Montes M, Hernández-Munain C, Suñé C. Prp40 pre-mRNA processing factor 40 homolog B (PRPF40B) associates with SF1 and U2AF65 and modulates alternative pre-mRNA splicing in vivo. RNA. 2015;21(3):438–457. | ||
Moses J, Goodchild A, Rivory LP. Intended transcriptional silencing with siRNA results in gene repression through sequence-specific off-targeting. RNA. 2010;16(2):430–441. | ||
Sánchez-Álvarez M, Goldstrohm AC, Garcia-Blanco MA, Suñé C. Human transcription elongation factor CA150 localizes to splicing factor-rich nuclear speckles and assembles transcription and splicing components into complexes through its amino and carboxyl regions. Mol Cell Biol. 2006;26(13):4998–5014. | ||
Bermejo M, Lopez-Huertas MR, Garcia-Perez J, et al. Dasatinib inhibits HIV-1 replication through the interference of SAMHD1 phosphorylation in CD4+ T cells. Biochem Pharmacol. 2016;106:30–45. | ||
Samadikhah HR, Majidi A, Nikkhah M, Hosseinkhani S. Preparation, characterization, and efficient transfection of cationic liposomes and nanomagnetic cationic liposomes. Int J Nanomedicine. 2011;6:2275–2283. | ||
Huang QD, Ou WJ, Chen H, et al. Novel cationic lipids possessing protonated cyclen and imidazolium salt for gene delivery. Eur J Pharm Biopharm. 2011;78(3):326–335. | ||
Biswas J, Bajaj A, Bhattacharya S. Membranes of cationic gemini lipids based on cholesterol with hydroxyl headgroups and their interactions with DNA and phospholipid. J Phys Chem B. 2011;115(3):478–486. | ||
Islam RU, Hean J, van Otterlo WA, de Koning CB, Arbuthnot P. Efficient nucleic acid transduction with lipoplexes containing novel piperazine- and polyamine-conjugated cholesterol derivatives. Bioorg Med Chem Lett. 2009;19(1):100–103. | ||
Pozzi D, Marchini C, Cardarelli F, et al. Transfection efficiency boost of cholesterol-containing lipoplexes. Biochim Biophys Acta. 2012;1818(9):2335–2343. | ||
Cortesi R, Campioni M, Ravani L, Drechsler M, Pinotti M, Esposito E. Cationic lipid nanosystems as carriers for nucleic acids. N Biotechnol. 2014;31(1):44–54. | ||
Xin Y, Huang M, Guo WW, Huang Q, Zhang LZ, Jiang G. Nano-based delivery of RNAi in cancer therapy. Mol Cancer. 2017;16(1):134. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.