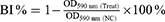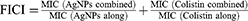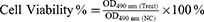Back to Journals » International Journal of Nanomedicine » Volume 18
Biosynthesis of Silver Nanoparticles Using the Biofilm Supernatant of Pseudomonas aeruginosa PA75 and Evaluation of Their Antibacterial, Antibiofilm, and Antitumor Activities
Authors Xia F , Tao X, Wang H, Shui J, Min C, Xia Y, Li J, Tang M, Liu Z, Hu Y, Luo H, Zou M
Received 14 March 2023
Accepted for publication 6 May 2023
Published 10 May 2023 Volume 2023:18 Pages 2485—2502
DOI https://doi.org/10.2147/IJN.S410314
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lijie Grace Zhang
Fengjun Xia,1,2,* Xiaoyan Tao,2,3,* Haichen Wang,1,2 Jian Shui,4 Changhang Min,1,2 Yubing Xia,1,2 Jun Li,1,2 Mengli Tang,1,2 ZhaoJun Liu,1,2 Yongmei Hu,1,2 Huidan Luo,1,2 Mingxiang Zou1,2
1National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, 410008, People’s Republic of China; 2Department of Clinical Laboratory, Xiangya Hospital, Central South University, Changsha, 410008, People’s Republic of China; 3Department of Laboratory Medicine and Sichuan Provincial Key Laboratory for Human Disease Gene Study, Sichuan Provincial People’s Hospital, University of Electronic Science and Technology of China, Chengdu, 610072, People’s Republic of China; 4Department of Clinical Laboratory, Changsha Central Hospital, Changsha, 410008, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Mingxiang Zou, National Clinical Research Center for Geriatric Disorders, Department of Clinical Laboratory, Xiangya Hospital, Central South University, Changsha, Hunan Province, 410008, People’s Republic of China, Tel/Fax +86-7384327440, Email [email protected]
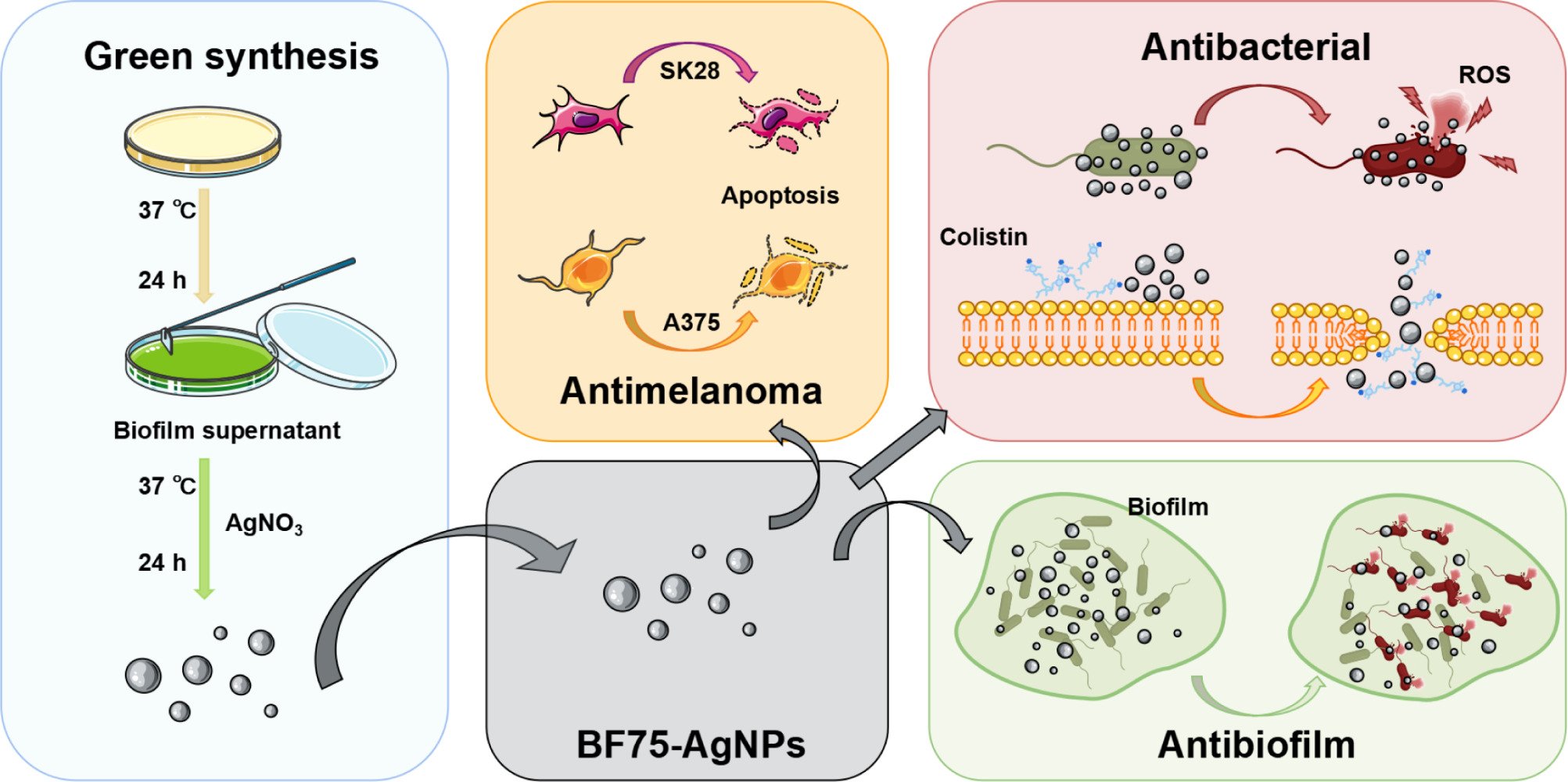
Purpose: As an under-explored biomaterial, bacterial biofilms have a wide range of applications in the green synthesis of nanomaterials. The biofilm supernatant of Pseudomonas aeruginosa PA75 was used to synthesize novel silver nanoparticles (AgNPs). BF75-AgNPs were found to possess several biological properties.
Methods: In this study, we biosynthesized BF75-AgNPs using biofilm supernatant as the reducing agent, stabilizer, and dispersant and investigated their biopotential in terms of antibacterial, antibiofilm, and antitumor activities.
Results: The synthesized BF75-AgNPs demonstrated a typical face-centered cubic crystal structure; they were well dispersed; and they were spherical with a size of 13.899 ± 4.036 nm. The average zeta potential of the BF75-AgNPs was − 31.0 ± 8.1 mV. The BF75-AgNPs exhibited strong antibacterial activities against the methicillin-resistant Staphylococcus aureus (MRSA), extended-spectrum beta-lactamase Escherichia coli (ESBL-EC), extensively drug-resistant Klebsiella pneumoniae (XDR-KP), and carbapenem-resistant Pseudomonas aeruginosa (CR-PA). Moreover, the BF75-AgNPs had a strong bactericidal effect on XDR-KP at 1/2 × MIC, and the expression level of reactive oxygen species (ROS) in bacteria was significantly increased. A synergistic effect was observed when the BF75-AgNPs and colistin were used for the co-treatment of two colistin-resistant XDR-KP strains, with fractional inhibitory concentration index (FICI) values of 0.281 and 0.187, respectively. Furthermore, the BF75-AgNPs demonstrated a strong biofilm inhibition activity and mature biofilm bactericidal activity against XDR-KP. The BF75-AgNPs also exhibited a strong antitumor activity against melanoma cells and low cytotoxicity against normal epidermal cells. In addition, the BF75-AgNPs increased the proportion of apoptotic cells in two melanoma cell lines, and the proportion of late apoptotic cells increased with BF75-AgNP concentration.
Conclusion: This study suggests that BF75-AgNPs synthesized from biofilm supernatant have broad prospects for antibacterial, antibiofilm, and antitumor applications.
Keywords: multidrug resistant, green synthesis, colistin, metallic silver, melanoma
Graphical Abstract:
Introduction
The emergence of multidrug-resistant (non-susceptible to three or more antibiotic classes) and extensively drug-resistant (non-susceptible to all but one or two antibiotic classes) pathogens poses a severe threat to public health.1,2 Approximately 4.95 million deaths worldwide were associated with bacterial antibiotic resistance in 2019.3 Several types of drug-resistant bacteria are predominant, including the methicillin-resistant Staphylococcus aureus (MRSA), the extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae, and carbapenem-resistant organisms (CROs).4,5 Colistin serves as a last-resort antibiotic against pathogens with extensive drug resistance. However, with the increasing use of colistin, colistin-resistant bacteria have appeared and gradually increased, leaving few antibiotics available for the treatment of resistant bacteria.6,7 Therefore, new antibacterial agents must be developed.
Silver nanoparticles (AgNPs) are metallic silver particles with diameters ranging from 1 to 100 nm. AgNPs exhibit excellent antibacterial effects, and they have been widely used as antibacterial agents.8,9 Physical, chemical, and biological methods can be used for the synthesis AgNPs. However, physical methods for AgNP synthesis usually require high energy consumption, and chemical methods produce toxic substances. Therefore, environmentally friendly methods for AgNP synthesis using biological materials, such as animals, plants, and microorganisms, are being widely studied.10–13 Green synthesis of AgNPs involves the use of living organisms such as algae, fungi, bacteria, and plants or secondary metabolites with specific biological functions extracted from them as raw materials to reduce Ag+ to stable AgNPs via redox reactions under certain conditions.14,15
The synthesis of AgNPs using bacterial-associated metabolites is rapidly developing owing to the rapid growth of bacteria, low nutritional requirements, and high abundance of secondary metabolites. Bacterial biofilms are microcolony aggregates formed by one or more bacteria on the surfaces of tissues or active materials.16 Biofilms are composed of proteins, extracellular polysaccharides, and/or extracellular DNA.17–19 Many studies have demonstrated that bacteria exhibit different metabolic processes in the biofilm and planktonic growth modes.20,21 The biosynthesis of AgNPs only in planktonic bacteria may result in difficulty in fully utilizing some secondary metabolites that have not yet been expressed.22 Pseudomonas aeruginosa is well known for its ability to form biofilms. In our previous study, we discovered that the biofilm supernatant of the extensively drug-resistant P. aeruginosa PA75 strain had strong antibacterial activity. However, the supernatant of PA75 growing in the planktonic state showed no antibacterial activity, indicating that the secondary metabolites of PA75 in the biofilm state have unique biological functions. However, few studies exist on the synthesis of AgNPs using biofilms.
Cancer, a major cause of death, is characterized by abnormal cell growth. According to the 2018 Global Cancer Statistics, the incidence and mortality rates of melanoma have increased in recent years.23 Surgical treatment of malignant melanoma is rarely successful, and local recurrence is common. Previous studies have confirmed the antitumor effects of AgNPs in vitro and in vivo.24 Unlike conventional cancer treatments, such as radiotherapy and chemotherapy, which are toxic to healthy cells, AgNPs have low toxicity. Therefore, AgNPs have considerable potential for applications in cancer treatment. Previous studies have revealed that green synthetic AgNPs can target the mitochondria of skin cancer A431 cells to cause the overproduction of reactive oxygen species (ROS); this discovery provides an alternative approach for anticancer therapy.25 Meanwhile, the ability of AgNPs to induce apoptosis in lung, breast, cervical, and non-melanoma skin cancer has been studied.26–29 However, AgNP-induced cell apoptosis in malignant melanoma has not been reported, and the apoptotic effect has not been verified.
Therefore, the current study aimed to biosynthesize AgNPs using biofilm supernatant and investigate their antibacterial, antibiofilm and antitumor activities.
Materials and Methods
Bacterial Strains and Identification
P. aeruginosa PA75 was isolated from a sputum specimen of Xiangya Hospital, China, in 2016 and identified via matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS, Zybio, China). Molecular identification of P. aeruginosa PA75 was accomplished by sequencing the 16S rDNA PCR products. The primers used for amplification were 27: F 5’- AGAGTTTGATCMTGGCTCAG -3’ and 1492: R 5’- CGGTTACCTTGTTACGACTT -3’.30 The amplified fragment was sequenced by Sangon Biotech (Shanghai, China), and the sequences were analyzed using BLAST. Sequence alignment was completed using ClustalW2, and a phylogenetic relationship was established using the neighbor-joining method.31,32
Staphylococcus epidermidis ATCC35984 was provided by the American Type Culture Collection (ATCC), and it served as the biofilm-forming strain in our study. Five isolates each of MRSA, ESBL E. coli (ESBL-EC), XDR-KP, and CR P. aeruginosa (CR-PA) were provided by the Department of Clinical Laboratory of Xiangya Hospital, Central South University. The bacteria were cultured in a Mueller Hinton (MH) broth at 37 °C for all experiments. The strains were identified using MALDI-TOF MS (Figures S1–S4). The antimicrobial susceptibility testing and crystal violet staining methods are described in Supplementary Material 1. The susceptibility profiles of the clinical strains are listed in Tables S1–S4. The results of antimicrobial susceptibility testing indicated that among the five XDR-KP strains, XDR-KP4 and XDR-KP5 were colistin resistant, and the results of crystal violet staining indicated that XDR-KP3, XDR-KP4, and XDR-KP5 were strong biofilm-forming strains (Figure S5).
Cell Culture
Human skin epithelial HaCaT cells and human melanoma cells (SK28 and A375) were obtained from the ATCC. HaCaT cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco, USA). SK28 and A375 cells were maintained in Roswell Park Memorial Institute 1640 medium (BasalMedia Technologies, China). All media were supplemented with 10% fetal bovine serum and 1% antibiotics (penicillin and streptomycin). The cells were cultured at 37 °C in 5% CO2.
Extraction of P. aeruginosa PA75 Biofilm Supernatant
The trypsin soy agar plate method was used to simulate the growth of biofilms according to previous studies.33,34 Briefly, overnight cultures of P. aeruginosa PA75 were adjusted to 1×106 CFU/mL with ultrapure water. Subsequently, 700 μL of the solution was evenly spread on a single tryptic soy agar (15 cm in diameter) and cultured for 24 h at 37 °C. The lawns on the surface of the four dishes were scraped into 30 mL of sterile ultrapure water with a sterile cell spatula and shaken vigorously. Finally, the mixture was centrifuged at 15,000 rpm for 15 min and filtered through a 0.22 μm filter membrane (Millipore, USA) to obtain the cell-free supernatant.
Bioactivity Verification of the P. aeruginosa PA75 Biofilm Supernatant
Bacterial Growth Curve
Four milliliters of bacterial suspension (1 × 108 CFU/mL) and 2 mL of PA75 biofilm supernatant were mixed and incubated at 37 °C with shaking (200 rpm). Bacterial growth was detected by measuring the absorbance at 600 nm after 0, 1, 2, 3, 4, 6, 8, 12, and 24 h. Sterilized ultrapure water served as the negative control. Three parallel tubes were used in each experiment.
Biofilm Inhibition Assay
A 96-well plate was used for the antibiofilm experiments according to a previous study.35 In each well, 100 μL of the bacterial suspension (1 × 108 CFU/mL) and 50 μL of the PA75 biofilm supernatant were added. Ultrapure water was used as the negative control. The plates were incubated at 37 °C for 8 h. The bacterial solution was discarded, and the cells were washed with normal saline three times to remove the bacteria. In each well, 150 μL of 0.1% crystal violet solution was added, and the cells were stained for 20 min at room temperature. After discarding the dye, the cells were washed three times with normal saline and decolorized with 150 μL of 95% ethanol for 10 min. Biofilm formation was quantified at OD590 nm using a microplate reader (TECAN, Switzerland). The inhibition rate of biofilm formation was calculated as follows:
where BI% indicates the biofilm inhibition rate, Treat indicates in the presence of biofilm supernatant, and NC indicates in the absence of biofilm supernatant.
Green Synthesis of BF75-AgNPs Using the PA75 Biofilm Supernatant
Twenty milliliters (25 mM) of filter-sterilized AgNO3 solution (Shanghai Fine Chemical Research Institute, China) was added to 80 mL of biofilm supernatant and incubated at 200 rpm for 24 h (37 °C). The synthesis of BF75-AgNPs was confirmed via visual observation of a color change. Biosynthesized BF75-AgNPs were collected via centrifugation at 15,000 rpm for 30 min. Finally, the BF75-AgNPs were washed with distilled water three times and freeze-dried to achieve a constant weight. The BF75-AgNPs were stored at 4 °C until further analysis.
Characterization of Green-Synthesized BF75-AgNPs
The optical properties of the BF75-AgNPs were studied via ultraviolet–visible (UV–vis) spectroscopy (TECAN, Austria) in the wavelength range of 320–700 nm after dispersion via ultrasonication. The particle size, size distribution, and zeta potential were determined using dynamic light scattering (DLS, Malvern, UK). The morphology, size, and distribution of the BF75-AgNPs were observed using transmission electron microscopy (TEM, Philip, Netherlands) and scanning electron microscopy (SEM, ZEISS, Germany). In addition, energy dispersive X-ray (EDX) spectroscopy was carried out to confirm the presence of elemental silver in the sample. Atomic force microscopy (AFM, Bruker, Germany) was used to observe the two-dimensional (2D) and three-dimensional (3D) morphologies of the particle surface and determine its height distribution. The crystal structure of the BF75-AgNPs was analyzed via X-ray diffraction (XRD, Bruker, Germany). A Fourier-transform infrared (FTIR) spectrometer (Thermo, USA) was used for analysis in the range of 400–4000 cm−1 with a 4 cm−1 resolution to determine various functional groups in the BF75-AgNPs.
Antibacterial Effects of the BF75-AgNPs
Minimum Inhibitory Concentration (MIC) and Minimal Bactericidal Concentration (MBC) Measurements
The antibacterial spectrum of the BF75-AgNPs was evaluated using MIC and MBC measurements. The MIC assay was conducted as previously described, with slight modifications.32 Briefly, bacteria (1 × 106 CFU/mL) were mixed with the BF75-AgNPs (the final concentration ranged from 0.98 to 250 µg/mL). The mixture was incubated at 37 °C, and the results were determined by observing the presence or absence of turbidity with transmitted light. The first well with no microbial growth was defined as the MIC (μg/mL). The mixture was separately spread on MH agar and incubated overnight at 37 °C. The lowest concentration at which growth was visibly inhibited was defined as the MBC. The MBC/MIC ratio was calculated to determine the mode of activity of the BF75-AgNPs. The mode was bactericidal when the score was <4; otherwise, it was bacteriostatic.36
Live/Dead Staining Assay and Field-Emission Scanning Electron Microscopy (FE-SEM)
To determine the antibacterial activity of the BF75-AgNPs, five XDR-KP strains were used for live/dead cell staining and FE-SEM analysis. After incubation at 37 °C for 16 h, the bacteria were washed three times and re-suspended in 1 × PBS (pH = 7.4). The bacterial solution was treated with the BF75-AgNPs at 1/2 × MIC for 4 h. Ultra-pure water served as a negative control. For live/dead staining, the treated suspension was stained using the Live/Dead BacLight Bacterial Viability Kit L7012 (Invitrogen, USA). Imaging was performed using a fluorescence microscope (Zena, Germany). For the FE-SEM analysis, the fixed and dehydrated bacterial solution samples were dried overnight at room temperature, coated with gold, and imaged via FE-SEM (Hitachi, Japan).
ROS Assay
After five XDR-KP strains were treated with the BF75-AgNPs at 1/2 × MIC for 4 h, the bacteria were cultured with DCFH-DA (Beyotime, China) in the dark for 1 h to determine ROS production. Changes in bacterial ROS expression were observed using fluorescence microscopy and analyzed via flow cytometry (Dxp Athena™, Cytek, USA).
Checkerboard Assay
A checkerboard assay was used to determine the synergistic effect of the BF75-AgNPs and colistin against two colistin-resistant KP strains.37 The initial concentrations of the BF75-AgNPs and colistin were 125 and 64 µg/mL, respectively. Two-fold serial dilutions of the BF75-AgNPs were prepared in horizontal rows, and two-fold serial dilutions of colistin were prepared in vertical rows using a 96-well plate. The plates were prepared well by well to obtain a single plate in which both the antimicrobial agents were cross-diluted. Colistin-resistant XDR-KP strains were cultured in the exponential phase in an MH broth medium. The final inoculum used was 5×105 CFU/mL. The plate was incubated at 37 °C for 18 h. Fractional inhibitory concentration index (FICI) values were defined as the lowest concentration at which a combination of the BF75-AgNPs and colistin inhibited bacterial growth. The FICI was calculated using the following formula:
The FICI values were interpreted as follows: FICI ≤ 0.5 (synergy), 0.5 < FICI ≤ 4.0 (no interaction), and FICI > 4.0 (antagonism).38,39
Antibiofilm Activity of the BF75-AgNPs
Effect of the BF75-AgNPs on Biofilm Formation
Strong biofilm-forming KP strains were used to verify the antibiofilm activity of the BF75-AgNPs. Briefly, the overnight culture was diluted with the respective medium to a final concentration of 1×106 CFU/mL. The inoculum was added to a 96-well plate and incubated at 37 °C for 6 h. The medium was then replaced with a fresh medium containing various concentrations of the BF75-AgNPs (7.5–120 μg/mL), and the samples were incubated at 37 °C for an additional 18 h (bringing the total age of the biofilm to 24 h). The inhibition rate of biofilm formation was calculated using Equation (1).
Effect of the BF75-AgNPs on Mature Biofilms
Strong biofilm-forming XDR-KP strains were seeded on 18 mm cell slides for biofilm culture. The bacterial colony counting assay and live/dead staining assay were performed as described in a previous study, with slight modifications.35 Briefly, an overnight grown bacterial culture was diluted with fresh MH broth to prepare a final inoculum of 1×106 CFU/mL. Approximately 200 µL of bacterial inoculum was loaded onto 18 mm cell slides and incubated at 37 °C without any disruption of biofilm formation. After 24 h, the culture medium was replaced with fresh MH broth containing sterile water, 60 μg/mL BF75-AgNPs, or 120 μg/mL BF75-AgNPs and incubated for 24 h.
After exposure to the BF75-AgNPs, each well of the biofilm was collected in 5 mL of sterile normal saline and homogenized via sonication. The homogenized biofilm suspension (100 µL) was serially diluted, plated on MH broth agar, and incubated overnight at 37 °C. The number of colonies was counted to determine the viability of the bacterial cells.
Mature biofilms treated with the BF75-AgNPs were stained using a Live/Dead BacLight Bacterial Viability Kit (Invitrogen, USA) after three rinses with saline, and the bacterial solution was imaged using a fluorescence microscope (Zeiss, Germany).
Evaluation of the Antitumor Activity of the BF75-AgNPs
3-[4,5-Dimethylthiazol-2-Yl]-2,5 Diphenyl Tetrazolium Bromide (MTT) Assay
The 50% inhibitory concentration (IC50) of the BF75-AgNPs was determined using an MTT assay (Beyotime, China).40 In brief, cells (1 × 104 cells/well) were seeded into 96-well plates. After incubation for 24 h, the cells were treated with various concentrations of AgNPs for 24 h. MTT was then added to each well, followed by incubation for an additional 4 h. Finally, 150 μL of dimethyl sulfoxide (DMSO) was added to each well and gently mixed. The OD490 nm was used to calculate the percentage of viable cells.
Treat indicates treatment with AgNPs, and NC indicates treatment with the same amount of ultrapure water.
Apoptosis Assay
To measure the percentage of cells undergoing apoptosis/necrosis, flow cytometry analysis using an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Staining Apoptosis Detection Kit (Beyotime, China) was performed. After the relevant stimulation, cells (1 × 104 cells/well) in each group were collected, washed with PBS, and stained with 10 μL of Annexin V-FITC and 5 μL of PI for 15 min. The stained cells were detected using a Cytek Dxp Athena flow cytometer (Cytek, USA).41
Statistical Analysis
All experiments were performed in triplicate. Data are presented as the mean ± standard deviation. Analyses were performed using IBM SPSS Statistics 20, GraphPad Prism 9, ImageJ, R 4.0.3, and FlowJo 10.6.2. Intergroup differences were estimated by one-way analysis of variance, followed by post-hoc multiple comparison (Tukey’s test). Values were considered statistically significant at P < 0.05.
Results and Discussion
Identification of P. aeruginosa PA75
MALDI-TOF-MS analysis revealed that the PA75 strain was P. aeruginosa (Figure 1A). The 16S rDNA PCR product fragment size of P. aeruginosa PA75 was 1458 bp (Figure 1B), and the sequence of 16S rDNA was submitted to GenBank (accession number ON241930). In the phylogenetic tree, P. aeruginosa PA75 clustered within the genus Pseudomonas and was most closely related to the P. aeruginosa DSM 50071 strain (accession number NR_117678.1, Figure 1C).
 |
Figure 1 Identification of P. aeruginosa PA75. (A) MALDI-TOF-MS of PA75; (B) Agarose electrophoresis of 16S rDNA PCR products of P. aeruginosa PA75; (C) the phylogenetic tree of P. aeruginosa PA75. |
Extraction of PA75 Biofilm Supernatant and Antibiofilm Activity
The P. aeruginosa PA75 biofilm supernatant was light green, clear, and transparent (Figure 2A). The growth curve assay results showed that the growth of S. epidermidis ATCC35984 in the group treated with the P. aeruginosa PA75 biofilm supernatant was significantly lower than that in the non-treated group (P < 0.001, Figure 2B). The biofilm inhibition assay showed that the OD590nm of S. epidermidis ATCC35984 in the treated group (0.126 ± 0.002) was significantly lower than that in the non-treated group (3.218 ± 0.028, P < 0.001), with an inhibition rate of 80.43% (Figure 2C). Therefore, we chose the P. aeruginosa PA75 biofilm supernatant as the biological material to synthesize AgNPs.
Green Synthesis of BF75-AgNPs
In this study, BF75-AgNPs were synthesized using P. aeruginosa PA75 biofilm supernatant. The color change from pale yellow to sepia preliminarily indicated the formation of AgNPs (Figure 3A). After freeze-drying and weighing, the concentration of the synthetic BF75-AgNPs stock was found to be 1225 μg/mL (Figure 3B). AgNPs have a special optical property, the surface plasmon resonance (SPR) effect, which causes them to exhibit specific absorption of natural light.42 The UV–vis characterization of the BF75-AgNPs exhibited a distinct absorption peak at 412 nm, suggesting the generation of BF75-AgNPs.10 The UV–vis absorption peak exhibited no specific change after 30 d at 4 °C, indicating that the BF75-AgNPs had stable properties (Figure 3C).
To determine the optimal conditions, four variables, including the reaction temperature (4, 25, 37, and 45 °C), reaction time (1–72 h), final AgNO3 concentration (1–7 mmol/L), and solution pH (5.0, 7.0, and 9.0) were adjusted, and the spectral changes were recorded. The UV–vis absorption peak gradually increased with increasing temperature (Figure S6) and time (Figure S7). The color of the reaction solution was the darkest at 48 h, and the UV–vis absorption peak reached a maximum at 72 h; however, their rate of change was the highest during the first 24 hours. The UV–vis absorption peak was the largest when the final concentration of AgNO3 was 5 mM (Figure S8). Under different pH conditions, the UV–vis absorption spectra were similar (Figure S9). In contrast to the previous study, the pH change had no significant effect on the synthesis of the BF75-AgNPs.43 The current study found that maximum yields were obtained at 37 °C after 24 h of incubation using 5 mM AgNO3. Changes in these key parameters can disturb the stability of BF75-AgNPs and agglomerate them. Therefore, the biofilm supernatant could be used as a reducing and dispersing agent, and the BF75-AgNPs were synthesized within 24 h without harsh temperature conditions and catalyst assistance.
Characterization of the BF75-AgNPs
The DLS results demonstrated that the average diameter of the BF75-AgNPs was 105.7 ± 40.1 nm, and their polydispersity index (PDI) was 0.168 (Figure 4A). The stability of the BF75-AgNPs was further analyzed via zeta potential measurements, and the average zeta potential was −31.0 ± 8.1 mV (Figure 4B). The PDI and zeta potential values confirmed the good dispersion and excellent stability of the BF75-AgNPs.44 TEM analysis showed that the BF75-AgNPs were spherical or nearly spherical with good dispersibility (Figure 4C).45 All BF75-AgNPs displayed in the TEM images were selected for particle size analysis, and the average particle size of the BF75-AgNPs was 13.899 ± 4.036 nm (Figure 4D).
SEM (Figure 5A) analysis also confirmed that the BF75-AgNPs were spherical or nearly spherical, with a small particle size and concentrated distribution. The EDX spectrometer of the BF75-AgNPs showed a signal at 3 keV corresponding to the silver atom (Figure 5B and C). Through AFM analysis, 2D and 3D surface topography images of BF75-AgNPs with sizes of 10.5–32.3 nm were constructed (Figure 5D). Corresponding to the silver standard card (Joint Committee Powder Diffraction Standards No. 04–0783), XRD analysis of the BF75-AgNPs revealed four typical silver diffraction peaks at 2θ angles of 38.34°, 44.02°, 64.53°, and 77.16° corresponding to the (111), (200), (220), and (311) planes, respectively (Figure 5E), indicating that the BF75-AgNPs had a typical face-centered cubic crystal structure. Weak diffraction peaks existed at 2θ angles of 32.22°, 46.25°, 54.76°, and 57.35° corresponding to the structure of an AgCl crystal, indicating the presence of a trace amount of AgCl crystals mixed in the BF75-AgNPs. FTIR analysis was used to examine the surface properties of the purified BF75-AgNPs. The FTIR spectrum revealed the presence of bands at 3271 cm−1 for the amide NH stretching vibration, 1646 cm−1 for the amide I C=O stretching vibration, 1538 cm−1 for the amide II C-N stretching vibration, and 1396 and 1234 cm−1 for the amide C-N stretching vibration (Figure 5F). Furthermore, we observed peaks at 2924 cm−1 for the C-H stretching vibration and 1454 cm−1 for the C-H bending vibration. The main peaks in the FTIR pattern suggested the existence of multiple amide bonds, indicating the presence of proteins as reducing and stabilizing agents in the biofilm supernatant. These active protein fractions may be adsorbed on the surfaces of the BF75-AgNPs via free amine groups, which promote their stability and participate in important biological functions.46
Antibacterial Effects of BF75-AgNPs
Multidrug-resistant bacteria were used to detect the broad-spectrum antibacterial activity of the BF75-AgNPs. The MIC values of gram-negative and gram-positive bacteria showed excellent consistency (7.81–15.63 μg/mL), while the MBC values against gram-negative bacteria (15.63–31.25 μg/mL) were significantly (P < 0.0001) lower than those against the five MRSA strains (62.50–250.00 μg/mL, Table 1). The bactericidal effect on gram-negative bacteria was better than that on gram-positive bacteria, which may be attributed to the different structures of the MRSA cell walls. Other studies have also demonstrated that the sensitivity of gram-negative bacteria to AgNPs is higher than that of gram-positive bacteria.47,48
 |
Table 1 Antibacterial Effects of BF75-AgNPs Against Four Different Multidrug-Resistant Strains |
The size of nanoparticles plays a crucial role in their antimicrobial activity because smaller nanoparticles are more easily internalized through cell membranes. The small size of the BF75-AgNPs allowed them to easily enter and destroy the bacterial cells. To determine the antibacterial activity of the BF75-AgNPs, five XDR-KP strains were used as the research strains for live/dead staining and FE-SEM analysis. Fluorescence microscopy demonstrated that the proportion of dead bacteria in the AgNP group was higher than that in the control group (Figure 6A). FE-SEM results showed shrinkage, deformation, and rupture of the cell membranes as well as leakage of the cellular contents of the five XDR-KP strains after BF75-AgNP treatment (Figure 6B). Although the antibacterial mechanism of AgNPs has not been fully elucidated, the three most likely antibacterial mechanisms are as follows: (a) AgNPs release free Ag+, which is absorbed by bacteria, and then disrupt bacterial ATP production and DNA replication; (b) AgNPs promote the formation of pores in the bacterial cell wall and directly destroy the permeability of the cell membrane, leading to the lysis and death of bacteria, and; (c) AgNPs and Ag+ induce bacterial ROS production and oxidative stress.49
In addition, AgNPs can increase the production of active oxidizing species in bacteria, which further damages cellular components and ultimately leads to bacterial death.35 Flow cytometry (Figure 7A) and fluorescence microscopy (Figure 7B) results demonstrated that the ROS expression of the five XDR-KP strains significantly increased after exposure to the BF75-AgNPs. Further analysis of the average fluorescence intensity detected via flow cytometry demonstrated that the expression levels of ROS in the five strains in the treated groups were significantly (P < 0.0001) higher than those in the control group (Figure 7C). In conclusion, this study suggests that BF75-AgNPs can achieve antibacterial activity by increasing the ROS level of bacteria and disturbing the bacterial structure.
As shown in Table 2, the checkerboard assay results indicated a synergistic effect between colistin and the BF75-AgNPs, with potential antibacterial activity against two XDR-KP strains (KP4 and KP5 with FICI values of 0.281 and 0.187, respectively). The sensitivity of colistin-resistant XDR-KP to colistin significantly increased (the maximum increase was 32 times). Therefore, the BF75-AgNPs synthesized in this study may be used as a new antibacterial agent for the treatment of colistin-resistant K. pneumoniae infection, with potential clinical application to decrease the incidence of colistin-resistant bacteria and reduce the side effects of colistin in patients with poor kidney function.
 |
Table 2 MICs of BF75-AgNPs and Colistin (Alone or Combination) and FICIs of BF75-AgNPs in Combination with Colistin |
Antibiofilm Activity of BF75-AgNPs
Mature biofilms are impenetrable to most antimicrobial agents and represent a major cause of various chronic infections that are resistant to antibiotics. XDR-KP strains with strong biofilm-forming abilities exist, and chronic infection caused by XDR-KP should not be ignored. Therefore, we tested the effect of the BF75-AgNPs on the biofilm-forming abilities and subsequent antimicrobial effects of strong biofilm-forming XDR-KP strains. When the concentration of the BF75-AgNPs exceeded 4 × MIC (60 μg/mL), the inhibition rate of the BF75-AgNPs on XDR-KP biofilm formation was greater than 80%, which was higher than previously reported levels (Figure 8A).50 When mature biofilms of KP3, KP4, and KP5 strains were treated with the BF75-AgNPs at concentrations of 60 and 120 μg/mL, the number of bacteria in the biofilms significantly decreased (Figure 8B). Meanwhile, the proportion of dead bacteria in the KP3, KP4, and KP5 strain biofilms significantly increased while the proportion of viable bacteria significantly decreased when the concentration of the BF75-AgNPs reached 120 μg/mL (Figure 8C). In conclusion, we found that BF75-AgNPs have a strong antibiofilm effect on XDR-KP strains, and the number of viable bacteria decrease with increasing BF75-AgNP concentration. The concentration dependence of AgNP antibiofilm activity had been confirmed in E. coli and P. aeruginosa in previous studies.47,48 In addition, the live/dead staining images confirmed the prominent destabilization effect of AgNPs on the XDR-KP strain.47
Evaluation of the Antitumor Activity of BF75-AgNPs
The cytotoxic potential of the BF75-AgNPs against SK28 and A375 human melanoma cells was evaluated using the MTT assay. The IC50 values of the BF75-AgNPs in SK28 and A375 cell lines were 4.75 and 7.76 μg/mL, respectively (Figure 9A). When the concentration of BF75-AgNPs was 4 μg/mL, the inhibitory rates of the BF75-AgNPs on SK28 and A375 melanoma cells were 50.24% and 20.24%, respectively. At 8 μg/mL, the inhibitory rates further increased to 60.24% and 56.26%. The cytotoxicity of the BF75-AgNPs in both melanoma cell lines was dose dependent. The BF75-AgNPs were tested on normal human keratinocyte HaCaT cells to determine their cytotoxicity. The IC50 value in the HaCaT cell lines was 54.37 μg/mL. When the concentration of the BF75-AgNPs was 10 μg/mL, the inhibition rate of the BF75-AgNPs on HaCaT normal epithelial cells was 15.59%. The results revealed that the cytotoxicity of the BF75-AgNPs to HaCaT cells was negligible at concentrations ≤ 10 μg/mL.
The morphological changes in cells treated with the BF75-AgNPs at 4 μg/mL were observed under a microscope (Figure 9B). Compared with the control group, the morphology of HaCaT cells treated with the BF75-AgNPs did not change significantly, while the SK28 and A375 melanoma cells in the treated group showed rounded and pyknotic cells. Cell morphology assays further verified the cytotoxicity of the BF75-AgNPs at 4 μg/mL in melanoma cell lines but not in normal epidermal cells. In addition, residual cell debris was observed in the treated SK28 cells after cell division. The difference between the cytotoxic effects of AgNPs on tumor cells and normal cells may be attributed to the fact that tumor cells are metabolically active and easily absorb surrounding AgNPs, whereas normal cells require time to phagocytose foreign substances using their cellular mechanisms.26
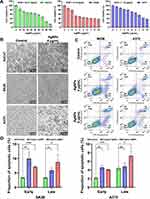
Annexin V/PI double staining was used to evaluate the effect of the BF75-AgNPs on apoptosis. To evaluate the apoptosis-inducing activity of the BF75-AgNPs in SK28 and A375 cells comprehensively, two concentrations covering the IC50 (4 and 8 μg/mL) were selected for the apoptosis assay. One of the three independent studies is shown in Figure 9C. Compared with the control group, the percentage of apoptotic cells in all BF75-AgNP-treated groups increased. The difference in apoptotic cells in the Q2 and Q3 quadrants of SK28 and A375 cells was further analyzed (Figure 9D). Compared with the control group, early apoptosis (P = 0.008 and P < 0.001) and late apoptosis (P = 0.009 and P = 0.026) of SK28 cells treated with BF75-AgNPs at 4 and 8 μg/mL increased significantly. For A375 cells, early apoptosis increased significantly at both concentrations (P = 0.002 and P < 0.001), whereas late apoptosis increased significantly at 8 μg/mL (P = 0.011), and the increase in late apoptosis at 4 μg/mL was not significant (P = 0.478). The decreased proportion of early apoptotic cells when the concentration of BF75-AgNPs was 8 μg/mL may be attributed to the induction of early apoptotic cells to late apoptotic cells with the increase in the BF75-AgNP concentration. These results indicated that BF75-AgNPs could induce early and late apoptosis in SK28 and A375 melanoma cell lines and that the high concentration of BF75-AgNPs had a stronger apoptosis-inducing effect on melanoma cells. Moreover, at a concentration of as low as 10 μg/mL, BF75-AgNPs can simultaneously exhibit antibacterial, synergistic, and antitumor effects and have low toxicity to normal cells. Currently, no such drugs with both antibacterial and antitumor effects are used in clinical practice, and the BF75-AgNPs synthesized in this study may fill this gap, giving BF75-AgNPs broader clinical application prospects.
Conclusions
In this study, the biofilm supernatant of P. aeruginosa PA75 was used as a reducing agent, stabilizer, and dispersant to facilitate the green synthesis of BF75-AgNPs. The BF75-AgNPs exhibited broad-spectrum utility against multidrug resistant strains, especially against gram-negative pathogens. The BF75-AgNPs had a strong bactericidal effect on XDR-KP cells, and the underlying mechanism may be an increase in bacterial ROS expression. The BF75-AgNPs also synergized with colistin in colistin-resistant XDR-KP. The BF75-AgNPs inhibited the formation of XDR-KP biofilms and had bactericidal effects on bacterial cells in the biofilm. In addition, the BF75-AgNPs showed low cytotoxicity to normal human epidermal cells but strong cytotoxicity to melanoma cells, with the mechanism attributed to cell apoptosis. The novel AgNPs synthesized in this study provide new directions for antibacterial and antibiofilm applications against bacteria with extensive drug resistance as well as antimelanoma applications.
Acknowledgments
All authors are grateful to their respective institutions for their support. We thank Dr. Qian Gao, Department of Clinical Laboratory, Xiangya Hospital, Changsha, China for kindly providing human normal keratinocyte HaCaT cells. We would like to thank Editage (www.editage.cn) for English language editing.
Funding
This work was funded by the Natural Science Funding of Hunan Province under Grant (Nos. 2020JJ5901, 2021JJ40980, and 2021JJ40624).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
2. Liu X, Wu Y, Zhu Y, et al. Emergence of colistin-resistant hypervirulent Klebsiella pneumoniae (CoR-HvKp) in China. Emerg Microbes Infect. 2022;11(1):648–661. doi:10.1080/22221751.2022.2036078
3. Collaborators AR, Ikuta KS, Sharara F. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi:10.1016/S0140-6736(21)02724-0
4. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi:10.1016/S1473-3099(17)30753-3
5. Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. Infectious Diseases Society of America 2022 guidance on the treatment of Extended-Spectrum β-Lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin Infect Dis. 2022;75(2):187–212. doi:10.1093/cid/ciac268
6. Zhang S, Abbas M, Rehman MU, et al. Updates on the global dissemination of colistin-resistant Escherichia coli: an emerging threat to public health. Sci Total Environ. 2021;799:149280. doi:10.1016/j.scitotenv.2021.149280
7. Huang PH, Chen WY, Chou SH, Wang FD, Lin YT. Risk factors for the development of colistin resistance during colistin treatment of carbapenem-resistant Klebsiella pneumoniae infections. Microbiol Spectr. 2022;10(3):e0038122. doi:10.1128/spectrum.00381-22
8. Noralian Z, Gashti MP, Moghaddam MR, Tayyeb H, Erfanian I. Ultrasonically developed silver/iota-carrageenan/cotton bionanocomposite as an efficient material for biomedical applications. Int J Biol Macromol. 2021;180:439–457. doi:10.1016/j.ijbiomac.2021.02.204
9. Mo F, Zhou Q, He Y. Nano-Ag: environmental applications and perspectives. Sci Total Environ. 2022;829:154644. doi:10.1016/j.scitotenv.2022.154644
10. Kulkarni RR, Shaiwale NS, Deobagkar DN, Deobagkar DD. Synthesis and extracellular accumulation of silver nanoparticles by employing radiation-resistant Deinococcus radiodurans, their characterization, and determination of bioactivity. Int J Nanomedicine. 2015;10:963–974. doi:10.2147/IJN.S72888
11. Elgorban AM, El-Samawaty AE-RM, Yassin MA, et al. Antifungal silver nanoparticles: synthesis, characterization and biological evaluation. Biotechnol Biotechnol Equip. 2016;30(1):56–62. doi:10.1080/13102818.2015.1106339
12. Khan M, Shaik MR, Adil SF, et al. Plant extracts as green reductants for the synthesis of silver nanoparticles: lessons from chemical synthesis. Dalton Trans. 2018;47(35):11988–12010. doi:10.1039/C8DT01152D
13. Mondal AH, Yadav D, Mitra S, Mukhopadhyay K. Biosynthesis of silver nanoparticles using culture supernatant of shewanella sp. ARY1 and their antibacterial activity. Int J Nanomedicine. 2020;15:8295–8310. doi:10.2147/IJN.S274535
14. Adil SF, Assal ME, Khan M, Al-Warthan A, Siddiqui MR, Liz-Marzán LM. Biogenic synthesis of metallic nanoparticles and prospects toward green chemistry. Dalton Trans. 2015;44(21):9709–9717. doi:10.1039/C4DT03222E
15. Marslin G, Siram K, Maqbool Q, et al. Secondary metabolites in the green synthesis of metallic nanoparticles. Materials. 2018;11(6):940–964. doi:10.3390/ma11060940
16. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi:10.1038/nrmicro2415
17. Davies DG, Chakrabarty AM, Geesey GG. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59(4):1181–1186. doi:10.1128/aem.59.4.1181-1186.1993
18. Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):1487. doi:10.1126/science.295.5559.1487
19. O’Neill E, Pozzi C, Houston P, et al. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190(11):3835–3850. doi:10.1128/JB.00167-08
20. Thöming JG, Häussler S. Pseudomonas aeruginosa is more tolerant under biofilm than under planktonic growth conditions: a multi-isolate survey. Front Cell Infect Microbiol. 2022;12:851784. doi:10.3389/fcimb.2022.851784
21. Waite RD, Papakonstantinopoulou A, Littler E, Curtis MA. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J Bacteriol. 2005;187(18):6571–6576. doi:10.1128/JB.187.18.6571-6576.2005
22. Gahlawat G, Shikha S, Chaddha BS, Chaudhuri SR, Mayilraj S, Choudhury AR. Microbial glycolipoprotein-capped silver nanoparticles as emerging antibacterial agents against cholera. Microb Cell Fact. 2016;15:25–38. doi:10.1186/s12934-016-0422-x
23. Khazaei Z, Ghorat F, Jarrahi AM, Adineh HA, Sohrabivafa M, Goodarzi E. Global incidence and mortality of skin cancer by histological subtype and its relationship with the human development index (HDI); an ecology study in 2018. WCRJ. 2019;6:e1265.
24. Kuang X, Wang Z, Luo Z, et al. Ag nanoparticles enhance immune checkpoint blockade efficacy by promoting of immune surveillance in melanoma. J Colloid Interface Sci. 2022;616:189–200. doi:10.1016/j.jcis.2022.02.050
25. Nayak D, Kumari M, Rajachandar S, Ashe S, Thathapudi NC, Nayak B. Biofilm impeding AgNPs target skin carcinoma by inducing mitochondrial membrane depolarization mediated through ROS production. ACS Appl Mater Interfaces. 2016;8(42):28538–28553. doi:10.1021/acsami.6b11391
26. Gholami N, Cohan RA, Razavi A, Bigdeli R, Dashbolaghi A, Asgary V. Cytotoxic and apoptotic properties of a novel nano-toxin formulation based on biologically synthesized silver nanoparticle loaded with recombinant truncated pseudomonas exotoxin A. J Cell Physiol. 2020;235(4):3711–3720. doi:10.1002/jcp.29265
27. Pasha A, Kumbhakar DV, Sana SS, et al. Role of biosynthesized Ag-NPs using aspergillus niger (MK503444.1) in antimicrobial, anti-cancer and anti-angiogenic activities. Front Pharmacol. 2021;12:812474. doi:10.3389/fphar.2021.812474
28. Nayak D, Thathapudi NC, Ashe S, Nayak B. Bioengineered ethosomes encapsulating AgNPs and Tasar silk sericin proteins for non melanoma skin carcinoma (NMSC) as an alternative therapeutics. Int J Pharm. 2021;596:120265. doi:10.1016/j.ijpharm.2021.120265
29. Jagtap RR, Garud A, Puranik SS, et al. Biofabrication of silver nanoparticles (AgNPs) using embelin for effective therapeutic management of lung cancer. Front Nutr. 2022;9:960674. doi:10.3389/fnut.2022.960674
30. Gao J, Wang Y, Wang CW, Lu BH. First report of bacterial root rot of ginseng caused by Pseudomonas aeruginosa in China. Plant Dis. 2014;98(11):1577. doi:10.1094/PDIS-03-14-0276-PDN
31. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi:10.1093/oxfordjournals.molbev.a040454
32. Cavassin ED, de Figueiredo LF, Otoch JP, et al. Comparison of methods to detect the in vitro activity of silver nanoparticles (AgNP) against multidrug resistant bacteria. J Nanobiotechnology. 2015;13:64–79. doi:10.1186/s12951-015-0120-6
33. Bendaoud M, Vinogradov E, Balashova NV, Kadouri DE, Kachlany SC, Kaplan JB. Broad-spectrum biofilm inhibition by Kingella kingae exopolysaccharide. J Bacteriol. 2011;193(15):3879–3886. doi:10.1128/JB.00311-11
34. Karwacki MT, Kadouri DE, Bendaoud M, et al. Antibiofilm activity of Actinobacillus pleuropneumoniae serotype 5 capsular polysaccharide. PLoS One. 2013;8(5):e63844. doi:10.1371/journal.pone.0063844
35. Singh P, Pandit S, Garnæs J, et al. Green synthesis of gold and silver nanoparticles from Cannabis sativa (industrial hemp) and their capacity for biofilm inhibition. Int J Nanomedicine. 2018;13:3571–3591. doi:10.2147/IJN.S157958
36. Konaté K, Mavoungou JF, Lepengué AN, et al. Antibacterial activity against β- lactamase producing Methicillin and Ampicillin-resistants Staphylococcus aureus: Fractional Inhibitory Concentration Index (FICI) determination. Ann Clin Microbiol Antimicrob. 2012;11:18–29. doi:10.1186/1476-0711-11-18
37. Allend SO, Garcia MO, da Cunha KF, et al. Biogenic silver nanoparticle (Bio-AgNP) has an antibacterial effect against carbapenem-resistant Acinetobacter baumannii with synergism and additivity when combined with polymyxin B. J Appl Microbiol. 2022;132(2):1036–1047. doi:10.1111/jam.15297
38. Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 2003;52(1):1. doi:10.1093/jac/dkg301
39. Wesseling CMJ, Martin NI. Synergy by perturbing the gram-negative outer membrane: opening the door for gram-positive specific antibiotics. ACS Infect Dis. 2022;8(9):1731–1757. doi:10.1021/acsinfecdis.2c00193
40. Ahmed T, Shahid M, Noman M, et al. Bioprospecting a native silver-resistant Bacillus safensis strain for green synthesis and subsequent antibacterial and anticancer activities of silver nanoparticles. J Adv Res. 2020;24:475–483. doi:10.1016/j.jare.2020.05.011
41. Almalki MA, Khalifa AYZ. Silver nanoparticles synthesis from Bacillus sp KFU36 and its anticancer effect in breast cancer MCF-7 cells via induction of apoptotic mechanism. J Photochem Photobiol B. 2020;204:111786. doi:10.1016/j.jphotobiol.2020.111786
42. Singh P, Kim YJ, Singh H, et al. Biosynthesis, characterization, and antimicrobial applications of silver nanoparticles. Int J Nanomedicine. 2015;10:2567–2577. doi:10.2147/IJN.S72313
43. Iqtedar M, Aslam M, Akhyar M, Shehzaad A, Abdullah R, Kaleem A. Extracellular biosynthesis, characterization, optimization of silver nanoparticles (AgNPs) using Bacillus mojavensis BTCB15 and its antimicrobial activity against multidrug resistant pathogens. Prep Biochem Biotechnol. 2019;49(2):136–142. doi:10.1080/10826068.2018.1550654
44. Khan T, Yasmin A, Townley HE. An evaluation of the activity of biologically synthesized silver nanoparticles against bacteria, fungi and mammalian cell lines. Colloids Surf B Biointerfaces. 2020;194:111156. doi:10.1016/j.colsurfb.2020.111156
45. Riaz Rajoka MS, Mehwish HM, Zhang H, et al. Antibacterial and antioxidant activity of exopolysaccharide mediated silver nanoparticle synthesized by Lactobacillus brevis isolated from Chinese koumiss. Colloids Surf B Biointerfaces. 2020;186:110734. doi:10.1016/j.colsurfb.2019.110734
46. Gurunathan S, Kalishwaralal K, Vaidyanathan R, et al. Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli. Colloids Surf B Biointerfaces. 2009;74(1):328–335. doi:10.1016/j.colsurfb.2009.07.048
47. Singh P, Pandit S, Jers C, Joshi AS, Garnæs J, Mijakovic I. Silver nanoparticles produced from Cedecea sp. exhibit antibiofilm activity and remarkable stability. Sci Rep. 2021;11(1):12619. doi:10.1038/s41598-021-92006-4
48. Zamanpour N, Mohammad Esmaeily A, Mashreghi M, Shahnavaz B, Reza Sharifmoghadam M, Kompany A. Application of a marine luminescent Vibrio sp. B4L for biosynthesis of silver nanoparticles with unique characteristics, biochemical properties, antibacterial and antibiofilm activities. Bioorg Chem. 2021;114:105102. doi:10.1016/j.bioorg.2021.105102
49. Prasath S, Palaniappan K. Is using nanosilver mattresses/pillows safe? A review of potential health implications of silver nanoparticles on human health. Environ Geochem Health. 2019;41(5):2295–2313. doi:10.1007/s10653-019-00240-7
50. Siddique MH, Aslam B, Imran M, et al. Effect of silver nanoparticles on biofilm formation and EPS production of multidrug-resistant Klebsiella pneumoniae. Biomed Res Int. 2020;2020:6398165. doi:10.1155/2020/6398165
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.