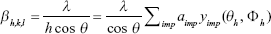Back to Journals » International Journal of Nanomedicine » Volume 14
Biomimetic hydroxyapatite nanocrystals are an active carrier for Salmonella bacteriophages
Authors Fulgione A, Ianniello F, Papaianni M, Contaldi F, Sgamma T , Giannini C, Pastore S , Velotta R , Della Ventura B, Roveri N, Lelli M, Capuano F, Capparelli R
Received 9 October 2018
Accepted for publication 15 January 2019
Published 28 March 2019 Volume 2019:14 Pages 2219—2232
DOI https://doi.org/10.2147/IJN.S190188
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Andrea Fulgione,1,* Flora Ianniello,1,* Marina Papaianni,1 Felice Contaldi,1 Tiziana Sgamma,2 Cinzia Giannini,3 Stella Pastore,3 Raffaele Velotta,4 Bartolomeo Della Ventura,5 Norberto Roveri,6 Marco Lelli,6 Federico Capuano,7 Rosanna Capparelli1
1Department of Agriculture, University of Naples “Federico II”, Portici, Naples, Italy; 2Biomolecular Technology Group, School of Allied Health Sciences, De Montfort University, Leicester, UK; 3Istituto di Cristallografia, Consiglio Nazionale delle Ricerche, Bari, Italy; 4Department of Physics “Ettore Pancini”, University of Naples “Federico II”, Portici, Naples, Italy; 5Department of Physics, Politecnico di Milano, Milano, Italy; 6Chemical Center Srl, Granarolo dell’Emilia, Bologna, Italy; 7Department of Food Microbiology, Istituto Zooprofilattico Sperimentale del Mezzogiorno, Portici, Naples, Italy
*These authors contributed equally to this work
Purpose: The use of bacteriophages represents a valid alternative to conventional antimicrobial treatments, overcoming the widespread bacterial antibiotic resistance phenomenon. In this work, we evaluated whether biomimetic hydroxyapatite (HA) nanocrystals are able to enhance some properties of bacteriophages. The final goal of this study was to demonstrate that biomimetic HA nanocrystals can be used for bacteriophage delivery in the context of bacterial infections, and contribute – at the same time – to enhance some of the biological properties of the same bacteriophages such as stability, preservation, antimicrobial activity, and so on.
Materials and methods: Phage isolation and characterization were carried out by using Mitomycin C and following double-layer agar technique. The biomimetic HA water suspension was synthesized in order to obtain nanocrystals with plate-like morphology and nanometric dimensions. The interaction of phages with the HA was investigated by dynamic light scattering and Zeta potential analyses. The cytotoxicity and intracellular killing activities of the phage–HA complex were evaluated in human hepatocellular carcinoma HepG2 cells. The bacterial inhibition capacity of the complex was assessed on chicken minced meat samples infected with Salmonella Rissen.
Results: Our data highlighted that the biomimetic HA nanocrystal–bacteriophage complex was more stable and more effective than phages alone in all tested experimental conditions.
Conclusion: Our results evidenced the important contribution of biomimetic HA nanocrystals: they act as an excellent carrier for bacteriophage delivery and enhance its biological characteristics. This study confirmed the significant role of the mineral HA when it is complexed with biological entities like bacteriophages, as it has been shown for molecules such as lactoferrin.
Keywords: biomimetic hydroxyapatite nanocrystals, Salmonella Rissen, bacteriophages, antibiotic resistance
Introduction
Inappropriate abuse of antibiotics has led to the development of drug-resistant microorganisms.1 Antibiotic resistance is a constantly evolving phenomenon and represents a serious problem with high death toll and a substantial economic impact worldwide; it complicates patient management and treatment strategy and prolongs hospital stays. In Europe antibiotic-resistant bacteria infect 4 million people every year.2,3 Therefore, therapies that can serve as an alternative to antibiotics need to be developed.
A valid and alternative approach to solve the widespread phenomenon of “antibiotic resistance” of different pathogens such as Salmonella,4 Staphylococcus aureus,5 Escherichia coli,6 and Streptococcus7 – in addition to well-known antimicrobial peptides8 – is the use of bacteriophages or phages. Bacteriophages are the most abundant viruses found in the biosphere;9,10 they grow quickly and exponentially, and they are efficacy since the fist interaction with bacteria.11 At the same time, phages have some disadvantages such as low stability over time and low resistance (or short half-life) in acidic environments such as that of the stomach.12,13
These limitations could be overcome by stabilizing the phages by combining them with nanoparticles of different materials such as carbon, silica, metal oxide, graphene, and hydroxyapatite (HA). HA, in particular, has been frequently used in several studies to stabilize, protect, and deliver molecules or radionuclides.14–22
HA represents the major components of bone, tooth, and cartilaginous tissues. It possesses several properties such as biocompatibility, biomimetic dimensions, osteoconductivity, and degradability, which make it suitable for several applications16,23 when combined with different biological molecules.19 Indeed, HA nanocrystals have been successfully employed to build bone scaffolds and implant coating materials and vehicles for drug targeting.24–29 Furthermore, HA nanocrystals have low toxicity and remarkable stability, as reported in our previous studies.30
In industrialized countries, infections acquired by food and water represent a big concern for public health, and about 50% of foodborne infections in humans are caused by Salmonella spp.31 Despite control measures and monitoring carried out by the healthcare authorities, cases of Salmonella contaminations are still very frequent.32
Salmonella serovars such as Enteritidis and Typhimurium are accountable for most of the salmonellosis cases.33 In recent years, infections by other serovars such as Kentucky and Rissen became more frequent; the latter Salmonella strain, especially, has been isolated from pork and chicken products and in human and swineherds gastrointestinal tract.34,35
Based on these considerations, we decided to investigate whether the biomimetic HA nanocrystals – which mimick the natural bone mineral – could interact with Salmonella bacteriophages and whether the newly developed complex could control Salmonella bacterial infection.
Materials and methods
Bacteria
All the Salmonella spp. strains used in this study were provided by the Istituto Zooprofilattico Sperimentale Del Mezzogiorno (Portici, Naples, Italy). Among these, S. Rissen was selected as reference strain for bacteriophage (phage) isolation.
Phage isolation
Phage (SR φ1) from S. Rissen was isolated as described by Capparelli et al.36 Briefly, S. Rissen was grown in 5 mL of Luria Bertani broth (LB; Sigma-Aldrich Co., St Louis, MO, USA) and, when the culture reached the exponential growth phase, Mitomycin C (1 μg/mL) was added. Then, the suspension was incubated at 37°C for 30 minutes. After incubation, it was centrifuged twice at 5,000 rpm for 10 minutes; each time the supernatant was discarded and the pellet resuspended in 5 mL of LB in order to remove any residue of Mitomycin C. Later, the suspension was incubated at 37°C for 4 hours. Then another centrifugation step was carried out but this time, the supernatant – containing bacteriophages – was collected, filtered through a 0.22 μm membrane, and screened for the presence of phages using the double-layer agar (DLA) method. The last step included a lower nutrient agar layer and an upper soft agar layer (4 mL of 0.7% bacteriological agar [Sigma-Aldrich Co.] mixed with 107 colony forming unit [CFU]/mL of bacterial strain used for phage isolation and previously filtered bacteriophage solution). After overnight incubation at 37°C, the plates were observed for the presence of clear zone (plaque formation) over the surface of the double agar plate. The phages isolated were stored at −20°C in buffer SM.37 However, the pellet was streaked onto Salmonella Chromogenic Agar Base (CM1007, Oxoid; Thermo Fisher Scientific, Waltham, MA, USA) to confirm the presence of Salmonella spp. bacteria.
The titer of phage, expressed as plaque forming units (PFUs), was evaluated by using the DLA technique as reported by Sambrook et al.38
Phage host range
Host range of phages was evaluated against 14 different Salmonella strains by the overlay method.39 Briefly, the test consisted of spotting 100 μL of SR φ1 on the surface of a double agar layer as reported above.
Multiplicity of infection (MOI) of phage
MOI is the ratio of virus particles to host cells.40 To establish the best MOI, S. Rissen strain was grown in LB at 37°C till 104 CFU/mL. Later, 100 μL of bacterial suspension was treated with 100 μL of SR φ1 at different ratios, ranging from 10−3 to 103 PFU/CFU, in a 96-well microplate. Positive and negative controls were represented by a mixture containing 100 μL of S. Rissen (104 CFU/mL) plus 100 μL of SM buffer or 100 μL of LB broth plus 100 μL of SM buffer, respectively. After 16–18 hours of incubation at 37°C, the optical density was measured at OD 600 nm to determine the optimal MOI. This last parameter shows that the lower phage titer is able to kill the majority of the bacteria, and it was used for all the subsequent experiments.
One-step growth curve
The “burst size” and phage life cycle were evaluated by the one-step growth assay. The culture of S. Rissen (108 CFU/mL), in exponential growth phase, was infected with phage according to the optimal MOI (previously selected). The mixture was incubated at 37°C for 5 minutes to favor phage adsorption, and later, it was serially diluted up to 10−4 in 20 mL of LB. The above mixture was incubated at 37°C for 90 minutes, and 100 μL of sample was taken every 5 minutes and plated, during the whole incubation period. Latent period was the interval between the beginning of the adsorption (not including 5 minutes pre-incubation) and the onset of the first burst (corresponding to the initial rise in phage titer). Burst size was the ratio between the final count of released phage particles and the initial amount of infected bacterial cells.
Electron microscopic analysis
SR φ1 (108 PFU/mL) was purified by CsCl density gradient ultracentrifugation and dialyzed against SM buffer overnight at 4°C.41 Phage particles were negatively stained with 2% of phosphotungstic acid (pH 7.2) for 20 minutes. Later, phages were observed by using a Philips EM 300 electron microscope.
Phage phylogenetic analysis
Phylogenetic analysis was performed using the maximum likelihood method.42 Data alignment was carried out with Blosum 65 (gap open penalty =11; gap extension penalty =3) and tree was built by using Jukes-Cantor and UPGMA models.
Phage genome sequencing, assembly, and annotation
Genomic DNA was quantified using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific); DNA purity was assessed with a Nanodrop (Thermo Fisher Scientific), and DNA size was determined with a 2200 Tape Station Instrument (Agilent Technologies, Santa Clara, CA, USA). Illumina libraries were produced starting from 1 μg of genomic DNA, which was sheared using the Covaris S220 instrument (Covaris Inc. Woburn, MA, USA) and the TruSeq DNA Sample Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer’s guidelines. Sequencing was performed on a NextSeq500 instrument with the 150-nt paired-end protocol (Illumina) according to the manufacturer’s guidelines. Illumina reads underwent quality filtering and trimming using Sickle and were quality corrected with Bayes Hammer before being assembled de novo. Genomes were assembled de novo from Illumina reads using SPAdes 2.9.0 with multiple k-mer combinations. The resultant contigs were scaffolded using SSPACE 3.0. Five micrograms of high-molecular-weight genomic DNA (peak >60 kb) was used to prepare ~20 kb-insert single-molecule real-time (SMRT)-bell libraries according to the manufacturer’s guidelines (Pacific Biosciences, Menlo Park, CA, USA). The library templates were sequenced using the SMRT sequencing technology on a PacBio RSII sequencer (Pacific Biosciences) at Macrogen, Inc (Korea). PacBio subreads were extracted using Bash5tools (version 0.8.0), filtrated, and assembled de novo using Falcon-Integrate with the suggested settings for bacterial genome. The assembled genome sequence was polished by Quiver v 0.9.2, and gene annotation was performed using RAST web service (http://rast.nmpdr.org/).43
Biomimetic HA nanocrystal synthesis and characterization
Biomimetic HA nanocrystals were produced as described by Nocerino et al.44 Briefly, HA nanocrystals were precipitated from an aqueous solution of (CH3COO)2Ca (75 mM) by slow addition (one drop per second) of an aqueous solution of H3PO4 (50 mM), keeping the pH constant at 10 (by the addition of (NH4)OH solution). The synthesis was performed at room temperature. After this last process, the suspension of HA was washed with distilled water in order to remove ammonium ions and favor the interaction between nanocrystals.
Transmission electron microscopy (TEM) investigation was carried out using a 1200 EX microscope, linked to X-ray analysis detectors and a 3010 UHR operating at 300 kV (JEOL Ltd, Tokyo, Japan). Few droplets of the samples (in ultrapure water) were deposited on perforated carbon foils supported on conventional copper microgrids. The surface area was determined using a Sorpty 1750 instrument (Carlo Erba Reagents S.r.l., Milan, Italy) using N2 absorption at 77 K.45
Synthesis of complex HA–SR φ1
The HA–SR φ1 complex was prepared by mixing 10 mL of HA (100 mg/mL) with 10 mL of SR φ1 (107 PFU/mL) under stirring wad. The suspension was stable, and no precipitation phenomena were observed during the synthesis process. Later, aliquots of 1 mL of the mixture were incubated – under shaking – at room temperature for the following time intervals: 30, 90, 180, and 300 minutes and 24 hours. After proper incubation, the sample was centrifuged, and the pellet was suspended in SM buffer. The concentration of the active phage particles was evaluated by the DLA method. After overnight incubation, the optimal incubation time was selected according to the results. Next, other tests were carried out by maintaining constant concentration of the HA but increasing the bacteriophage titer. The results showed that, by increasing the phage titer, the concentration of the active phage particles estimated by the DLA method, after the optimal time of incubation, remained constant at 107 PFU/mL (the titer used for the above complex synthesis) (data not shown). Based on these data, we evidenced that the maximum loading capacity of the HA used for this study was 107 PFU/mL of bacteriophages.
Complex HA–SR φ1 characterization
Zeta potential, dynamic light scattering (DLS) analysis, and pH stability
SR φ1, HA–SR φ1, and HA were analyzed for the measurement of zeta potential and DLS in appropriate disposable folded capillary cells (DTS1070; Malvern Instruments, Malvern, UK) using a Zetasizer Nano ZS (Malvern Instruments). For the DLS analysis, the instrument was, in addition, equipped with a 633 nm He–Ne laser and an avalanche photodiode detector placed at a detection angle of 173°. Each analysis was carried out in triplicate for three independent experiments. The analysis temperature was 25°C and about 1 mL of sample (at pH value of 7) was used for the test. The results were examined, and, for each sample, the zeta potential average and DLS measurement value were determined.
To estimate the stability of SR φ1 and HA–SR φ1 over time, the titer of phage – or mixed with HA – was evaluated for 2 successive months, at weekly intervals. During this time, the samples were stored at +4°C.46
In addition, the effects of an acid or alkaline pH, on SR φ1 or HA–SR φ1 complex, were evaluated by mimicking different pH conditions (ranging from 2 to 10). Briefly, the phage suspensions were incubated at 37°C for 1 hour in buffer SM at the following pH values: 2, 4, 7, and 10. After incubation, the phage titer was estimated by the DLA method as reported by Jun et al.47 Each assay was performed in triplicate.
X-ray analysis
X-ray powder diffraction (XPD) patterns were obtained at room temperature by using a Rigaku RINT2500 rotating anode laboratory diffractometer (50 kV, 200 mA) equipped with the silicon strip Rigaku D/teX Ultra detector. An asymmetric Johansson Ge (111) crystal was used to select the monochromatic Cu Kα1 radiation (λ=1.54056 Å). The measurements were carried out in transmission geometry by introducing the samples in a Lindemann glass capillary of 0.3 mm diameter. The XPD patterns were first indexed by the program QUALX48 by matching the XPD patterns with the ICSD database. They were further analyzed by using a whole-profile Rietveld-based fitting program49 to determine the crystalline domain size as follows:
- The instrumental resolution function (IRF) was evaluated by fitting the XPD pattern of a LaB6 NIST standard recorded under the same experimental conditions as those used for measuring the samples. The IRF data file was provided separately to the program in order to allow subsequent refinement of the diffraction patterns of the samples.
- The crystal structure of the samples, once determined, was input into the program and refined. The inhomogeneous peak broadening of the diffraction peaks was described by a phenomenological model based on a modified Scherrer formula:
|
where βh,k,l represent the size contribution to the integral width of the (h,k,l) reflection and are real spherical harmonics. After refinement of the αimp coefficients, the program calculates the apparent size of the crystal domains along each reciprocal lattice vector (h,k,l) direction.
Cytotoxicity trials
MTT assay
Human hepatocellular carcinoma HepG2 (HB-8065; American Type Culture Collection [ATCC], Manassas, VA, USA) cells were grown in minimal essential media (MEM) plus 10% FBS, 2 mM glutamine, 1% nonessential amino acids (NEAA), 100 U/mL penicillin, and 100 μg/mL streptomycin (all from Thermo Fisher Scientific). Cells (200 μL at 30,000 cells/well) were placed in a 96-well plate and were treated with SR φ1 (107 PFU/mL), HA–SR φ1 (100 mg/mL plus 107 PFU/mL), or HA (100 mg/mL) for 24, 48, and 72 hours, respectively.
The positive and negative controls contained 10% DMSO or PBS, respectively. All the conditions tested were performed in triplicate. An aliquot of 20 μL of MTT dissolved in PBS at a concentration of 5 mg/mL was added to each well. After 2 hours, the supernatant was discarded and 150 μL of isopropanol was added. The plate was incubated at 37°C for 30 minutes and then, the optical density was measured at 560 nm.
Lactate dehydrogenase (LDH) assay
An LDH assay was performed using a CytoTox 96 Non-Radio cytotoxicity assay kit (Promega Corporation, Fitchburg, WI, USA) at 24, 48, and 72 hours according to the manufacturer’s protocol. The LDH levels were evaluated for all the conditions tested for the MTT assay.
Internalization of fluorescent HA–SR φ1
In order to evaluate the internalization of the complex, HA nanoparticles were labeled by fluorescein isothiocyanate (FITC). Briefly, HA (100 mg/mL) was mixed with 20 mL of 3-aminopropyltriethoxysilane (Sigma-Aldrich Co.) in 100 mL of anhydrous ethanol and stirred at 74°C for 3 hours. Later, fluorescein (6 μg/mL; Sigma-Aldrich Co.) was added and continued to react at 74°C for about 20 hours in the dark. The mixture was centrifuged at 5,000 rpm for 1 minute and the pellet was rinsed three times with anhydrous ethanol and deionized water, and later suspended in SM buffer.50 Subsequently, SR φ1 (107 PFU/mL) was added to the HA previously labeled. An aliquot of 10 μL of the complex HA–SR φ1 at different dilutions (1:1,000, 1:100, and 1:10) was added to HepG2 cells (1.0×105 per well) for 24 hours. The control cells were treated with 10 μL of SM buffer. After treatment, the medium was discarded, and cells were rinsed twice with PBS and fixed in 4% paraformaldehyde for 10 minutes at room temperature. DAPI was used to counterstain nuclei. Slices were observed using a Zeiss LSM 710 Confocal Laser Scanning Microscope (Carl Zeiss MicroImaging GmbH). Samples were vertically scanned with a 63× or 40× (1.40 NA) Plan-Apochromat oil-immersion objective. Images were obtained with Zeiss ZEN Confocal Software (Carl Zeiss MicroImaging GmbH).
Intracellular killing activity
HepG2 cells were placed in a 24-well plate (0.2×106 cells/well), incubated overnight (37°C, 5% CO2) in MEM supplemented with 10% FBS (Thermo Fisher Scientific). The experiment was carried out in two different ways in order to evaluate different types of antimicrobial activity of the complex according to different infection processes. First, the cells were infected with S. Rissen (108 CFU/mL) for 1 hour. The medium was discarded, and the cells were washed with PBS (three times) and incubated in MEM plus 10% FBS and gentamicin (12.5 μg/mL)36 at 37°C in 5% CO2 for 3 hours. Later, the cells were treated with SR φ1 (107 PFU/mL), HA–SR φ1 (100 mg/mL plus 107 PFU/mL), or HA (100 mg/mL) for 24 hours as reported by McLaughlin et al51 and Withanage et al.52
In the second infection process, the cells were infected with S. Rissen (108 CFU/mL) and simultaneously treated with SR φ1 (107 PFU/mL), HA–SR φ1 (100 mg/mL plus 107 PFU/mL), or HA (100 mg/mL) for 1 hour. The cells were then treated with gentamicin as described above. After 3 hours of incubation at 37°C in 5% CO2, the medium was discarded, and the cells were incubated in MEM containing 10% FBS for 24 hours.
The cells of both experiments were lysed with Tween 20 (0.05%) and incubated at 37°C in 5% CO2 for 4 hours, and each lysate was then serially diluted in PBS and plated on XLT4 (Sigma-Aldrich Co.). The plates were incubated at 37°C overnight and evaluated for the presence of bacteria.
Both the above experiments were carried out using mouse macrophage cell line J774A.1 (TIB-67; ATCC) instead of HepG2 cells. J774A.1 cells were grown in DMEM supplemented with 10% FBS, 2 mM glutamine, 1% NEAA, 100 U/mL penicillin, and 100 μg/mL streptomycin (all from Thermo Fisher Scientific). Cells were maintained in a humidified, 37°C, 5% CO2 incubator.
Bacterial reduction assay on chicken minced meat
S. Rissen was processed as reported by Sukumaran et al.53 For the test, 250 g of chicken minced meat was divided into five equal parts. Four sections were infected with S. Rissen (103 CFU/mL) and incubated at room temperature according to ISO 16140-2: 2016;54 while the last one was treated only with PBS (negative control). After 30 minutes, three of the four infected parts were treated respectively with SR φ1 (108 PFU/mL), HA–SR φ1 (100 mg/mL plus 108 PFU/mL), or HA (100 mg/mL). All samples were stored at 4°C for 7 days and tested for the presence of the bacteria every 24 hours. In particular, at each time point, 5 g of meat was taken from each sample and homogenized in 45 mL of 0.1% buffered peptone water (AES Laboratoire Groupe, France) at 200 rpm for 1 minute, using the Stomacher 400 Circulator (Seward Ltd, Bohemia, NY, USA). Subsequently, 100 μL of sample was plated on XLT4 agar and incubated at 37°C for 24 hours. The efficacy of each treatment was evaluated by counting the CFU formed on the plate after the incubation time.
Results
Phage isolation and characterization
Phage excision from S. Rissen was induced by Mitomycin C procedure.36,55 The SR φ1 host range was tested against 14 Salmonella strains, each belonging to a different serovar, and it was active against 11 out of 14 selected strains (Table 1). The MOI test allowed us to select the value of 0.1 as optimal MOI (Figure S1). This value has been used for all the later experiments. The one-step growth analysis highlighted that the latent period of SR φ1 was 60 minutes (Figure S2) and that its burst size was 54 PFU/cell.
  | Table 1 Phage host range against different Salmonella strains |
TEM analysis of SR φ1 allowed us to classify the phage as a member of the Podoviridae family, according to a previous publication.10 In particular, TEM analysis revealed that the tail length and head diameter were 16±2.0 nm (mean ± SD) and 57±1.0 nm (mean ± SD), respectively; and the total length was 73±2.0 nm (mean ± SD) (Figure 1).
  | Figure 1 Electron microscopic analysis of phage. SR φ1 observed with a TEM analysis. Scale bar, 100 nm. |
Comparative genomic analysis of phage
DNA sequencing yielded a total of 2,199,543 reads (660 Mb) and an average coverage of 13,200×. Phage SR φ1 genome consists of 51,738 bp with a G+C content of 48.4%. The genomic sequence of SR φ1 described in this study has been deposited in the GenBank database (accession number: KY709687). Open reading frames (ORFs) with a length of at least 38 amino acids were selected. A total of 622 ORFs were predicted to be present within the genome. However, only 87 ORFs (13.98%) were predicted to be functional based on gene predictions and annotation of the genome (Figure 2). Concrete gene information such as positions, directions, sizes, and putative functions of SR φ1 coding DNA sequences (CDSs) are shown in Table S1. The genome contains 87 predicted CDSs, of which 70 matched with already identified phage genes. Identified genes included 30 genes affecting bacteriophage physiology, 12 phage structures, 10 DNA replication, and three bacterial lysis. The remaining 32 CDSs encoded hypothetical proteins. Phage SR φ1-predicted genome did not show genes coding for toxins, antibiotic resistance, or Salmonella virulence.
To gain an insight into the characteristics of the phage SR φ1 genome, we performed a phylogenetic analysis, comparing its genome to the predicted genomes of other 37 fully sequenced phages. The phylogenetic analysis confirmed that phage φ1 is a Podoviridae, specifically showing 19 homologous genes with PHAGE_Salmon_SPN9CC (NC017985) (Figure 3).
Biomimetic HA nanocrystal characterization
The biomimetic HA nanocrystals used in this study had a composition very close to that of the human body.56
The TEM analysis revealed the length of HA nanocrystals (~30–40 nm) and its plate-like morphology (Figure S3). The high reactivity of HA is ascribed to its amorphous surface16 and to its high surface area of about 110 m2/g, which is only slightly lower than that of biological nanocrystals (120 m2/g).30
Moreover, the degree of crystallinity and the presence of carbonate ions in the structure of biomimetic HA (data not shown) clearly confirm that the HA selected for this study is – structurally – very similar to that of the bone, not only in the size of the nanocrystals but also in ionic substitution.
Preparation and characterization of the complex (HA–SR φ1)
The interaction of SR φ1 with HA, carried out as reported in Patent IT102017000050733,57 was evaluated at different incubation time intervals. The results showed that SR φ1 has interacted with HA already after 30 minutes. Furthermore, the graph showed that the amount of complexes SR φ1–HA increased over time (Figure 4A). Based on this result, we considered overnight incubation to be the optimal time interval for the complex synthesis.
The interaction between HA and SR φ1 was studied also through the zeta potential analysis, in which SR φ1 was negatively charged (−11.28±1.16) while HA was positively charged (2.9±0.9 mV). Therefore, the SR φ1 and HA complex showed a positive zeta potential of 0.9±1.60 mV (Figure 4B).
In addition, the stability of the SR φ1 and the HA–SR φ1 complex was assessed after the complex synthesis for about 2 months at 7 days intervals. Stability for long periods is essential for using phages in several biocontrol applications.58 The titer of the phage SR φ1 alone started to decrease already after 1 week; instead, when the phage was complexed with HA, its titer was stable for up to 6 weeks without any variation at the different time points analyzed (Figure 4C).
The analysis of the HA–SR φ1 complex stability at different pH values (ranging from 2 to 10) assessed the strong stability of the complex, compared to that of SR φ1 alone (Figure 4D). Further characterization of the complex was carried out by DLS analysis. During this test, no rapid aggregation phenomenon was observed. As shown in Figure 4E, the diameter of HA was 300 nm while only diameter of SR φ1 was smaller (about 200 nm) than the HA. When HA and SR φ1 were complexed, the estimated hydrodynamic diameter was about 400 nm, thereby confirming the interaction between these two elements as reported by Wang at al.59
XPD analysis showed that HA nanocrystals60 have a hexagonal crystalline structure (structure parameters are reported in Table S2). XPD patterns were further studied by using a whole-profile Rietveld-based fitting program49 as shown in Figure S4. The Rietveld analysis allowed us to determine cell parameters and crystalline domain size along the [002] and [110] crystallographic directions (summarized in Table S3). This analysis also showed the structural variation of the HA crystalline domain size with or without SR φ1 (Figure 5, black and red dots, respectively). When the HA percentage was increased, the samples without SR φ1 did not show any change in the crystalline domain size along [002] and [110] directions. Instead, when the HA amount was increased, in the case of the complex (HA–SR φ1), the crystalline domain size decreased (Figure 5).
Cytotoxicity trials
To evaluate the cytotoxic effect of SR φ1, HA, and the complex HA–SR φ1, a cell viability test was carried out by evaluating the reduction of MTT to formazan in human hepatocellular carcinoma HepG2 cells. The results showed that none of the treatments affected the cell vitality up to 72 hours (Figure S5A).
Moreover, the amount of the extracellular enzyme LDH, a cell death indicator,61 was assessed under the same experimental conditions described above. Again, based on the amount of LHD produced, we concluded that none of the treatments produced cytotoxic effects (Figure S5B).
Internalization of fluorescent complex
As Salmonella spp. is an intracellular pathogen, the capacity of the complex to penetrate HepG2 eukaryotic cells was evaluated by laser scanning confocal microscopy analysis. Figure 6 evidences the cell uptake of the complex. In particular, the complex HA–SR φ1 (HA colored in green) was internalized into the cytoplasm of the HepG2 cells after 24 hours of treatment. Nuclei were counterstained with DAPI dye (blue). No fluorescent signal was detected in the control cells.
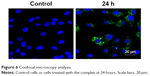  | Figure 6 Confocal microscopy analysis. |
Intracellular killing activity
Once the ability of the complex to penetrate host cells was determined, the intracellular killing activity was gauged in two different experimental conditions: the HA–SR φ1 complex was administered at 1 hour postinfection or together with Salmonella bacteria. The results showed that 1 hour postinfection, only HA–SR φ1 showed lytic activity against bacteria, reducing the bacterial load significantly (Figure 7A) while the effect of phages alone was minimal. In the second experiment, in which all the treatments were carried out simultaneously with the bacteria infection, both SR φ1 and HA–SR φ1 showed comparable lytic activity (Figure 7B). The data reported in Figure 7A evidenced that 1 hour after the infection with S. Rissen, the presence of biomimetic HA favored the uptake of phages into the HepG2 cells, allowing phages to kill also the intracellular bacteria.
Furthermore, Figure 7B showed that the biomimetic HA nanocrystals did not compromise the lytic activity of the phages, when the complex was applied concurrently with the bacteria. The trend observed in HepG2 cells was also observed for murine macrophage cell line J774A.1 (data not shown).
Bacterial reduction assay on chicken minced meat
The efficacy of the complex HA–SR φ1 to reduce Salmonella contamination was investigated in chicken minced meat which is considered one of the meat categories intended to be cooked before consumption (together with mechanically separated meat and meat preparations) with the highest level of noncompliance, as reported by European Food Safety Authority (EFSA).62 S. Rissen colonies, in the sample treated with SR φ1, were reduced by 0.3 log CFU/g, while the sample treated with only HA was contaminated very similar to that of the positive control (5.5 log CFU/g). Instead, in the case of treatments with HA–SR φ1, the bacterial load of S. Rissen was reduced by 3 log CFU/g (Figure 8).
Discussion
Different material nanoparticles such as carbon, silica, metal oxide, graphene, and HA have been used in several studies to stabilize, protect, deliver, or enhance some biological properties of molecules.14–16,30,44
The characteristics of HA used in this study (such as composition, structure, size, and morphology) and the results obtained allow us to confirm that this mineral, due to its features, was able to chemically interact not only with molecules but also with biological structures like bacteriophages. Its low degree of crystallinity and the presence of carbonate ions in the crystal structure are other important characteristics that make HA extremely reactive in biological systems, and particularly suitable to interact and transport the bacteriophages, as reported in this study.
The bacteriophage SR φ1 – isolated in this study – showed an efficient bacteriolytic activity against S. Livingstone, S. Infantis, S. Potsdam, S. Thompson, S. Mbandaka, S. Winston, S. Montevideo, S. Virchow, S. Ohio, S. Jerusalem, S. Inganda, S. Wil, S. Typhimurium, and S. Enteritidis as shown in Table 1. These results evidenced that it could be used as an antimicrobial agent against the strains of Salmonella spp. selected for this study. Further investigations are needed to evaluate whether the phage SR φ1 could show antimicrobial activity against other Salmonella strains and serovars.
Based on these results, we evaluated whether the use of HA could improve some properties of the bacteriophage. First, we used the mineral HA, which did not affect the cell viability and was not toxic for human cells (Figure S5).
The complex HA–SR φ1 showed enhanced lytic activity against S. Rissen than SR φ1 alone, thus indicating that the interaction with HA increased the antibacterial activity of the bacteriophage. Furthermore, the mineral contributed to stabilize the phage activity over time and at different pH values, making the complex suitable for long-term treatment and effective for controlling infection in harsh niches, showing acid pH (such as stomach, urinary tract, etc), or in different physiological environments of healthy tissues (including blood).
The interaction of HA with phage is a good strategy to overcome the problem related to the reduction of bacteriophage viability in acidic conditions.12
The zeta potential analysis and DLS measurements demonstrated the interaction of the phage with HA. In fact, Figure 4B showed the behavior of the zeta potential of SR φ1 and HA alone and when they were complexed. In the latter case, neutralization of the negative surface charges of SR φ1 was observable. Because the charges of two elements are reversed, neutralization of the surface charges could be principally due to electrostatic interactions. Furthermore, the results of the DLS analysis allow us to hypothesize that when the two elements are complexed, the HA acts as a scaffold for the phages.
These results are also supported by Wang et al.59 Indeed, they demonstrated the ability of the bacteriophages to bind calcium ions, or non-stoichiometric HAs with positive zeta potential.59 The HA used in this study, just like the ones mentioned above, has a positive zeta potential, a non-stoichiometric Ca/P ratio; all these characteristics induce the interaction of the HA with proteins and carboxyl groups of the phage capsid by the development of an electrostatic bond.
Some studies have shown the capacity of HA to permeate the cell membrane through the energy-dependent process of clathrin-mediated endocytosis63 or phagocytosis64 but have, also, highlighted that the permeabilization process is influenced by the equilibrium of multiple features such as dimension, charge, shape, and surface area.24,64–66
It has been reported that the bacteriophages are also able to bypass the cell membrane.67
Based on these evidences, we tested the capacity of the complex, and in particular the contribution of the mineral HA to penetrate into the eukaryotic cells. The results demonstrated that, in the same infection condition, the presence of the mineral increases the number of phages that are able to penetrate into cells more efficiently and consequently kill intracellular bacteria.36 This mineral could be a good candidate in the use of phage therapy against obligate or facultative intracellular bacteria like Mycobacterium spp., Chlamydia spp., Rickettsia spp. or Salmonella spp., Listeria spp., and Brucella spp.68
In addition, another experiment was carried out on HepG2 cells or J774A.1 cells not infected with bacteria. The results confirmed the ability of the HA to enter into eukaryotic cells and the capacity of bacteriophage to bypass the cell membrane (both reported in literature) and, at the same time, highlighted the role of HA in increasing the number of phages that pass through the cell membrane (data not shown) although there was no infection.
The last test, carried out to evaluate the efficacy of the complex to control bacterial infection in a food matrix such as chicken minced meat and for long time, showed the ability of the complex to reduce bacterial load also in the case of food contamination.
Conclusion
The main drawback in using phages are their lack of stability over time and their low activity against intracellular infections due to their low efficiency in penetrating eukaryotic cells. In addition, phages do not tolerate the low pH present in the stomach and when used in food processing their activity can be compromised. In this study, these problems were successfully addressed by complexing phages with HA (nontoxic mineral for humans). This complex is stable, allows phages to enter eukaryotic cells more efficiently than phages alone, and, at the same time, the complexed phages were stable at very low pH. These results evidence the important contribution of HA making it a promising approach to overcome problems which could emerge when biological entities are used.
One of the most important problems in phage therapy is the application of phages as biocontrol agents against contamination in food.69 This drawback has been addressed, and it has been resolved by carrying out a test on infected meat. The results showed how HA enhances the lytic activity of phage to control bacterial meat infection. Strikingly, the complex is able to control Salmonella infection in food.
Furthermore, the approach proposed here (use of biomimetic HA nanocrystals as a carrier for bacteriophages) can be extended to different fields of interest such as biomedical, agricultural, and other commercial applications.
Acknowledgments
The authors wish to thank Francesco Baldassarre and Chiara Colletti for their assistance in this research.
Author contributions
RC, FCa, and NR have made major contributions to the conception of the study; AF, FI, MP, FCo, TS, CG, SP, RV, BDV, and ML contributed to the acquisition, analysis, or interpretation of the data; AF, FI, MP, CG, and BDV performed the experiments; RC wrote the manuscript. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
AF, FI, NR, ML, FCo, and RC are the inventors of the Patent IT102017000050733 presented to the Italian Ministry of Economic Development on May 10, 2017. All the other authors report no conflicts of interest in this work.
References
Ferri M, Ranucci E, Romagnoli P, Giaccone V. Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr. 2017;57(13):2857–2876. | ||
Holmes AH, Moore LSP, Sundsfjord A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. The Lancet. 2016;387(10014):176–187. | ||
Berkowitz FE. Antibiotic resistance in bacteria. South Med J. 1995;88(8):797–804. | ||
Capuano F, Mancusi A, Capparelli R, Esposito S, Proroga YT. Characterization of drug resistance and virulotypes of Salmonella strains isolated from food and humans. Foodborne Pathog Dis. 2013;10(11):963–968. | ||
Capparelli R, Nocerino N, Lanzetta R, et al. Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS One. 2010;5(7):e11720. | ||
Poirel L, Madec JY, Lupo A, et al. Antimicrobial Resistance in Escherichia coli. Microbiol Spectr. 2018;6(4). | ||
Marco S, Rullo R, Albino A, Masullo M, de Vendittis E, Amato M. The thioredoxin system in the dental caries pathogen Streptococcus mutans and the food-industry bacterium Streptococcus thermophilus. Biochimie. 2013;95(11):2145–2156. | ||
Capparelli R, De Chiara F, Nocerino N, et al. New perspectives for natural antimicrobial peptides: application as antinflammatory drugs in a murine model. BMC Immunol. 2012;13(1):61. | ||
Doss J, Culbertson K, Hahn D, Camacho J, Barekzi N. A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Viruses. 2017;9(3):50. | ||
Jurczak-Kurek A, Gąsior T, Nejman-Faleńczyk B, et al. Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated from urban sewage. Sci Rep. 2016;6(1):34338. | ||
Inal JM. Phage therapy: a reappraisal of bacteriophages as antibiotics. Arch Immunol Ther Exp. 2003;51(4):237–244. | ||
Ma Y, Pacan JC, Wang Q, et al. Microencapsulation of bacteriophage felix O1 into chitosan-alginate microspheres for oral delivery. Appl Environ Microbiol. 2008;74(15):4799–4805. | ||
Colom J, Cano-Sarabia M, Otero J, et al. Microencapsulation with alginate/CaCO3: A strategy for improved phage therapy. Sci Rep. 2017;7(1):41441. | ||
Chowdhury S, Yusof F, Salim WW, Sulaiman N, Faruck MO. An overview of drug delivery vehicles for cancer treatment: nanocarriers and nanoparticles including photovoltaic nanoparticles. J Photochem Photobiol B. 2016;164:151–159. | ||
Krishnamoorthy K, Jeyasubramanian K, Premanathan M, Subbiah G, Shin HS, Kim SJ. Graphene oxide nanopaint. Carbon. 2014;72:328–337. | ||
Roveri N, Palazzo B, Iafisco M. The role of biomimetism in developing nanostructured inorganic matrices for drug delivery. Expert Opin Drug Deliv. 2008;5(8):861–877. | ||
Benedetti M, De Castro F, Romano A, et al. Adsorption of the cis-[Pt(NH3)2(P2O7)](2-) (phosphaplatin) on hydroxyapatite nanocrystals as a smart way to selectively release activated cis-[Pt(NH3)2Cl2] (cisplatin) in tumor tissues. J Inorg Biochem. 2016;157:73–79. | ||
Benedetti M, Antonucci D, De Castro F, et al. Metalated nucleotide chemisorption on hydroxyapatite. J Inorg Biochem. 2015;153:279–283. | ||
Lelli M, Roveri N, Marzano C, et al. Hydroxyapatite nanocrystals as a smart, pH sensitive, delivery system for kiteplatin. Dalton Trans. 2016;45(33):13187–13195. | ||
Rimola A, Sakhno Y, Bertinetti L, Lelli M, Martra G, Ugliengo P. Toward a surface science model for biology: glycine adsorption on nanohydroxyapatite with well-defined surfaces. J Phys Chem Lett. 2011;2(12):1390–1394. | ||
Cai Y, Gao T, Fu S, Sun P. Development of zoledronic acid functionalized hydroxyapatite loaded polymeric nanoparticles for the treatment of osteoporosis. Exp Ther Med. 2018;16(2):704–710. | ||
Chakraborty S, Das T, Banerjee S, Sarma HD, Venkatesh M. Preparation and preliminary biological evaluation of 177Lu-labelled hydroxyapatite as a promising agent for radiation synovectomy of small joints. Nucl Med Commun. 2006;27(8):661–668. | ||
Macchetta A, Turner IG, Bowen CR. Fabrication of HA/TCP scaffolds with a graded and porous structure using a camphene-based freeze-casting method. Acta Biomater. 2009;5(4):1319–1327. | ||
Uskoković V, Uskoković DP. Nanosized hydroxyapatite and other calcium phosphates: chemistry of formation and application as drug and gene delivery agents. J Biomed Mater Res B Appl Biomater. 2011;96B(1):152–191. | ||
V Dorozhkin S. Nanodimensional and nanocrystalline calcium orthophosphates. AJBE. 2012;2(3):48–97. | ||
Fox K, Tran PA, Tran N. Recent advances in research applications of nanophase hydroxyapatite. Chemphyschem. 2012;13(10):2495–2506. | ||
Rodríguez-Ruiz I, Delgado-López JM, Durán-Olivencia MA, et al. pH-responsive delivery of doxorubicin from citrate-apatite nanocrystals with tailored carbonate content. Langmuir. 2013;29(26):8213–8221. | ||
Kozempel J, Vlk M, Málková E, et al. Prospective carriers of 223Ra for targeted alpha particle therapy. J Radioanal Nucl Chem. 2015;304(1):443–447. | ||
Palazzo B, Iafisco M, Laforgia M, et al. Biomimetic hydroxyapatite–drug nanocrystals as potential bone substitutes with antitumor drug delivery properties. Adv Funct Mater. 2007;17(13):2180–2188. | ||
Fulgione A, Nocerino N, Iannaccone M, et al. Lactoferrin adsorbed onto biomimetic hydroxyapatite nanocrystals controlling – in vivo – the helicobacter pylori infection. PLoS One. 2016;11(7):e0158646. | ||
Beuchat LR. Ecological factors influencing survival and growth of human pathogens on raw fruits and vegetables. Microbes Infect. 2002;4(4):413–423. | ||
Finstad S, O’Bryan CA, Marcy JA, Crandall PG, Ricke SC. Salmonella and broiler processing in the United States: relationship to foodborne salmonellosis. Food Res Int. 2012;45(2):789–794. | ||
Herikstad H, Motarjemi Y, Tauxe RV. Salmonella surveillance: a global survey of public health serotyping. Epidemiol Infect. 2002;129(1):1–8. | ||
Oliveira CJB, Carvalho LFOS, Fernandes SA, Tavechio AT, Menezes CCP, Domingues FJ. Antimicrobial resistance of Salmonella serotypes isolated from slaughter-age pigs and environmental samples. Microb Drug Resist. 2002;8(4):407–411. | ||
Angkititrakul S, Chomvarin C, Chaita T, Kanistanon K, Waethewutajarn S. Epidemiology of antimicrobial resistance in Salmonella isolated from pork, chicken meat and humans in Thailand. Southeast Asian J Trop Med Public Health. 2005;36(6):1510–1515. | ||
Capparelli R, Parlato M, Borriello G, Salvatore P, Iannelli D. Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob Agents Chemother. 2007;51(8):2765–2773. | ||
SM buffer. Cold Spring Harb Protoc. 2006;2006(1):pdb.rec8111. doi:10.1101/pdb.rec8111. | ||
Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. | ||
Zinno P, Devirgiliis C, Ercolini D, Ongeng D, Mauriello G. Bacteriophage P22 to challenge Salmonella in foods. Int J Food Microbiol. 2014;191:69–74. | ||
Birge EA. Bacterial and Bacteriophage Genetics. New York, NY: Springer New York; 2000. | ||
Nasukawa T, Uchiyama J, Taharaguchi S, et al. Virus purification by CsCl density gradient using General centrifugation. Arch Virol. 2017;162(11):3523–3528. | ||
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. | ||
Aziz RK, Bartels D, Best AA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. | ||
Nocerino N, Fulgione A, Iannaccone M, et al. Biological activity of lactoferrin-functionalized biomimetic hydroxyapatite nanocrystals. Int J Nanomedicine. 2014;9:1175–1184. | ||
Brunauer S, Emmett PH, Teller E. Adsorption of gases in multimolecular layers. J Am Chem Soc. 1938;60(2):309–319. | ||
Mattila S, Ruotsalainen P, Jalasvuori M. On-demand isolation of bacteriophages against drug-resistant bacteria for personalized phage therapy. Front Microbiol. 2015;6(96):1271. | ||
Jun JW, Kim JH, Shin SP, Han JE, Chai JY, Park SC. Characterization and complete genome sequence of the Shigella bacteriophage pSf-1. Res Microbiol. 2013;164(10):979–986. | ||
Altomare L, Fare’ S. Cells response to topographic and chemical micropatterns. J Appl Biomater Biomech. 2008;6(3):132–143. | ||
Fernández-Díaz MT, Martínez JL, Rodríguez-Carvajal J, et al. Metamagnetism in single-crystal Pr2NiO4. Phys Rev B Condens Matter. 1993;47(10):5834–5840. | ||
Cui X, Liang T, Liu C, Yuan Y, Qian J. Correlation of particle properties with cytotoxicity and cellular uptake of hydroxyapatite nanoparticles in human gastric cancer cells. Mater Sci Eng C Mater Biol Appl. 2016;67:453–460. | ||
Mclaughlin LM, Govoni GR, Gerke C, et al. The Salmonella SPI2 effector SseI mediates long-term systemic infection by modulating host cell migration. PLoS Pathog. 2009;5(11):e1000671. | ||
Withanage GSK, Mastroeni P, Brooks HJ, Maskell DJ, Mcconnell I. Oxidative and nitrosative responses of the chicken macrophage cell line MQ-NCSU to experimental Salmonella infection. Br Poult Sci. 2005;46(3):261–267. | ||
Sukumaran AT, Nannapaneni R, Kiess A, Sharma CS. Reduction of Salmonella on chicken meat and chicken skin by combined or sequential application of lytic bacteriophage with chemical antimicrobials. Int J Food Microbiol. 2015;207:8–15. | ||
ISO 16140-2:2016 – Microbiology of the food chain – Method validation – Part 2: Protocol for the validation of alternative (proprietary) methods against a reference method. Available from: https://www.iso.org/standard/54870.html. Accessed November 29, 2018. | ||
Kaneko J, Kimura T, Kawakami Y, Tomita T, Kamio Y. Panton-Valentine leukocidin genes in a phage-like particle isolated from mitomycin C-treated Staphylococcus aureus V8 (ATCC 49775). Biosci Biotechnol Biochem. 1997;61(11):1960–1962. | ||
Palazzo B, Walsh D, Iafisco M, et al. Amino acid synergetic effect on structure, morphology and surface properties of biomimetic apatite nanocrystals. Acta Biomater. 2009;5(4):1241–1252. | ||
Capparelli R, Ianniello F, Capuano F, et al. Complesso comprendente almeno un calcio fosfato ED almeno un virus. 2017. | ||
Goode D, Allen VM, Barrow PA. Reduction of experimental Salmonella and Campylobacter contamination of chicken skin by application of lytic bacteriophages. Appl Environ Microbiol. 2003;69(8):5032–5036. | ||
Wang F, Cao B, Mao C. Bacteriophage bundles with Pre-Aligned Ca initiate the oriented nucleation and growth of hydroxylapatite. Chem Mater. 2010;22(12):3630–3636. | ||
Kay MI, Young RA, Posner AS. Crystal structure of hydroxyapatite. Nature. 1964;204(4963):1050–1052. | ||
Awad WA, Aschenbach JR, Zentek J. Cytotoxicity and metabolic stress induced by deoxynivalenol in the porcine intestinal IPEC-J2 cell line. J Anim Physiol Anim Nutr. 2012;96(4):709–716. | ||
The European Union summary report on trends and sources of zoonoses, zoonotic agents and food–borne outbreaks in 2016. EFSA J. 2017;15(12):5077. | ||
Bauer IW, Li SP, Han YC, Yuan L, Yin MZ. Internalization of hydroxyapatite nanoparticles in liver cancer cells. J Mater Sci Mater Med. 2008;19(3):1091–1095. | ||
Motskin M, Wright DM, Muller K, et al. Hydroxyapatite nano and microparticles: correlation of particle properties with cytotoxicity and biostability. Biomaterials. 2009;30(19):3307–3317. | ||
Yang X, Li Y, Liu X, Zhang R, Feng Q. In vitro uptake of hydroxyapatite nanoparticles and their effect on osteogenic differentiation of human mesenchymal stem cells. Stem Cells Int. 2018;2018(34):1–10. | ||
Yin M, Xu W, Cui B, et al. Effects of the interaction between hydroxyapatite nanoparticles and hepatoma cells. J Wuhan Univ Technol Mat Sci Edit. 2014;29(3):635–642. | ||
Nguyen S, Baker K, Padman BS, et al. Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. MBio. 2017;8(6):6. | ||
Silva MT. Classical labeling of bacterial pathogens according to their lifestyle in the host: inconsistencies and alternatives. Front Microbiol. 2012;3:71. | ||
Cooper IR. A review of current methods using bacteriophages in live animals, food and animal products intended for human consumption. J Microbiol Methods. 2016;130:38–47. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.