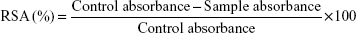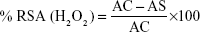Back to Journals » International Journal of Nanomedicine » Volume 11
Biogenesis of silver nanoparticles using endophytic fungus Pestalotiopsis microspora and evaluation of their antioxidant and anticancer activities
Authors Netala VR, Bethu MS, Pushpalatha B, Baki VB, Aishwarya S, Rao JV, Tartte V
Received 16 May 2016
Accepted for publication 22 July 2016
Published 31 October 2016 Volume 2016:11 Pages 5683—5696
DOI https://doi.org/10.2147/IJN.S112857
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Vasudeva Reddy Netala,1 Murali Satyanarayana Bethu,2,3 Bobbu Pushpalatha,1 Vijaya Bhaskar Baki,4 Sani Aishwarya,1 J Venkateswara Rao,2 Vijaya Tartte5
1Department of Biotechnology, Sri Venkateswara University, Tirupati, 2Biology Division, Indian Institute of Chemical Technology, Hyderabad, 3Academy of Scientific and Innovative Research, New Delhi, 4Department of Bioinformatics, 5Department of Botany, Sri Venkateswara University, Tirupati, India
Abstract: An endophytic fungal strain isolated from the leaves of Gymnema sylvestre was identified as Pestalotiopsis microspora VJ1/VS1 based on nucleotide sequencing of internal transcribed spacer region (ITS 1-5.8S-ITS 2) of 18S rRNA gene (NCBI accession number KX213894). In this study, an efficient and ecofriendly approach has been reported for the synthesis of silver nanoparticles (AgNPs) using aqueous culture filtrate of P. microspora. Ultraviolet-visible analysis confirmed the synthesis of AgNPs by showing characteristic absorption peak at 435 nm. Fourier transform infrared spectroscopy analysis revealed the presence of phenolic compounds and proteins in the fungal filtrate, which are plausibly involved in the biosynthesis and capping of AgNPs. Transmission electron microscopy (TEM) showed that the AgNPs were spherical in shape of 2–10 nm in size. Selected area electron diffraction and X-ray diffraction studies determined the crystalline nature of AgNPs with face-centered cubic (FCC) lattice phase. Dynamic light scattering analysis showed that the biosynthesized AgNPs possess high negative zeta potential value of -35.7 mV. Biosynthesized AgNPs were proved to be potential antioxidants by showing effective radical scavenging activity against 2,2'-diphenyl-1-picrylhydrazyl and H2O2 radicals with IC50 values of 76.95±2.96 and 94.95±2.18 µg/mL, respectively. The biosynthesized AgNPs exhibited significant cytotoxic effects against B16F10 (mouse melanoma, IC50 =26.43±3.41 µg/mL), SKOV3 (human ovarian carcinoma, IC50 =16.24±2.48 µg/mL), A549 (human lung adenocarcinoma, IC50 =39.83±3.74 µg/mL), and PC3 (human prostate carcinoma, IC50 =27.71±2.89 µg/mL) cells. The biosynthesized AgNPs were found to be biocompatible toward normal cells (Chinese hamster ovary cell line, IC50 =438.53±4.2 µg/mL). Cytological observations on most susceptible SKOV3 cells revealed concentration-dependent apoptotic changes that include cell membrane blebbing, cell shrinkage, pyknotic nuclei, karyorrhexis followed by destructive fragmentation of nuclei. The results together in this study strongly provided a base for the development of potential and versatile biomedical applications of biosynthesized AgNPs in the near future.
Keywords: AgNPs, Pestalotiopsis microspora, 18S rRNA, SKOV3, B16F10, AO-EB staining
Introduction
Over the last decade, majority of the researchers shifted their focus to nanometer-sized particles, particularly metal nanoparticles because of their controllable size and shape, ease of synthesis, and strong optical properties.1 Metal nanoparticles have become the prime choice in almost every branch of science including physics, chemistry, computer science, biology, and biomedicine. Owing to their large surface-to-volume ratio, spatial confinement, surface energy, and reduced imperfections, metallic nanoparticles especially silver nanoparticles (AgNPs) possess unique physicochemical properties such as optical, electronic, optoelectronic, thermal, and catalytic properties.1–4 The physicochemical properties of AgNPs are mainly determined by their shape, size, dielectric properties, dispersity, and crystalline nature.5,6 Hence the synthesis of AgNPs with controlled size and shape has gained significant interest in the field of nanotechnology. Because of these diverse physicochemical properties, AgNPs have been found to possess innumerable applications in various fields including optical receptors,3 sensors,7 catalysts in chemical reactions,4 signal enhancers in surface-enhanced Raman spectroscopy-based enzyme immunoassay,8 tissue engineering,9 DNA detection and delivery,10 antioxidant,11,12 cytotoxic,13,14 and antimicrobial agents.15–17
Various physical and chemical methods have been reported for the synthesis of AgNPs including microwave-assisted,18 γ-irradiation-assisted,19 thermal decomposition,20 laser ablation,21 lithography,22 polyol process,23 and chemical reduction methods. But all of these methods involve application of hazardous chemicals and environmental and biological risks and are also laborious and expensive. Biological approaches are simple, rapid, and cost-effective and involve the synthesis of nontoxic, clean, and biocompatible AgNPs. The vast and diverse fungal flora on this earth has attracted scientists around the world a significant interest to explore it for the synthesis of AgNPs. Fungal-mediated synthesis of AgNPs has recently emerged as a novel platform in the field of nanobiotechnology. Compared to bacteria, fungi are more advantageous because of fast growing rate, very easy to culture, and can be maintained in the laboratory. As fungi being eukaryotes, secrete more amounts of enzymes and proteins, significantly increase productivity, and ensure protein coating and high stability along with preventing agglomeration of the nanoparticles. Various fungal extracts including Alternaria solani, Penicillium funiculosum,24 Aspergillus terreus,25 Amylomyces rouxii,26 Coriolus versicolor,27 Fusarium solani,28 Fusarium oxysporum, 29 Humicola sp.,30 Puccinia graminis,31 Aspergillus niger,32 Aspergillus versicolor,33 Schizophyllum radiatum,34 Trichoderma viridae,35 Alternaria alternata,36 and Phaenerochaete chrysosporium37 have been extensively used for the biosynthesis of AgNPs.
Endophytic fungi are the intriguing group of fungal species that colonize living and healthy tissues of plants without causing any apparent symptoms of disease. Endophytic fungi produce natural bioactive compounds that are considered to be alternative sources for plant-producing bioactive compounds. The bioactive compounds from endophytic fungi possess antimicrobial,38 antioxidant,38,39 immunomodulating, and anticancer activities.40,41 The exploration of endophytic fungi for the synthesis of metal nanoparticles could be another great advantage. Javed et al26 synthesized the AgNPs of 5–27 nm in size using the endophytic fungus A. rouxii strain KSU-09 isolated from the roots of date palm Phoenix dactylifera, and the biosynthesized AgNPs showed antimicrobial activity against Shigella dysenteriae type I, Staphylococcus aureus, Citrobacter sp., Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, Candida albicans, and F. oxysporum. Devi et al24 biosynthesized the AgNPs of 5–20 nm in size using the endophytic fungi A. solani GS1 and P. funiculosum GS2 isolated from a medicinal plant Gloriosa superba, and the synthesized AgNPs showed antimicrobial activity against bacterial strains Streptococcus pyogenes, E. coli, and Enterococcus faecalis and a fungal strain C. albicans. Asad et al30 showed the anticancer activity of AgNPs synthesized using fungus Humicola sp., and AgNPs exhibited anticancer activity against MDA-MB-231 (human breast carcinoma) cell line. Netala et al33 prepared the AgNPs of 3–40 nm in size using endophytic fungus A. versicolor isolated from a medicinal plant Centella asiatica and showed the free radical scavenging activity (RSA) of AgNPs against 2,2′-diphenyl-1-picrylhydrazyl (DPPH) radicals with an IC50 value of 60.64 μg/mL. AgNPs also showed effective antimicrobial activity against S. aureus, Streptococcus pneumonia, P. aeruginosa, Klebsiella pneumonia, C. albicans, and Candida nonalbicans.
In the present study, the biosynthesis of AgNPs using extracellular filtrate of endophytic fungus Pestalotiopsis microspora isolated from the healthy leaf tissues of Gymnema sylvestre, an important medicinal plant that harbors many endophytic fungi, was reported. The AgNPs were characterized using different techniques including ultraviolet-visible (UV-Vis), Fourier transform infrared spectroscopy (FTIR), transmission electron microscopy (TEM), selected area electron diffraction (SAED), X-ray diffraction (XRD), particle size analysis, and eeta potential measurements. Biosynthesized AgNPs exhibit multifunctional biological activities including antimicrobial activity against both bacterial and fungal pathogens, free RSA against DPPH and hydrogen peroxide (H2O2) radicals, and anticancer activity against different cancer cell lines – B16F10 (mouse melanoma), SKOV3 (human ovarian carcinoma), PC3 (human prostate carcinoma), and A549 (human lung adenocarcinoma).
Materials and methods
Collection of plant materials
Mature leaves of the G. sylvestre were collected from Tirumala hills, Eastern Ghats, India, and authenticated with taxonomist, Department of Botany, Sri Venkateswara University, Tirupati, Andhra Pradesh, India. A voucher specimen (SVUBHGS-15) was deposited in the herbarium of the department.
Isolation of endophytic fungi
Leaves were first washed under running tap water and then with teepol solution. Then the leaves were rinsed thoroughly with Milli-Q water. In laminar air flow chamber, the leaves were surface sterilized with 10% H2O2 and then with 80% alcohol. Finally, the leaves were rinsed thoroughly with Milli-Q water for 3–4 times and then were transferred onto blotting paper and cut into small segments. The leaf segments were transferred aseptically onto the potato dextrose agar (PDA) plates. Plates were incubated at 25°C±2°C for 10 days. After 10 days, fungal mycelia grown on the surface of the PDA medium were picked and transferred onto fresh PDA plates. Identification of the pure fungal cultures was carried out by molecular characterization through 18S rRNA gene amplification and sequencing.
Molecular characterization of endophytic fungus
DNA extraction
The fungal mycelia grown on surface of PDA were harvested, placed into 1 mL Millipore water, frozen in liquid nitrogen, and disrupted using mortar and pestle. The extraction of genomic DNA was carried out using HiPurA fungal DNA isolation kit (Himedia, Mumbai, India).
Polymerase chain reaction amplification and Sanger’s dideoxy nucleotide sequencing of 18S rRNA gene
For the amplification of 18S rRNA gene from the extracted genomic DNA, fungal domain specific primers ITS1-5′-TCCGTAGGTGAACCTGCGG-3′ (forward primer) and ITS4-5′-TCCTCCGCTTATTGATATGC-3′ (reverse primer) were used. Amplification was performed in a 50 μL reaction mixture containing 50 ng of template DNA, 20 pmol of each primer, 200 μM each deoxy nucleoside triphosphate, 0.4 U of Taq DNA polymerase, and 1.5 mM MgCl2 in a CG palm cycler (Genetix Biotech Asia, Bangalore, India). The amplification cycles consisted of initial denaturation at 94°C for 5 minutes followed by 30 cycles of 94°C for 45 s, annealing at 55°C for 1 minute, extension at 72°C for 1 minute, and a final extension at 72°C for 7 minutes. The amplification was confirmed by running the amplified product in 1.2% w/v agarose gel electrophoresis with ethidium bromide staining and documented by gel documentation system (UVDI; Major Science, Saratoga, CA, USA). The polymerase chain reaction (PCR) products were purified using gel extraction kit. Sequencing was carried out using Sanger’s method at Yaazh Xenomics, Chennai, India. Homology search of the sequences obtained was carried out using BLASTn at NCBI GenBank (http://www.ncbi.nlm.nih.gov). Multiple sequence alignment of the obtained sequences and closely related sequences retrieved from NCBI GenBank was performed using ClustalW2 with default parameters. Phylogenetic tree was constructed by the neighbor-joining (NJ) method with nucleotide pairwise genetic distance corrections. Bootstrap test of phylogeny was carried out to check the reliability of tree topology as a percentage of 1,000 replications. All branches with <70% bootstrap support were collapsed.
Biosynthesis of AgNPs
Biosynthesis of AgNPs was carried out according to the method described by Jaidev et al.32 The fungal isolate P. microspora was cultured in 100 mL of potato dextrose broth. Then the medium was incubated at 25°C±2°C in shaking incubator (Labline, Chennai, India) at a speed of 100 rpm. After 6 days of incubation, fungal biomass was separated, macerated, transferred to 100 mL of sterile double-distilled water, boiled, and filtered. Experimental solution was prepared by adding 10 mL of fungal culture filtrate to 90 mL of 1 mM AgNO3 solution and incubated in the dark for 24 hours at room temperature.
Characterization of AgNPs
UV-Vis spectrum was recorded on a UV-Vis double beam spectrophotometer (Model: Spectro UV 2060 Plus; Analytical Technologies Ltd, Baroda, India) from 200 to 700 nm with the resolution of 1 nm to confirm the biosynthesis of AgNPs. The colloidal solution of AgNPs was purified by repeated centrifugation at 15,000 rpm for 10 min followed by redispersion of the pellet of AgNPs into sterile double-distilled water. The centrifugation and redispersion processes were repeated thrice. The final pellet obtained was air dried to get the pure powder of AgNPs and was used for further characterizations. FTIR spectrum was recorded on Alpha interferometer (Bruker, Zurich, Switzerland) at a resolution of 2 cm−1. XRD analysis was carried out to study the crystalline structure of the AgNPs using Cu Kα radiation source on an Ultima IV X-ray powder diffractometer (Rigaku Ltd., Tokyo, Japan). TEM analysis was carried out to determine the shape and size of the biosynthesized AgNPs. The sample for TEM analysis was prepared by putting a drop of the AgNPs solution on carbon-coated copper grids and allowed to dryness. TEM analysis was done using FEI Tecnai F12 (Philips Optics Ltd, Eindhoven, the Netherlands) operated at 100 kV. SAED pattern of the AgNPs was also studied by TEM. Particle size analysis and zeta potential value (surface charge) were measured by using dynamic light scattering (DLS) system (SZ-100; Horiba Scientific, Stanmore, UK).
In vitro antioxidant activity
DPPH free radical scavenging assay
Free RSA of biosynthesized AgNPs was measured in vitro by DPPH radical scavenging assay.42 Approximately 4 mg of DPPH was dissolved in 100 mL of methanol and stored at 4°C until required. Approximately 2 mL of this stock solution was added to 1 mL of methanol solution containing test sample of different concentrations (25, 50, 75, and 100 μg/mL). Ascorbic acid was used as standard. The DPPH RSA was measured by taking the absorbance at 517 nm.
|
The concentration of AgNPs required to scavenge 50% of the radicals (IC50) was determined from the linear regression curve.
H2O2 radical scavenging assay
H2O2 RSA of the biosynthesized AgNPs was carried out according to the method of Patel et al.43 In this assay, different concentrations (25, 50, 75, and 100 μg/mL) of tested samples were used, and ascorbic acid was used as standard. The percentage of RSA was calculated using the following formula:
|
IC50 values are calculated using linear regression curve.
Cell lines and cell culture
All cancer cell lines B16F10 (mouse melanoma), SKOV3 (human ovarian carcinoma cells), PC3 (human prostate carcinoma), and A549 (human lung adenocarcinoma) and Chinese hamster ovary (CHO) cell line were obtained from National Centre for Cellular Sciences, Pune, India. Cells were cultured either in Roswell Park Memorial Institute (RPMI) medium-1640 (PC3) or Dulbecco’s Modified Eagle’s (DME) medium (CHO, B16F10, SKOV3, A549) supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 1 mM NaHCO3, 2 mM glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin. All cell lines were incubated at 37°C in a humidified atmosphere of 5% CO2.
Test concentrations
Initially, a stock solution of AgNPs was prepared in a suitable media (DMEM and RPMI-1640) with a final concentration of 8 mg/mL. Working solutions were prepared by serial dilutions with an appropriate culture media. AgNPs effect on different cell lines were tested using required concentrations ranging from 10 to 200 μg/mL.
Cytotoxicity study
Cytotoxicty was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay, according to the method of Mosmann.44 Briefly, the cells (1×104) were seeded in each well containing 100 μL of medium in 96-well plates. After overnight incubation (at 37°C in a humidified atmosphere of 5% CO2), exactly 100 μL of media with different test concentrations (10–200 μg/mL) were added to the cell suspension. The viability of cells was assessed after 24 h, by adding 10 μL of MTT (5 mg/mL) per well and incubated at 37°C for additional 3 h. The medium was discarded, and formazan blue that was formed in the cells was dissolved in 100 μL of dimethyl sulfoxide. The intensity of color formation was measured at 570 nm in a spectrophotometer (Spectra MAX Plus; Molecular Devices, Sunnyvale, CA, USA) supported by SOFTmax® PRO-5.4 (Molecular Devices). The percent inhibition of cell viability was determined with reference to the control values (without test compound). Data were generated from five separate experiments, and the mean values were subjected to linear regression analysis for the best straight-line fit. The IC50 concentrations (50% decrease in cell number as compared with that of the control values) were calculated using the respective regression equations.
Apoptosis assay
Experiments to study the mode of action of AgNPS were further extended using the most sensitive cell line (SKOV3). Based on the preliminary investigations, test concentrations of 10–40 μg/mL were used to study the morphological alterations.
Morphological analyses
Microscopic analysis
Cell shrinkage, membrane blebbing, and formation of apoptotic bodies were evaluated for 12 h (high magnification 400×) using a light microscope (Nikon Eclipse TS100; Tokyo, Japan). Cells undergoing apoptosis exhibit an increase in chromatin condensation. Morphologically, the apoptotic nuclei become smaller compared to those of normal cells; also, they become hyperfluorescent when labeled with some nuclear stains. Cells (2×104 cells/mL in a 24-well tissue culture plate) were either left untreated or stimulated with test compound (10, 20, 30, and 40 μg/mL) for 12 hours. The nuclear morphology was observed under fluorescence microscope (Optika Microscopes, Ponteranica, Italy) using respective filters.
Acridine orange/ethidium bromide staining
Cell death (apoptosis or necrosis) was analyzed by measuring the permeability of cell membrane with acridine orange (AO) and ethidium bromide (EB) double-staining assay. A cell suspension that had been exposed for 12 h was mixed with 5 μL of flurochrome solution in DMEM medium at 37°C for 10 minutes which contains 100 μg/mL AO and 100 μg/mL EB. Later, cells were washed with phosphate-buffered saline (PBS pH 7.4) twice and visualized under a blue filter. Live cells are permeable to AO but not to EB and stained green, and dead cells are permeable to both AO and EB, and EB stained DNA red. Thus this staining assay is easier to determine apoptotic cells or necrotic cells. Digital images were taken with fluorescence microscope.
Hoechst staining
The bisbenzimide dye Hoechst 33342 (Sigma aldrich, Hyderabad, India) was used to study the condensed chromatin of apoptotic cells, which binds to adenosine/thymidine (AT) at minor groove of DNA and stains brightly. However, normal cells (looser chromatin) exhibit moderate staining. The rate of apoptotic cell formation was monitored after 12 hours post exposure in treated cells and compared with untreated cells (control). Briefly 12 hours post exposure cells (SKOV3) were harvested by centrifugation (110 rpm for 10 minutes at 4°C), and the obtained pellet was resuspended in 1 mL PBS. Each sample was stained with 10 μL Hoechst solution (dissolved in double-distilled water at a stock concentration of 0.1 mg/mL) for 5 minutes at 37°C in the dark. The dye was removed by centrifugation (1,000 rpm for 10 minutes at 4°C) and washed with PBS. Condensed chromatin was visualized under blue filter with a magnification of 400×.
Results and discussion
An endophytic fungus isolated from the healthy and mature leaves of G. sylvestre was identified on the basis of the phenotypic characters like colony morphology, mycelia, and hyphal pattern. In this study, fungal isolate contained 5-celled conidia with equally pigmented cells (concolorous) and unbranched apical appendages (bisetulatae) and was identified as Pestalotiopsis sp. To identify the organism at species level, nucleotide sequencing of 18S rRNA gene was carried out. Genomic DNA isolated from this culture was subjected to PCR for amplification of 18 S rRNA gene using ITS primers. The amplified products resulted in 610 bp upon Sanger dideoxy nucleotide sequencing. The sequence obtained was subjected to BLAST search for homology and identification. The fungal sequence was identified as P. microspora VJ1/VS1. The sequence has been deposited in NCBI nucleotide database and the accession number has been assigned as KX213894. The phylogenetic tree constructed using NJ method with the closely related sequences obtained from the NCBI Genbank was shown in Figure 1. The correct identification of endophytic fungi by phenotypic characters is a major problem, as many of them are pleomorphic and exhibit different anamorphs in planta and in vitro. Hence, most of the recent studies use molecular identification through ITS sequencing of 18S rRNA gene. Furthermore, the authentication of the identified species should be carried out by comparing the obtained sequences to those available in NCBI data.
P. microspora belongs to the phylum Ascomycetes and produces many bioactive compounds of pharmaceutical and biomedical importance.38,39 Bioactive compounds including ambuic acid, pestalopyrone, and hydroxyl pestalopyrone are active against human pathogens, and jesterone and hydroxyl jesterone are active against plant pathogens.38–41 The exploration of the P. microspora for the synthesis of AgNPs could be another important advantage of this fungus. The biosynthesis of AgNPs using aqueous culture filtrate of P. microspora was investigated. After 24 hours of incubation, the reaction solution turned to dark brown color which indicated the synthesis of AgNPs. It is well known that AgNPs exhibit a dark brown color in aqueous solution due to surface plasmon resonance (SPR) phenomenon which in turn due to electromagnetic waves were excited in the visible region.1,2,33
UV-Vis analysis
To confirm the biosynthesis of AgNPs from fungal culture filtrate, UV-Vis analysis was carried out. UV-Vis spectrum showed a strong absorption peak at 435 nm (Figure 2) due to the surface plasmon (it describes the collective excitation of conduction electrons in a metal) excitation. The fungal extract does not show any such peak. The peak between 380 and 460 nm is the characteristic peak for the AgNPs synthesized. The peak is due to the surface plasmon vibrations exhibited by AgNPs in the visible range. The SPR peak could be responsible for spherical shape of the biosynthesized AgNPs. The intensity and peak position of the SPR peak depends on the size, morphology, and dielectric properties of the metal nanoparticles.5,6 The applications of metal nanoparticles have attracted a significant interest as a novel platform for nanobiotechnology and nanomedicine due to convenient surface bioconjugation with molecular biomarkers and remarkable optical properties related to the localized plasmon resonance.1–4
FTIR analysis
FTIR analysis was performed to reveal the biomolecules responsible for bioreduction of silver ions (Ag+) into AgNPs (Ag0) and capping of AgNPs synthesized. FTIR spectra (Figure 3) showed peaks at 3,333, 2,122, 1,635, 1,400, 1,066, and 699 cm−1. The peak at 3,333 cm−1 corresponds to stretching vibrations of O-H bonds of alcohols or phenols.45 The peak at 1,635 cm−1 is attributed to the N-H bend of primary amines.46 The peak at 1,400 cm−1 is due to symmetrical C-O stretching of carboxyl groups.47 The peak at 1,066 cm−1 is responsible for OH deformation of alcoholic and phenolic OH groups. The peak at 699 cm−1 is due to aromatic >C-H bond.48 FTIR results clearly revealed the presence of phenolic compounds (flavanoids/triterpenoids) and proteins that are plausibly involved in the biosynthesis of AgNPs and also that proteins play an important role in the stabilization of AgNPs by capping, which in turn prevent agglomeration and stabilization of formulation. Chemical synthesis procedures involve toxic chemicals (polymers/surfactants) as capping agents which limit the applications of AgNPs in biological systems as these are hazardous and noncompatible. Separate capping agents have to be avoided as they are mostly hazardous chemicals. This is the main advantage of biomimetic approaches over chemical synthesis approaches.
  | Figure 3 FTIR spectrum of biosynthesized AgNPs. |
XRD analysis
Crystalline nature of the biosynthesized AgNPs was determined by XRD analysis. XRD pattern (Figure 4) confirmed the face-centered cubic (FCC) lattice of silver by showing diffraction peaks at 38.22°, 44.38°, 64.56°, and 77.60° corresponding to the planes (111), (200), (220), and (311), respectively. XRD results are in agreement with standard JCPDS data (File No 87-0597). The results of XRD pattern showed that the synthesized AgNPs were crystalline in nature with FCC phase and were consistent with earlier reports for the AgNPs synthesized using fungi.24–27
  | Figure 4 XRD pattern of biosynthesized AgNPs. |
TEM analysis
TEM analysis was performed to determine the size and morphology of the AgNPs. TEM micrograph (Figure 5A) showed mono dispersed AgNPs sized between 2 and 10 nm with spherical shape and without any aggregation. The size was further confirmed by particle size analysis by DLS. SAED pattern (Figure 5B) showed the diffraction rings corresponding to (111), (200), (220), and (311) planes of the FCC crystal lattice and hence confirmed their highly crystalline nature. The crystalline nature was further confirmed by XRD analysis. TEM observations are in line with many earlier reports for AgNPs synthesized using various fungal extracts.24–30
Particle size analysis by DLS
Particle size analysis was employed to calculate the hydrodynamic radii or size of the biosynthesized AgNPs. The sizes of biosynthesized AgNPs were between 2 and 10 nm with an average hydrodynamic radius of 2.8 nm (Figure 5C). Results of particle size analysis are consistent with TEM results, and the difference in their size suggests that the AgNPs are capped by proteins and polyphenolic compounds of different molecular weights. Proteins and flavonoids conferred stability by capping, and the stability was further confirmed by zeta potential measurements.
Zeta potential measurement
Zeta potential is the key parameter to reveal the stability of biosynthesized AgNPs in a colloidal dispersion. Zeta potential indicates the degree of electrostatic repulsive forces present between similarly charged and adjacent nanoparticles in a colloidal dispersion. For the nanoparticles, high negative zeta potential value confers high dispersity, good colloidal nature, long-term stability, and without any aggregation. The high stability of AgNPs is very crucial for their biomedical applications. The zeta potential value for the biosynthesized AgNPs in this study was found to be −35.7 mV (Figure 5D). This high negative value confirms electrostatic repulsion among the AgNPs and thereby prevents the nanoparticles from agglomeration in the medium which in turn leads to long-term stability of the formulation.
Table 1 showed the comparison of the characteristic features of the AgNPs synthesized using different fungal species reported earlier.
  | Table 1 Comparison of the characteristic features of AgNPs biosynthesized by different fungal extracts |
In vitro antioxidant activity
In vitro antioxidant activity of the biosynthesized AgNPs was checked by DPPH free radical scavenging assay. This method is dependent on the reduction of DPPH radical to the nonradical form DPPH–H in the presence of a hydrogen-donating antioxidant. The DPPH RSA (%) values of the aqueous culture filtrate, biosynthesized AgNPs, and standard ascorbic acid were represented in Figure 6A. The RSA of the AgNPs was increased with the increasing concentration, and the maximum RSA of 55.84%±1.31% was observed at highest test concentration of 100 μg/mL used in this assay, while at the same concentration the maximum RSA of the aqueous fungal culture filtrate was found to be 33.37%±1.12% which is ~40% less compared to that of AgNPs. IC50 concentrations (the concentration of the sample required to scavenge 50% radicals) of the aqueous fungal culture filtrate and biosynthesized AgNPs were found to be 182.89±3.43 μg/mL and 76.95±2.96 μg/mL, respectively.
The biosynthesized AgNPs also exhibited effective scavenging activity against H2O2 radicals (Figure 6B), and the maximum scavenging activity of 51.14%±1.78% was observed at the highest concentration of 100 μg/mL used in this assay. Whereas for fungal extracellular filtrate, the maximum scavenging activity recorded was 31.28%±1.63%. The IC50 values were also determined using linear regression curve. The IC50 values for the AgNPs and aqueous fungal culture filtrate were found to be 94.95±2.18 and 184.83±4.04 μg/mL, respectively. The lower IC50 value of the biosynthesized AgNPs indicates their effective free RSA. Based on our results, it is concluded that the biosynthesized AgNPs showed very good antioxidant activity. The excellent RSA of AgNPs allowed them to be used as antioxidants and as ingredients of antioxidant formulations in the biomedical and pharmaceutical fields.
Cytotoxicity study
It is very crucial to check the biocompatibility of AgNPs for their successful application in biomedicine and diagnostic fields. The cytotoxicity of AgNPs was also checked against normal CHO cell lines to check their biocompatibility, as safety is the main concern, and toxicity against normal cell lines can limit their therapeutic applications. In this study, AgNPs could not show any inhibition against normal cell lines at lower concentrations. We checked the effect of AgNPs at 100, 200, 300, 400, and 500 μg/mL concentrations. Cell viability of normal cells is decreased with an increasing concentration of AgNPs (Figure S1). The IC50 value of AgNPs against normal CHO cells was determined as 438.53±4.2 μg/mL.
The cytotoxic activity of AgNPs was tested against four human cancer cell lines B16F10 (mouse melanoma), SKOV3 (human ovarian carcinoma), A549 (human lung adenocarcinoma), and PC3 (human prostate carcinoma) by measuring the number of live cells after 24 h of treatment (MTT assay). Cell viability of the tested cancer cells is decreased with an increasing concentration of AgNPs (Figure 7). Only 9.87%±1.34% (in case of B16F10), 5.68%±1.10% (in case of SKOV3), 17.87%±2.19% (in case of A549), and 14.35%±2.31% (in case of PC3) cells are viable (or live) at the highest concentration of AgNPs (200 μg/mL) used in this study. IC50 (the concentration that results in a 50% decrease in cell number as compared with that of the control cultures in the absence of an inhibitor) values are determined. AgNPs exhibited potent antiproliferative activity and inhibited cell growth on B16F10, SKOV3, A549, and PC3 cells with cytotoxic potencies (IC50 values) of 26.43±3.41, 16.24±2.48, 39.83±3.74, and 27.71±2.89 μg/mL, respectively. As such based on the IC50 values, AgNPs showed more toxic effect on SKOV3 and were found to be 1.63-, 1.70-, and 2.45-fold greater potency than B16F10, PC3, and A549, respectively. However, the respective activities on B16F10, SKOV3, A549, and PC3 cells were comparatively 6.41-, 1.51-, 3.71-, and 24.30-fold less than the standard drug etoposide (Table 2), respectively.
Morphological changes and apoptosis of cells after exposure to AgNPs were studied on most susceptible SKOV3 cells using light microscopy, AO/EB dual staining, and Hoechst staining assays. Cytological observations revealed concentration-dependent apoptotic morphology in the exposed SKOV3 cells, stained with Hoechst 33342 and AO/EB. When exposed to AgNPs at concentrations of 20 and 40 μg/mL for 12 h, the treated SKOV3 cells showed distinct morphological changes (apoptotic features) indicating unhealthy cells, whereas cells in the control group appeared to be normal. As the AgNPs concentration increases, the exposed cells appeared to be clustered and exhibited several morphological alterations (cell membrane blebbing, cell shrinkage, nuclear condensation/pyknotic nuclei, destructive fragmentation of nuclei/karyorrhexis, etc.) which eventually lead to cell death (Figure 8). AO/EB staining is a common method to visualize the apoptotic features. AO is a vital dye that can penetrate normal cells and can exhibit green fluorescence in live cells, whereas EB stains only cells that have lost cell membrane integrity. Late apoptotic cells exhibit orange fluorescence as they incorporated EB. In our study, late apoptotic cells with irregular distribution of chromatin (karyorrhexis) and destructive fragmentation of the nuclei were indicated in yellow circles. Late apoptotic cells with condensed chromatin and their nuclei were indicated in red circles. Our results are consistent with earlier studies and confirm that the induction of apoptosis is mainly by features such as membrane blebbing, reduced and shrunken cells, condensed nuclei, and DNA fragmentation.49 Various reports suggest that cytotoxic activity of AgNPs toward cancer cells is attributed due to the production of reactive oxygen species (ROS) like peroxides, superoxides, and singlet oxygen species which in turn lead to cellular damage.50–52 Increased production of ROS, especially superoxides uncouple the electron transport chain and oxidative potential in mitochondria led to decreased adenosine triphosphate production which in turn leads to DNA damage.53,54 Several studies reported that active caspase-3 expression in cells treated with AgNPs mediate the apoptotic features. AgNPs could induce oxidative stresses that lead to the induction of apoptosis pathway via upregulation of P53 pathway.49–51
Anticancer activities of AgNPs might be related to the bioproducts of the fungal species that adhered to the outer surface of AgNPs. There were reports that the torreyanic acid, a natural bioactive compound from P. microspora, showed cytotoxicity against 25 different cancer cell lines with an average IC50 value of 9.4 μg/mL. Isopestacin and Pestalotiopsis are natural compounds from P. microspora and possess potent anticancer, antioxidant, and antifungal activities. FTIR results indicated that the bioproducts of fungal filtrate were coated on AgNPs which might be responsible for bioactivities of AgNPs.
In this study, the biosynthesized AgNPs were 2–10 nm in size with an average diameter of 2.8 nm. The smaller sizes of the nanomaterials have the advantage of high permeability and retention property. Small-sized nanoparticles can be easily uptaken by cells through endocytosis and other processes. As a result, they can enhance their therapeutic efficiency due to enhanced accumulation, effective diffusion, and deep penetration in the tumor matrix. Nanoparticles sized between 30 and 100 nm cannot diffuse effectively and cannot penetrate deeply into the tumor matrix, and they tend to accumulate at the blood vessel wall and can only reach first few layers of the tumor matrix. As a result, they cannot eradicate the tumor cells that are far from blood vessels. Eliminating the tumor cells near the blood vessels may halt the growth of the tumors but cannot eradicate the tumorigenic cells completely.
There are some reports on the anticancer activity of AgNPs synthesized from different fungal extracts. Asad et al30 reported that AgNPs synthesized from fungus Humicola sp. showed cytotoxicity against MDA-MB-231 human breast carcinoma cell line. El-Sonbaty54 reported that AgNPs from Agaricus bisporus showed cytotoxicity against human breast cancer cell line (MCF-7) with IC50 value of 50 μg/mL. In this study, P. microspora-mediated green synthesis of AgNPs selectively inhibits cell proliferation in the following cancer cell lines (B16F10, SKOV3, A549, and PC3; IC50 values of 26.43, 16.24, 39.83, and 27.71 μg/mL, respectively), but no significant effect was observed on normal CHO cells. Previous studies indicated that the secondary metabolites of fungus have exhibited potent anticancerous properties. Our FTIR analysis also clearly indicated that the secondary metabolites of fungus attached (capped) to the AgNPs showed significant anti cancerous properties.
Based on this result, it is evident that biosynthesized AgNPs are biocompatible to normal cells and cytotoxic to cancer cells and hence can be employed as future anticancer agents or drug delivery systems. Table 3 showed the characteristic features and bioactivities of AgNPs synthesized in this study.
Conclusion
The present study reports the simple and environmental benign approach for the biosynthesis of AgNPs using aqueous culture filtrate of P. microspora, an endophytic fungus isolated from the leaves of G. sylvestre. Fungal culture filtrate mediated the successful synthesis of spherical-shaped AgNPs of 2–10 nm in size. The AgNPs synthesized were very stable and biocompatible. The biosynthesized AgNPs exhibited effective antioxidant and anticancer activities. The biosynthesized AgNPs have proved to be potent cytotoxic agents against B16F10, SKOV3, PC3, and A549. The plausible apoptotic changes were also explained against the most susceptible SKOV3 cell lines. The results of cytotoxic studies demonstrated that the biosynthesized AgNPs can be used as anticancer agents in the near future.
Acknowledgment
The authors are thankful to Dr B Sreedhar, Scientist-F, IICT, Hyderabad, Telangana, India, for providing TEM facilities and his valuable suggestions in preparing the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Kreibig U, Vollmer M. Optical Properties of Metal Clusters. Berlin, New York: Springer; 1995:483–525. | ||
Ravindran A, Preethy C, Khan SS. Biofunctionalized silver nanoparticles: advances and prospects. Colloids Surf B Biointerfaces. 2013;105:342–352. | ||
Moreira LM, Carvalho EA, Bell MJV, et al. Thermo-optical properties of silver and gold nanofluids. J Therm Anal Calorim. 2013;114:557–564. | ||
Edison T, Sethuraman M. Instant green synthesis of silver nanoparticles using Terminalia chebula fruit extract and evaluation of their catalytic activity on reduction of Methylene Blue. Process Biochem. 2012;47:1351–1357. | ||
Kelly KL, Coronado E, Zhao LL, Schatz GC. The optical properties of metal nanoparticles: the influence of size shape, and dielectric environment. J Phys Chem B. 2003;107:668–677. | ||
Kilin DS, Prezhdo OV, Xia YN. Shape-controlled synthesis of silver nanoparticles: Ab initiostudy of preferential surface coordination with citric acid. Chem Phys Lett. 2008;458:113–116. | ||
Parisa S, Leila S, Zahra RM, Arezou S, Roya R, Bahareh S. A penicillin biosensor by using silver nanoparticles. Int J Electrochem Sci. 2014;9:6201–6212. | ||
Jiajie L, Liu H, Caihong H, et al. Aggregated silver nanoparticles based surface-enhanced raman scattering enzyme-linked immunosorbent assay for ultrasensitive detection of protein biomarkers and small molecules. Anal Chem. 2015;87:5790–5796. | ||
Chinnasamy G, Reddy JV, Bhaarathy V, Ramaktishna S, Srinivasan DK. Biocomposite nanofibrous strategies for the controlled release of biomolecules for skin tissue regeneration. Int J Nanomedicine. 2014;9:4709–4722. | ||
Chen XJ, Sanchez GBL, Qian ZX, Park SJ. Noble metal nanoparticles in DNA detection and delivery. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4:273–290. | ||
Bobbu PL, Netala VR, Aishwarya S, Reddy IRM, Kotakadi VS, Tartte V. Rapid synthesis of silver nanoparticles using aqueous leaf extract of Achyranthes aspera and study of their antimicrobial and free radical scavenging activities. Int J Pharm Pharm Sci. 2016;8:341–346. | ||
Ramamurthy CH, Padma M, Samadanam ID, et al. The extra cellular synthesis of gold and silver nanoparticles and their free radical scavenging and antibacterial properties. Colloids Surf B Biointerfaces. 2013;102:808–815. | ||
Arokiyaraj S, Arasu MV, Vincent S, et al. Rapid green synthesis of silver nanoparticles from Chrysanthemum indicum L and its antibacterial and cytotoxic effects: an in vitro study. Int J Nanomedicine. 2014;9:379–388. | ||
Arunachalam KD, Arun LB, Annamalai SK, Arunachalam AM. Potential anticancer properties of bioactive compounds of Gymnema sylvestre and its biofunctionalized silver nanoparticles. Int J Nanomedicine. 2015;10:31–41. | ||
Khan M, Khan M, Adil SF, et al. Green synthesis of silver nanoparticles mediated by Pulicaria glutinosa extract. Int J Nanomedicine. 2013;8:1507–1516. | ||
Pasupuleti VR, Prasad, Shiekh RA, et al. Biogenic silver nanoparticles using Rhinacanthus nasutus leaf extract: synthesis, spectral analysis, and antimicrobial studies. Int J Nanomedicine. 2013;8:3355–3364. | ||
Kasi M, Senthilkumar B, Duraisamy S, Saleh AS. Biosynthesis of silver nanoparticles using Acacia leucophloea extract and their antibacterial activity. Int J Nanomedicine. 2014;9:2431–2438. | ||
Zhao X, Xia Y, Li Q, et al. Microwave-assisted synthesis of silver nanoparticles using sodium alginate and their antibacterial activity. Colloids Surf A Physicochem Eng Aspects. 2014;444:180–188. | ||
Rojas JV, Castano CH. Radiation-assisted synthesis of iridium and rhodium nanoparticles supported on polyvinylpyrrolidone. J Radioanal Nucl Chem. 2014;302:555–561. | ||
Togashi T, Saito K, Matsuda Y, et al. Synthesis of water-dispersible silver nanoparticles by thermal decomposition of water-soluble silver oxalate precursors. J Nanosci Nanotechnol. 2014;14:6022–6027. | ||
Valverde-Alva MA, Garcia-Fernandez T, Villagran-Muniz M, et al. Synthesis of silver nanoparticles by laser ablation in ethanol: a pulsed photoacoustic study. Appl Surf Sci. 2015;355:341–349. | ||
Jensen TR, Malinsky MD, Haynes CL, Duyne RPV. Nanosphere lithography: tunable localized surface plasmon resonance spectra of silver nanoparticles. J Phys Chem B. 2000;104:10549–10556. | ||
Byeon JH, Kim YW. A novel polyol method to synthesize colloidal silver nanoparticles by ultrasonic irradiation. Ultrason Sonochem. 2012;19:209–215. | ||
Devi SL, Donalad AB, Joshi SR. Studies on biosynthesis of antimicrobial silver nanoparticles using endophytic fungi isolated from the ethno-medicinal plant Gloriosa superba L. P Natl A Sci India B. 2014;84:1091–1099. | ||
Li G, Dan H, Yongqing Q, et al. Fungus-mediated green synthesis of silver nanoparticles using Aspergillus terreus. Int J Mol Sci. 2012;13:466–476. | ||
Javed M, Dwivedi S, Singh BR, et al. Production of antimicrobial silver nanoparticles in water extracts of the fungus Amylomyces rouxii strain KSU-09. Biores Technol. 2010;101:8772–8776. | ||
Rashmi S, Preeti V. Biomimetic synthesis and characterization of protein capped silver nanoparticles. Biores Technol. 2009;100:501–504. | ||
Rafie MH, Shaheen TI, Mohamed AA, Hebeish A. Bio-synthesis and applications of silver nanoparticles onto cotton fabrics. Carbohydr Polym. 2012;90:915–920. | ||
Ahmad A, Mukherjee P, Senapati S, et al. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium oxysporum. Colloids Surf B. 2003;28:313–318. | ||
Asad S, Supriya S, Gopal CK, Absar A. Biological synthesis of silver nanoparticles using the fungus Humicola sp. and evaluation of their cytotoxicity using normal and cancer cell lines. Spectrochim Acta A Mol Biomol Spectrosc. 2013;114:144–147. | ||
Kirthi AV, Rahuman AA, Chidambaram J, et al. Novel approach to synthesis silver nanoparticles using plant pathogenic fungi, Puccinia graminis. Mat Lett. 2012;81:69–72. | ||
Jaidev LR, Narasimha G. Fungal mediated biosynthesis of silver nanoparticles, characterization and antimicrobial activity. Colloids Surf B. 2010;81:430–433. | ||
Netala VR, Kotakadi VS, Bobbu PL, Gaddam SA, Tartte V. Endophytic fungal isolate mediated biosynthesis of silver nanoparticles and their free radical scavenging activity and anti microbial studies. 3 Biotech. 2016;6(132):1–9. | ||
Metuku RP, Pabba S, Burra S, Himabindu N, Gudikandula K, Singara Charya MA. Biosynthesis of silver nanoparticles from Schizophyllum radiatum HE 863742.1: their characterization and antimicrobial activity. 3 Biotech. 2014;4:227–234. | ||
Fayaz AM, Balaji K, Kalaichelvan PT, Venkatesan R. Fungal based synthesis of silver nanoparticles – an effect of temperature on the size of particles. Colloids Surf B. 2009;74:123–126. | ||
Monali G, Jayendra K, Avinash I, Aniket G, Mahendra R. Fungus-mediated synthesis of silver nanoparticles and their activity against pathogenic fungi in combination with fluconoazole. Nanomedicine. 2009;5:382–386. | ||
Vigneshwaran N, Kathe AA, Varadarajan PV, Nachane RP, Balasubramanya RH. Biomimetics of silver nanoparticles by white rot fungus Phaenerochaete chrysosporium. Colloids Surf B. 2006;53: 55–59. | ||
Strobel GA, Ford EJ, Worapong JK, et al. Ispoestacin, an isobenzofuranone from Pestalotiopsis microspora, possessing antifungal and antioxidant activities. Phytochemistry. 2002;60:179–183. | ||
Ding G, Li Y, Fu S, Liu S, Wei J, Che Y. Ambuic acid and torreyanic acid derivatives from the endolichenic fungus Pestalotiopsis sp. J Nat Prod. 2009;72:182–186. | ||
Lee JC, Lobkovsky E, Pliam NB, Strobel G, Clardy J. Subglutinols A and B: immunosuppressive compounds from the endophytic fungus Fusarium subglutinans. J Org Chem. 1995;60:7076–7077. | ||
Li JY, Strobel GA. Jesterone and hydroxy-jesterone antioomycetcyclohexenenone epoxides from the endophytic fungus Pestalotiopsis jesteri. Phytochemistry. 2001;57:261–265. | ||
Aluru R, Duvvuru G, Netala VR, Tartte V, Deville A, Bodo B. Structure elucidation and antioxidant activity of the phenolic compounds from Rhynchosia suaveolens. Nat Prod Comm. 2015;10:609–611. | ||
Avani P, Amit P, Patel NM. Determination of polyphenols and free radical scavenging activity of Tephrosia purpurea linn leaves (Leguminosae). Pharmacogn Res. 2010;2:152–158. | ||
Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55-63. | ||
Thirunavoukkarasu M, Balaji U, Behera S, Panda PK, Mishra BK. Biosynthesis of silver nanoparticle from leaf extract of Desmodium gangeticum (L.) DC. and its biomedical potential. Spectrochim Acta Part A. 2013;116:424–427. | ||
Seyedeh MG, Sepideh H, Shojaosadati SA. Green synthesis of silver nanoparticles by a novel method: comparative study of their properties. Carbohydr Polym. 2012;89:467–472. | ||
Kora AJ, Sashidhar RB, Arunachalam J. Aqueous extract of gum olibanum (Boswellia serrata): a reductant and stabilizer for the biosynthesis of antibacterial silver nanoparticles. Process Biochem. 2012;47:1516–1520. | ||
Litvin VA, Minaev BF. Spectroscopy study of silver nanoparticles fabrication using synthetic humic substances and their antimicrobial activity. Spectrochim Acta Part A. 2013;108:115–122. | ||
Krishnaraj C, Muthukumaran P, Ramachandran R, Balakumaran MD, Kalaichelvan PT. Acalypha indica Linn: biogenic synthesis of silver and gold nanoparticles and their cytotoxic effects against MDA-MB-231, human breast cancer cells. Biotechnol Rep. 2014;4:42–49. | ||
Mukherjee S, Chowdhury D, Rajesh K, et al. Potential theranostics application of bio-synthesized silver nanoparticles (4-in-1 system). Theranostics. 2014;4(3):316–335. | ||
Pallab S, Chattopadhyay A, Ghosh SS. Induction of apoptosis in cancer cells at low silver nanoparticle concentrations using chitosan nanocarrier. ACS Appl Mater Interfaces. 2011;3:218–228. | ||
Gurunathan S, Jegadeesh R, Sri NAM, Priscilla AJ, Sabaratnam V. Green synthesis of silver nanoparticles using Ganoderma neo-japonicum Imazeki: a potential cytotoxic agent against breast cancer cells. Int J Nanomedicine. 2013;8:4399–4413. | ||
Mukherjee S, Mamatha D, Sumahitha P, Rajesh K, Vishnu SB, Patra CR. Green chemistry approach for the synthesis of gold nanoconjugates that induces the inhibition of cancer cells proliferation through induction of oxidative stress and their in vivo toxicity study. J Mater Chem B. 2015;3:3820–3830. | ||
El-Sonbaty SM. Fungus-mediated synthesis of silver nanoparticles and evaluation of antitumor activity. Cancer Nanotechnol. 2013;4:73–79. |
Supplementary material
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.