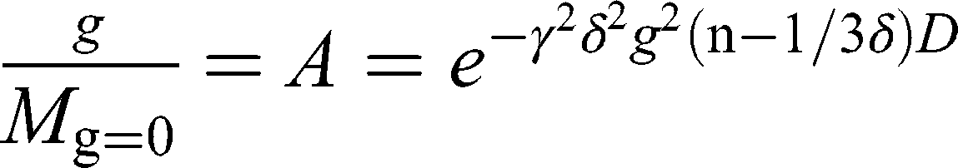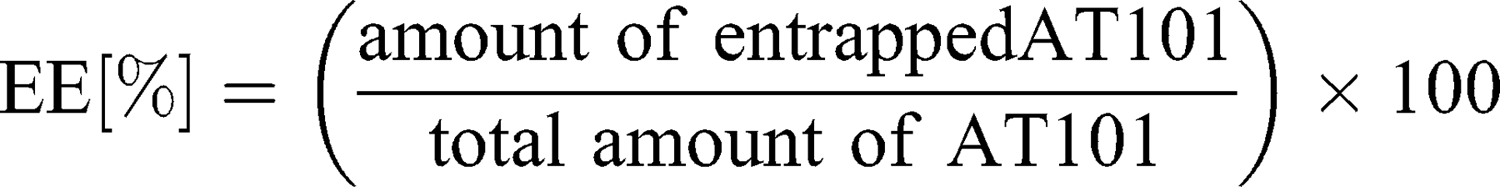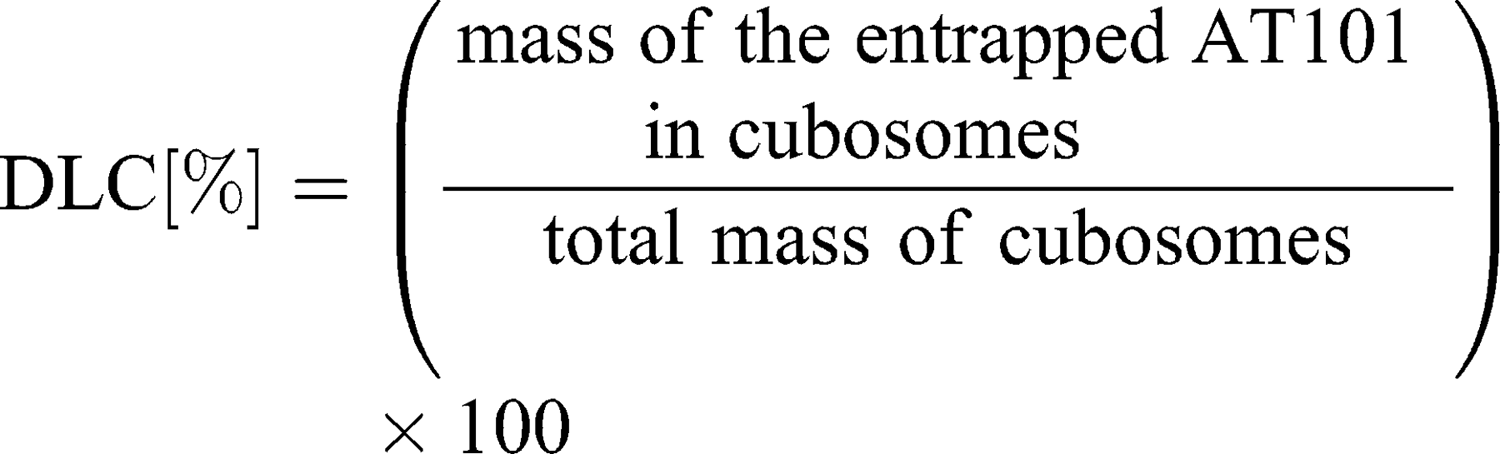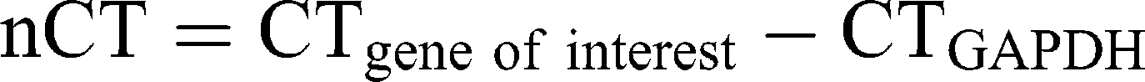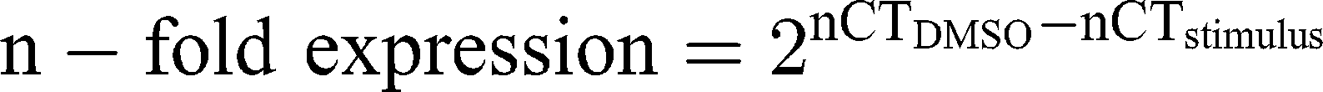Back to Journals » International Journal of Nanomedicine » Volume 15
AT101-Loaded Cubosomes as an Alternative for Improved Glioblastoma Therapy
Authors Flak DK, Adamski V, Nowaczyk G , Szutkowski K , Synowitz M, Jurga S , Held-Feindt J
Received 29 May 2020
Accepted for publication 14 July 2020
Published 5 October 2020 Volume 2020:15 Pages 7415—7431
DOI https://doi.org/10.2147/IJN.S265061
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Dorota K Flak,1 Vivian Adamski,2 Grzegorz Nowaczyk,1 Kosma Szutkowski,1 Michael Synowitz,2 Stefan Jurga,1 Janka Held-Feindt2
1NanoBioMedical Centre, Adam Mickiewicz University Poznań, Poznań, Poland; 2Department of Neurosurgery, University Hospital Schleswig-Holstein, Campus Kiel, Kiel, Germany
Correspondence: Dorota K Flak NanoBioMedical Centre
Adam Mickiewicz University Poznań, Wszechnicy Piastowskiej 3, Poznań PL 61614, Poland
Tel +48 8296713
Email [email protected]
Introduction: AT101, the R-(-)-enantiomer of the cottonseed-derived polyphenol gossypol, is a promising drug in glioblastoma multiforme (GBM) therapy due to its ability to trigger autophagic cell death but also to facilitate apoptosis in tumor cells. It does have some limitations such as poor solubility in water-based media and consequent low bioavailability, which affect its response rate during treatment. To overcome this drawback and to improve the anti-cancer potential of AT101, the use of cubosome-based formulation for AT101 drug delivery has been proposed. This is the first report on the use of cubosomes as AT101 drug carriers in GBM cells.
Materials and Methods: Cubosomes loaded with AT101 were prepared from glyceryl monooleate (GMO) and the surfactant Pluronic F-127 using the top–down approach. The drug was introduced into the lipid prior to dispersion. Prepared formulations were then subjected to complex physicochemical and biological characterization.
Results: Formulations of AT101-loaded cubosomes were highly stable colloids with a high drug entrapment efficiency (97.7%) and a continuous, sustained drug release approaching 35% over 72 h. Using selective and sensitive NMR diffusometry, the drug was shown to be efficiently bound to the lipid-based cubosomes. In vitro imaging studies showed the high efficiency of cubosomal nanoparticles uptake into GBM cells, as well as their marked ability to penetrate into tumor spheroids. Treatment of GBM cells with the AT101-loaded cubosomes, but not with the free drug, induced cytoskeletal rearrangement and shortening of actin fibers. The prepared nanoparticles revealed stronger in vitro cytotoxic effects against GBM cells (A172 and LN229 cell lines), than against normal brain cells (SVGA and HMC3 cell lines).
Conclusion: The results indicate that GMO-AT101 cubosome formulations are a promising basic tool for alternative approaches to GBM treatment.
Keywords: cubosome, lipid nanoparticles, glyceryl monooleate, drug delivery, GBM therapy, NMR diffusometry
Introduction
In recent years, the interest in novel and efficient drug delivery platforms has been directed toward highly curved nanoparticle (NP) systems formed from more exotic cellular membrane-like lyotropic phases. Under consideration are cubosomes formed by bicontinuous cubic phases followed by hexosomes formed by hexagonal phases of, eg, glyceryl monooleate or phytantriol.1,2 The potential success of these lipid-based NPs as drug delivery systems relies mainly on their high degree of biocompatibility and biodegradability, the presence of an extensive system of water channels, their physical properties including an amphiphilic lipid character and related drug (hydrophilic, hydrophobic, amphiphilic) loading/release efficiencies, as well as on the ease in producing NP. The pharmaceutical applications of cubosomes and hexosomes are still in the development stage compared with the well-established liposomes, but due to their unique properties and the promising research results with lipid-based NPs they are expected to expand.
Cubosomes are colloidal NPs formed mainly by a steric or electrostatic stabilization process modulated by the self-assembly of surfactant-like lipids forming the non-lamellar phases in excess of water and the presence of a stabilizer.2,3 The internal structure of cubosomes consists of a membrane bilayer forming a lattice structure type network with two continuous intertwinings, but non-intersecting aqueous channels – 3D well-ordered, bicontinuous cubic phase (V2).4 Their extensive system of water channels is responsible for the diffusion of their cargo, such as therapeutics with different polarities.3 In addition, highly curved lipid-based cubosomes have higher membrane surface-to-volume ratios, variations in membrane stress and increased loading capacity, compared with, eg, simple vesicle-like NPs (eg, liposomes), which have been the most exploited lipid assemblies in drug delivery systems so far.5,6 However, the relatively simple structure of vesicle-like NPs does not entirely mimic the complexity of cell membranes, which affects their interaction with treated cells, as well as their drug loading and release profiles. Therefore, liposome-based formulations suffer not only from premature drug leakage and fast release rate but also from a low permeability through the cell membrane and solid tumors, and a lack of targeting.7,8 The larger membrane surface area of cubosomes compared with simple vesicle-like NPs results in a significantly larger number of lipid molecules per particle. This enables not only a larger drug payload (with different polarities) but also controllable drug release. Moreover, cubosomes can be potentially used as efficient drug carriers due to their increased chemical and physical stability. However, to fully exploit the benefits of such nanomedicines, their size, shape, composition, biocompatibility and other properties related to drug delivery must be optimized.
Gliomas represent the majority of primary brain tumors in adults and the most malignant form, glioblastoma multiforme (GBM), accounts for more than 15% of all intracranial tumors and has a median survival time of 7–15 months from the time of diagnosis. The current standard treatment includes maximal safe surgical resection, followed by radiation with concurrent temozolomide (TMZ, Temodal®), an oral alkylating chemotherapy agent, and then adjuvant chemotherapy with TMZ. Due to the high degree of invasiveness and the fact that tumor cells infiltrate the surrounding brain tissue, surgical resection of the GBM mass is not curative, and the disease progresses or recurs. The improvements in GBM treatment are greatly limited by the inability to deliver chemotherapeutics to the brain tissue through the blood–brain barrier (BBB), which prevents toxins as well as many essential drugs from reaching the brain tissue. Moreover, despite the enhanced permeability and leakiness of glioma-associated vessels, effective chemotherapeutic delivery is limited by their heterogeneous distribution and the paucity of tumor cells around them, as well as by the altered dynamics of the cerebrospinal fluid.9,10 In addition to the discovery of new therapeutics and the development of enhanced treatment plans, nanoparticle-based drug delivery systems are being developed to help address this major hurdle in effective GBM chemotherapy treatment.11
Currently, AT101, the R-(-)-enantiomer of the naturally occurring cottonseed-derived polyphenol gossypol, is being studied as a promising drug for GBM therapy.12,13 AT101 is able to trigger autophagic cell death,14,15 and binds competitively to hydrophobic surface grooves of pro-survival Bcl-2 family members, counteracting their protective effects and facilitating apoptosis in tumor cells.13 Gossypol, as well as curcumin, bind to the 18 kDa translocator protein, previously known as the peripheral-type benzodiazepine receptor localized in the mitochondrial outer membrane, which is a key player in apoptotic signaling and cancer development.16 Recently, Mehner et al were able to show that AT101 induced cell death in GBM cells through regulation of the Akt-signaling pathway and the mitochondrial membrane potential.17 Furthermore, combined gossypol/TMZ treatment was associated with inhibition of tumor-associated angiogenesis, invasion and clonogenic growth in GBMs,18,19 AT101 potentiated cell death induced by TMZ, and sequentially applied single and combined TMZ and AT101 treatment strategies exhibited higher cytotoxicity and better tumor growth control compared with single TMZ treatment in a GBM in vitro model. They were also less harmful for human astrocytes.20,21 However, although well tolerated, gossypol had only a low, albeit a measurable response rate in patients with recurrent gliomas22 indicating that AT101 has some limitations such as, eg, poor solubility in water-based media and low bioavailability.
The use of a dedicated nanocarrier has been suggested in order to fully utilize the anti-cancer potential of AT101, but there have been only few attempts, so far, to proceed with such nanomedicine approaches. The first was liposome-based delivery of gossypol.23 Liposome formulations consisting of TPGS (D-alpha-tocopheryl polyethylene glycol succinate) or mPEG-DSPE (distearoylphosphatidylethanoloamine) modified with cholesterol and egg phosphatidylcholine exhibited high encapsulation efficiencies of over 90%, a drug concentration in the liposomes of more than 2 mg/mL, and a high stability. The authors claimed an improvement of the drug delivery system in terms of lower drug toxicity and better tolerated drug formulations for oral administration. The obtained increased IC50 values for the liposome-based formulations compared with the free drug in treatment of MCF-7 and KB cells indicated that the drug cytotoxicity was reduced by the encapsulation into liposomes. Another report focused on polymeric nanoparticles made from methoxypolyethylene glycol-maleimide (mPEG-Mal) and gossypol.24 The nanoparticles exhibited a favorable anti-cancer activity in human prostate cancer PC-3 cells via induction of apoptosis, with no toxicity of blank samples in a concentration comparable to that of the free drug. Another approach involved the use of gossypol-capped mitoxantrone-loaded mesoporous SiO2 nanoparticles for the coordinated and controlled release of two anti-cancer drugs. The prepared formulations exhibited preferential toxicity towards malignant breast cancer cells (MDA-MB-231).25 However, no studies have yet focused on the preparation of highly effective AT101-loaded cubosomes and their possibly enhanced therapeutic effects in the treatment of GBM. Thus, the intention of this study was to develop cubosome-based formulations for AT101 drug delivery, to analyze their physicochemical characteristics, and to investigate their uptake and biological effects in two different GBM cell lines.
Materials and Methods
Preparation of Blank and AT101-Encapsulating Cubosomal Nanoparticles
Cubosomes were prepared using the established top–down approach.3,26 Glyceryl monooleate (GMO, 90%, IOI Olea GmbH, Hamburg, DE) was used as the structure-forming lipid, and Pluronic F-127 (Merck, Darmstadt, DE) as the surfactant stabilizing the cubosome dispersion. For blank cubosomes, GMO was mixed with Pluronic F-127 in a w:w ratio of 1:0.25 and melted at 40°C until a viscous, transparent mixture was obtained. For drug-encapsulating nanoparticles, AT101 (Tocris Bioscience, Bristol, UK) was diluted in ethanol (99.8%, Avantor Performance Materials, Poland S.A.) and added to the mixture of lipid and surfactant to give a 10% (w/w) concentration. This was stirred until a homogenous mixture was obtained. The ethanol was evaporated from the mixture under vacuum overnight. Pre-warmed phosphate-buffered saline (PBS; pH 7.4) was added to the above mixture to obtain a concentration of 20 mg/mL (2% (w/w)) of GMO, and the solution was mixed and left overnight at room temperature (RT) for hydration to form a bulk cubic gel. To obtain a stable cubosome dispersion, the formed cubic gel was homogenized with a Branson Sonifier 250 (Emerson Electric Company, St Louis, MO, USA) at 60 W output power with a 2s ON and 2s OFF mode for a total of 15 min until a milky dispersion resulted. The homogenization was run in a jacketed vessel cooled with water. Prior to further analyses, the cubosome dispersions were allowed to stabilize at room temperature for 24 h.
Characterization of Cubosomal Nanoparticles
Transmission Electron Microscopy (TEM) – Morphology and Structure
Particle size and morphology were assessed by cryogenic transmission electron microscopy (cryo-TEM) with a JEM-1400 transmission electron microscope (Jeol, Akishima, JPN) operating at 120 kV. The samples were diluted with H2O(d) (1:1 (v/v) ratio). Then, 3−4 μL aliquots were deposited on lacey/carbon grids previously treated with plasma (Ted Pella, Inc, Redding, CA, USA) and freeze-dried at −170°C with the Cryoplunge 3 System (Gatan, Inc, Pleasanton, CA, USA). The temperature in the climate chamber was kept at about 22ºC and the humidity was maintained above 90% to prevent excessive evaporation. In order to verify the structure of the obtained liquid crystalline nanoparticles, a fast Fourier transform of selected images was performed, and a reverse transform was then used to obtain a filtered image showing the periodic, internal structure of cubosomes in more detail.
Dynamic Light Scattering (DLS) and Zeta Potential Measurements – Size and Surface Charge of Cubosomal Nanoparticles in Dispersions
The particle size distribution (Z-average, distribution by number and intensity) along with the polydispersity index (PdI), the zeta potential (ζ) of prepared cubosomes, as well as their stability over time and temperature, were measured on Zetasizer Nano-ZS (Malvern Instruments, Malvern, UK) based on the non-invasive dynamic light scattering method (DLS-NIBS) using an angle of 173° and electrophoretic light scattering (ELS), respectively. For the measurements, pure GMO cubosomes, as well as AT101-loaded cubosomes were stored either at RT or at 37°C and diluted in H2Od to 40 μg/mL prior to the measurement. Results are given as the mean and standard deviation (SD) of three individual analyses. In order to investigate the stability of the prepared cubosomes, the measurements were repeated at intervals over 17 days.
Nuclear Magnetic Resonance (NMR) – Diffusometry
NMR spectra were obtained using the 14 T Agilent NMR spectrometer DD2 and the dedicated diffusion probe DOTY DSI-1372 (28 T/m). Self-diffusion coefficients were determined using the “dbppste_wg” pulse sequence. 1H NMR spectra for cubosome samples in water were obtained using the “water_3919_watergate” pulse sequence. 1H NMR spectra for AT101 in methanol (99.8%, Avantor Performance Materials Poland SA) were obtained using “dpfg_water” using excitation sculpting pulse for selective excitation of two methanol resonance lines. All sequences mentioned above are based on Pulsed Gradient Spin Echo (PGSE) technique. The frequency selective rf pulse for double-resonance methanol suppression was designed using the pbox tool embedded in VnmrJ software. Finally, the NMR data were analyzed using MestReNova 12.0.4 software. Self-diffusion coefficients were derived from the Stejskal-Tanner equation27 describing the attenuation of the NMR signal in the presence of magnetic field gradient pulses:
where 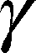 is the gyromagnetic ratio for protons,
is the gyromagnetic ratio for protons, 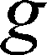 is the amplitude of the magnetic field gradient pulse,
is the amplitude of the magnetic field gradient pulse, 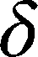 is the magnetic field gradient duration,
is the magnetic field gradient duration, 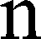 is the diffusion time and
is the diffusion time and 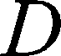 is the self-diffusion coefficient. The data were plotted as
is the self-diffusion coefficient. The data were plotted as 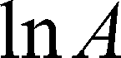 ln A vs g2so that a single diffusion coefficient would result in a linear dependence. For the distribution of diffusion coefficients, which resulted in the non-linear dependence of ln A(g2) the Bayesian DOSY (diffusion-ordered spectroscopy) transform was performed using MestReNova software.
ln A vs g2so that a single diffusion coefficient would result in a linear dependence. For the distribution of diffusion coefficients, which resulted in the non-linear dependence of ln A(g2) the Bayesian DOSY (diffusion-ordered spectroscopy) transform was performed using MestReNova software.
AT101 Drug Entrapment and Release Studies
To separate unbound drug from the cubosome particles, ultrafiltration centrifugation was performed prior to the entrapment efficiency determination. Five hundred microliters of drug-loaded cubosomes were filtrated through an Amicon® Ultra-0.5 or −15 centrifugal filter unit (Merck) with a 10,000 molecular weight cut-off (MWCO) at 14,000 g for 30 min, according to the manufacturer’s protocol. During the centrifugation, the unbound AT101 was filtrated, while loaded cubosomes remained inside the filter unit. To recover the cubosomes, the filters were placed upside down in a fresh Eppendorf tube® and centrifuged at 1000 g for 2 min. The concentration of unbound AT101 in the filtrate was determined spectrophotometrically at λ=290 nm with the LAMBDA 950 UV/Vis/NIR spectrophotometer (Perkin Elmer, Waltham, MA, USA). As proof of principle, the recovered cubosomes were disrupted with ethanol, and the concentration of encapsulated AT101 was analyzed in the same manner. The entrapment efficiency (EE%) and drug loading capacity (DLC%) were calculated with the following formulas:
The release of AT101 from the GMO cubosomes was determined by dialysis. For this purpose, 500 μL of AT101-loaded cubosome dispersion was filled into a Spectra/Por® 2 Dialysis Membrane Tubing (12,000–14,000 MWCO, VWR International, Radnor, PA, USA), which had been previously equilibrated with 0.1 M NaOH and then rinsed and stored in distilled water overnight. The filled membrane tube was then closed tightly and submerged in 20 mL of an artificial cerebrospinal fluid (aCSF) solution (124 mM NaCl, 5 mM KCl, 26 mM NaHCO3, 1.3 mM MgCl2, 2 mM CaCl2, 10 mM D-Glucose, 0.05% (w/v) BSA in H2O) at 37°C which was stirred continuously for 72 h. A 500 μL sample of the aCSF solution with the released drug was drawn after 3, 6, 24, 48 and 72 h and replaced with fresh solution. The AT101 concentration in the samples was determined spectrophotometrically as described above.
Cell Culture
The human glioblastoma cell lines A172 (ECACC 880624218) and LN229 (ATCC-CRL-2611) were purchased from the European Collection of Cell Cultures (ECACC, Salisbury, UK) or the American Type Culture Collection (ATCC, Manassas, Virginia, USA). The human fetal astrocyte cell line SVGA was kindly provided by the group of Christine Hanssen Rinaldo, University Hospital of North Norway29 with the permission of WJ Altwood.28 The human embryonic microglia cell line HMC3 (ATCC-CRL-3304) was obtained from ATCC. All cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM, ThermoFisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco® Qualified, ThermoFisher Scientific), 100 units/mL penicillin-streptomycin (ThermoFisher Scientific) and 2 mM L-glutamine (ThermoFisher Scientific), and grown under standard cell culture conditions at 37°C in humidified atmosphere containing 5% CO2. Furthermore, the cells were routinely checked for Mycoplasma contamination by bisbenzimide staining and for identity by Short Tandem Repeat profiling at the Department of Forensic Medicine (Kiel, DE) employing the Powerplex HS Genotyping Kit (Promega, Madison, WC, USA) and the 3500 Genetic Analyser (ThermoFisher Scientific).
In vitro Viability Studies
The cytotoxic effect of AT101-loaded GMO cubosomes was assessed by determining cell viability and compared with those of free drug and control. This was evaluated in two different GBM cell lines (A172 and LN229), as well as in normal cells of the central nervous system, such as astrocytes (SVGA) and microglia (HMC3). For the experiments, 6.0×103 cells/well were seeded into 96-well culture plates. To firstly estimate the toxicity of blank (empty) cubosomes the seeded cells were treated in triplicate with GMO cubosomes at concentrations increasing from 13 to 200 μg/mL. The cells were then treated with various concentrations of AT101-loaded GMO cubosomes, and the effect was compared with equal concentrations of the free drug, as well as with blank cubosomes and dimethylsulfoxide (DMSO) treated cells as a control. After 72 h of stimulation, the cell viability was determined by the colorimetric WST-1 assay (Hoffmann-La Roche, Basel, CH) according to the manufacturer’s instructions. Briefly, 10 μL of WST-1 solution was added to each well of 96-wells plate containing 100 µL medium and further incubated for 90 min at standard cell culture conditions. Afterwards, the absorbance was measured with the Infinite M Plex plate reader (Tecan Group, Männedorf, CH) at λ=450 nm and compared to λ=595 nm. Prior to measurement, the plate was orbitally shaken for 50 sec. The relative cell viability [%] was expressed as a percentage relative to the DMSO control. Data are reported as mean ± SD of experiments performed in triplicates.
Uptake of Cubosomal Nanoparticles – 2D and 3D Cell Culture Models
For the uptake studies, the AT101-loaded cubosomes were fluorescently labeled. For this purpose, GMO cubosomes (20 mg/mL) encapsulating AT101 were incubated with 5 µg/mL Nile Red (Merck) at 37°C overnight in the dark under constant agitation at 300 rpm. Unbound Nile Red molecules were removed by ultrafiltration centrifugation with Amicon® Ultra-4 Centrifugal Filter Devices (10000 MWCO) according to manufacturer’s instructions. Briefly, 250 μL aliquots of stained cubosomes were applied to the filter membrane and centrifuged at 3000 x g for 30 min. Fluorescently labeled nanoparticles were recovered from the filter and re-dispersed up to the initial volume of PBS. For visualization of cubosomal nanoparticle uptake in a 2D cell culture model, 1.5×105 A172 or LN229 cells were seeded onto coverslips. The uptake of cubosomes was also investigated in 3D tumor spheroids, which more closely resemble the tumor environment. For this purpose, 1.0x104 A172 or LN229 cells were cultured in hanging drops for two days. Formed spheroids were subsequently grown in six-well plates covered with 1% agarose for further six days. For staining, the spheroids were attached to coverslips overnight. Next, the cell layers and spheroids were stimulated with 26 µg/mL fluorescently marked AT101-loaded GMO cubosomes for 24 h. The 2D cells were then fixed and permeabilized with 4% (w/v) paraformaldehyde (Merck) for 10 min at RT. Afterwards, the coverslip was treated with 0.1% (v/v) Triton X-100 (Merck) for five minutes at RT. Unspecific binding sites were blocked with 1% (w/v) bovine serum albumin (BSA, Serva Electrophoresis GmbH, Heidelberg, DE) for 60 min at RT. Subsequently, the plasma membrane was stained with β-catenin (1:100, BD Biosciences, Franklin Lakes, NJ, USA) for 60 min at 37°C and fluorescently labeled with Alexa Fluor® 488 (1:1000, ThermoFisher Scientific) for 60 min at 37°C. Finally, the nuclei were stained with Hoechst 33,342 (1:100, ThermoFisher Scientific) for 10 min at RT, and the fixed cells were embedded in Immu-Mount™ solution (ThermoFisher Scientific). In contrast, the 3D spheroids were stained with Alexa Fluor®-conjugated Concanavalin A (1:20, ThermoFisher Scientific) for 30 min at 37°C without fixation in Live Cell Imaging Solution (ThermoFisher Scientific). Images were acquired by the AxioObserver.Z1 microscope (Carl Zeiss AG, Oberkochen, DE) using the ZEN2 software.
Effect of Cubosomal Nanoparticles on Cytoskeleton Behavior
The cytoskeletal rearrangement of actin filaments in cells was investigated after their stimulation with AT101-loaded cubosomes. For this purpose, 1.5×105 A172 or LN229 cells were seeded either onto coverslips or into six-well culture plates. After stimulating the cells with 26 µg/mL AT101-loaded GMO cubosomes or with the corresponding amount of free AT101 and controls for 6 h, the cells were fixed in 4% (w/v) paraformaldehyde and treated with 0.1% Triton X-100 as described above. Unspecific binding sites were blocked with 1% (w/v) BSA for 20 min at RT. Actin filaments were stained with Phalloidin-Atto 488 (1:75, Merck) for 20 min at RT. Hoechst staining allowed for nuclei identification. The cells were finally embedded in Immu-Mount™ solution, and images were acquired with the AxioObserver.Z1 microscope.
Effect of Cubosomal Nanoparticles on mRNA Expression
Ribonucleic acid (RNA) was isolated from A172 and LN229 cells, stimulated with 26 µg/mL AT101-loaded cubosomes, free AT101 and controls, respectively, for 24 h, using the TRIzol™ Reagent (ThermoFisher Scientific) according to the manufacturer’s instructions. DNase digestion, cDNA synthesis, and quantitative reverse transcription–polymerase chain reaction (qRT-PCR) were performed, as described earlier,30 using the TaqMan™ primer-probes (ThermoFisher Scientific) β-actin (ACTB, Hs01060665_g1) and glycerinaldehyde 3-phosphate dehydrogenase (GAPDH, Hs99999905_m1). Threshold cycles (CT) were determined, and ∆CT values of each sample were calculated as:
The induction of gene expression by stimulation is displayed as relative gene expression.
Statistical Analysis
The data were analyzed using the GraphPad Prism 5 Software (GraphPad Software, San Diego, CA, USA) using repeated one-way analysis of variance (ANOVA) with Tukey post-test. Statistical significance is marked with asterisks depending on the p-value: * p<0.05, ** p<0.01, *** p<0.001. The differences between the mean ζ potential and particles size values were analyzed using Students t-test, where p<0.05 indicated statistical significance.
Results and Discussion
Preparation and Characterization of Cubosomal Nanoparticles
Observing the temperature-composition phase diagram of a monoolein (GMO)/water system,31 pure (GMO) and drug-loaded cubosomes (GMO-AT101) were prepared in a top–down approach. The concentrations of the lipid GMO and the surfactant F-127 were chosen to be 10 wt% and 2.5 wt%, respectively, with respect to the final dispersion weight, while the concentration of the anti-cancer drug AT101 was 10 wt% with respect to the GMO content in the dispersion (ie, 0.025 wt% with respect to the final dispersion weight). The chosen GMO:F-127 ratio has been shown to yield the most stable cubosome dispersions.26 Prepared cubosome dispersion appeared homogenously milky white with pure cubosomes and semi-opaque yellowish with AT101-loaded cubosomes.
Cryo-TEM images presented in Figure 1 revealed that the obtained nanoparticles were cube-shaped with a clearly resolved inner periodic arrangement. AT101 drug loading affected the formation of the nanoparticles, as the cubic nanoparticles are accompanied by a fraction of smaller vesicle-like nanoparticles (liposomes). The fast Fourier transform images and inverse fast Fourier transform (FFT and iFFT inset in the image panels) both confirmed the formation of nanoparticles with periodical arrangement. Referring to the distance calculated from the insets in Figure 1 between interdependent networks, which corresponds directly to half a unit cell, it was found that the lattice constants for GMO and GMO-AT101 cubosomes were 17.4 and 16.6 nm, respectively. This result indicated that AT101 was able to influence the internal structure of the cubosomes, as well as, potentially their stabilization mechanism. Additionally, it was assumed that the presence of vesicle-like structures was attributed to the effect of the drug loading and its presence in the dispersion. Competition with the surfactant while forming nanostructures might probably occur. The excess of surfactant-like molecules can sterically stabilize the nanoparticles, preventing them from fusing into more exotic structures, such as cubosomes, which has been observed previously in similar systems.26,32,33
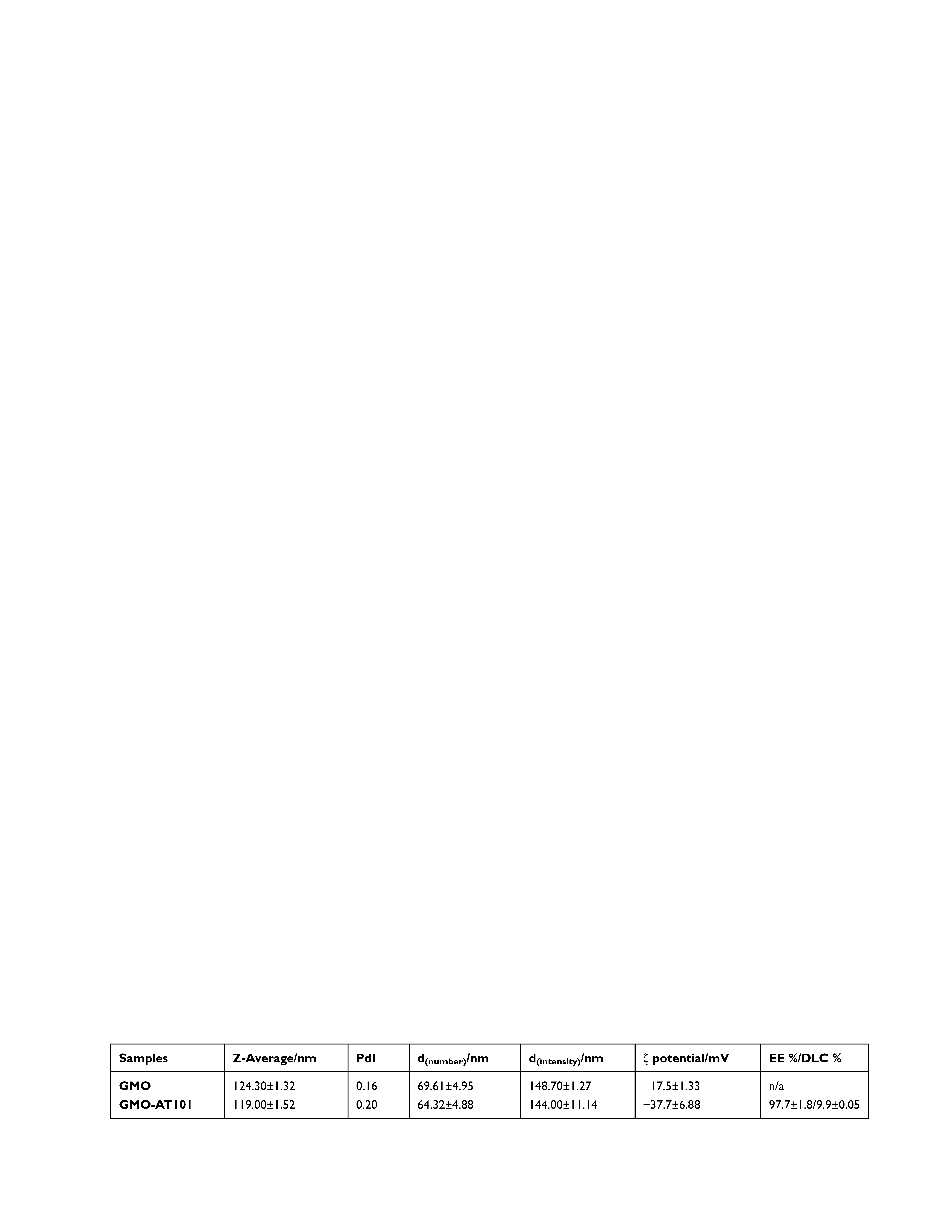
Particle size and zeta potential results of prepared cubosomal nanoparticle dispersions are reported in Table 1 and Figure 2, which summarizes the Z-average as the cumulant mean particle size from the distribution of the particle size (hydrodynamic diameter) measurements weighted by number (d(number)) and intensity (d(intensity)), as well as the polydispersity index (PdI), and the ζ potential.
Prepared and equilibrated cubosomal nanoparticles GMO and GMO-AT101 have diameters (Z-average) of 124.3 nm and 119.0 nm, respectively. It was observed that drug loading had little impact on particle size, which was slightly smaller than the unloaded ones. The particle size distribution of the smaller particle size of GMO-AT101cubosomes can be related to the presence of small vesicle-like nanostructures as observed by the cryo-TEM. The drug impact on nanoparticles is more noticeable in the case of the ζ potential. Pure and drug-loaded cubosomes are, generally, negatively charged. However, AT101-loading causes a significant increase in the absolute value of the ζ potential from −17.5 mV to −37.7 mV (p < 0.05). The high absolute value of the ζ potential indicates that loaded GMO-AT101 cubosomes form stable dispersions, which can be considered as an asset of this drug delivery system.34 This can be related to the reduction of the interfacial tension between GMO and water (according to the emulsification theory) resulting mainly in the formation of smaller cubosomal nanoparticles, but also in a contribution of vesicle-like structures (observed by the cryo-TEM, Figure 1), generally considered to form stable solutions.35 Similar ζ potential values have been observed previously for lipid-based drug delivery systems: −18.9 mV for monoolein-based cubosomes,36 or between −22.8 and −29.4 mV for monoolein-based cubosomes loaded with fluorescent dyes or pure cubosomes, respectively.37 On the other hand, liposomes loaded with AT101 exhibited a ζ potential between −26.01 mV and −58.37 mV, depending on their chemical composition (used different emulsifiers).23
Results on Z-average, d(number) and PdI confirm the observation that prepared cubosome dispersions were stable over time with regard to particle size, particularly when stored at RT. Storing at 37°C induced particle size instability in unloaded GMO cubosomes, while drug loading significantly affected cubosome nanoparticle behavior in dispersion, and prevented nanoparticles from a size increase over the time. Moreover, the obtained PdI values (Figure 2C), were generally lower than 0.5 with the exception of two points that had insignificantly higher values (ie, 0.550 for GMO_RT after one day and 0.501 for GMO-AT101_37° C after ten days). This indicates that both dispersions have a monomodal particle size distribution under the given storage condition (Figure 2E and F). ζ potential values obtained over the time of storage and for both storage temperatures (Figure 2D) also showed that AT101 loading indeed improved the colloidal stability of the cubosomes. The absolute ζ potential values for GMO-AT101 stored at RT and 37°C remained higher (between −31.2 mV and −42.6 mV), compared with unloaded GMO formulations (between −17.9 mV and −27.2 mV). These results are in agreement with the cryo-TEM analysis, which also indicated the effect of the drug on the structure and morphology of formed cubosomes on the structure, morphology of formed cubosomes, as well as the presence of vesicle-like particles, due to the higher content of surfactant-like molecules, ie, AT101 and the Pluronic F-127 in the formulation.
The determined drug entrapment efficiency (EE%) and drug loading capacity (DLC%) in prepared cubosomes after ultrafiltration centrifugation were 97.7%, and 9.9%, respectively, which is high compared with previously reported systems. Nasr et al achieved an EE% of 31.2% with 5-fluorouracil in similar monoolein/F-127-based cubosomes.38 In turn, Zeng et al reported an EE of 75% and a DLC of 0.45% in a paclitaxel-loaded mixed system of cubosome/hexosome nanoparticles.39 Considering AT101 loading in liposomes, Zhai et al reported an EE between approximately 79–97% depending on their chemical composition.23 The high EE in the studied GMO-AT101 system can be attributed to the fact that AT101 as water-insoluble hydrophobic drug embeds in the lipid bilayer in between hydrophobic alkyl chains of the GMO. In contrast, hydrophilic drugs (eg, 5-fluorouracil) are entrapped less efficiently in the lipid bilayer, but remain in water channels, where they are weakly adsorbed. However, the interaction and bonding of drug molecules with the lipids in nanoparticles also affect their drug release efficiency.2,40 As presented in Figure 3, the drug release profile of aCFS at 37°C shows that the entrapment of the hydrophobic AT101 drug into a lipid bilayer prevented its rapid premature leakage. This is related to the limited diffusion of the drug in the aqueous channels and therefore to its continuous and sustained release over a longer period. This is described kinetically by the Higuchi model by plotting the cumulative amount of released AT101 versus the square root of time.41,42 However, due to the strong interaction with the lipid bilayer, the overall drug release reaches 34.79% over 72 h. Since AT101 is not significantly further converted under physiological conditions,43 drug release has been investigated only up to 72 h. From the therapy point of view, such release behavior can be considered as an advantage, and a potential of cubosomes for slow release drug delivery systems has been already reported.44,45 By comparison, liposome-based nanocarriers of AT101 attained a release of between 25% and 60% (in FBS solution) after 48 h depending on the chemical composition of the liposomes.23 However, the high degree of drug release in this case has been related to the modification of liposomes with the emulsifiers, such as TPGS and DSPE-PEG, which affected the drug entrapment efficiency via their solubilizing action. Such modifications could be further considered for the improvement of cubosome-based drug delivery systems. In turn, a mixed cubosome/hexosome-based system for paclitaxel delivery reached a release of approx. 70% in 72 h.39 Other reports on cubosome–based drug delivery systems of, eg, 5-fluorouracil38 and doxorubicin36 showed a rapid drug release of almost 100% after 5 h. However, a significant release has also been observed during the preparation, which undoubtedly interfered with the use of these systems over longer time periods in the form of dispersions, but only in the form of a cubic gel, as a semi-product for final use. Thus, a prolonged time of drug release is preferable, as it may allow for the sustained drug accumulation at the tumor site. Yet, the observed EE and DLC of the investigated GMO-AT101 system are still very high, and therefore ensure the therapeutic efficacy, which will be presented below.
We resolved the translational motion of each of the compounds in the cubosome-water dispersion and investigated whether AT101 was loaded into the cubosome particles using high-resolution diffusion NMR. The 1H NMR spectra for AT101 in methanol is shown in Figure 4A, the 1H NMR spectrum for GMO is shown in Figure 4B, and that for AT101 in H2O/GMO is shown in Figure 4C. The resonance lines for AT101 molecules are observed between 6.5 and 8.5 ppm (Figure 4A and C). Since the GMO/H2O sample did yield distinctive resonance lines in the aforementioned spectral region, the appearance of new resonance lines in this region was attributed to AT101 entrapped in GMO particles. Accordingly, this region of chemical shifts was further analyzed in the Fourier transformed 2D PGSE data as shown in Figure 4D–F. The particular interest was on whether AT101 was bound inside the cubosome particle. For this reason, the bound-unbound analysis was employed, similar to one described previously by Szutkowski et al.46 Although the self-diffusion coefficients of the colloidal dispersion components were measured taking into account all possible chemical groups existing in the system, only the region from 6.5 to 8.5 ppm corresponding to AT101 molecules, and the line at 3.5 ppm assigned to GMO were further analyzed in details. The PGSE NMR Fourier-processed 2D data along with the Stejskal-Tanner27 fits are shown in Figure 5.
 |
Figure 5 The detailed analysis of 2D PGSE data from Figure 4E-F. (A) AT101 diffusion decay from Figure 4D, the self-diffusion coefficient is shown in the figure. (B) The diffusion decay from Figure 4E, the distribution of self-diffusion coefficients of GMO was obtained from 2D Bayesian DOSY display shown as an inset. (C) The diffusion decay of AT101 in a GMO/water system taken from line intensity at 8 ppm, the self-diffusion coefficient corresponds to the diffusivity of GMO. (D) Overlaid data points from AT101 in GMO (B) and GMO (C). |
The self-diffusion of AT101 in methanol is derived from the slope of the ln A(g2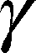 ) plot. The single self-diffusion coefficient is characterized by a linear dependence. Accordingly, the self-diffusion coefficient of AT101 was around 1.9×10−9 m2/s. The analysis of diffusion decay for GMO revealed that the size distribution of GMO was responsible for the non-linear dependence of the ln A(g2
) plot. The single self-diffusion coefficient is characterized by a linear dependence. Accordingly, the self-diffusion coefficient of AT101 was around 1.9×10−9 m2/s. The analysis of diffusion decay for GMO revealed that the size distribution of GMO was responsible for the non-linear dependence of the ln A(g2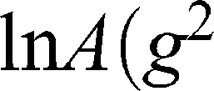 ) decay.
) decay.
Further studies revealed that the self-diffusion of GMO varied between 4.3 and 6.7×10−12 m2/s resulting in a distribution of the hydrodynamic radii between 30 and 46 nm (calculated from Einstein-Smoluchowski equation). Finally, the information about the AT101 entrapped in GMO cubosomes was obtained from data shown in Figure 5C, where the self-diffusion coefficient of AT101 is nearly the same as that obtained for GMO (Figure 5D). This fact clear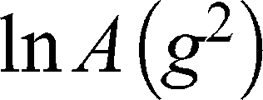 ly indicated that these two components, ie, AT101 and GMO, diffuse in colloidal dispersion as a combined construct of drug and lipid nanoparticle.
ly indicated that these two components, ie, AT101 and GMO, diffuse in colloidal dispersion as a combined construct of drug and lipid nanoparticle.
GMO-AT101 Cubosomes Penetrate into 3D Tumor Spheroids and are Taken Up by GBM Cells
The cytotoxicity of GMO-AT101 cubosomes was evaluated and compared with that of the free drug. Since the prepared nanoparticles in this study consisted of lipid subunits that formed structures closely resembling the plasma membrane structures of cells, one can assume that they would be efficiently incorporated into the cells giving an increased activity. Penetration of the nanoparticles into 3D tumor spheroids and their uptake into cells was confirmed by Nile Red labeling of cubosomes. Cell margins were defined with either concanavalin A in the case of spheroids, or β-catenin for fixed cell layers (Figure 6). Indeed, GMO-AT101 cubosomes were taken up by LN229 and A172 spheroids (~780 µm and ~535 µm, respectively) after 24 h of incubation. Beyond that, nanoparticles were strongly incorporated by single glioma cells (Figure 6, indicated by white arrows), probably due to internalization processes associated with endocytic pathways, which are known to play a pivotal role in the uptake of cubosomal nanoparticles.47 The large uptake of cubosomes into cells might yield an enhanced cytotoxic effect of AT101. Another study showed good penetration of cubosomes into brain tissue in vivo,48 thereby ensuring that they would reach tumor cells at distant tumor sites and with the tumor core guaranteeing to additionally reach tumor cells at distant tumor sites and within the tumor core.
Encapsulated AT101 Exhibits Stronger Cytotoxic Effects and More Extensive Rearrangement of Actin Fibers in GBM Cells Than Free AT101
Cubosomes consisting of GMO:F-127 are known to be biocompatible in HepG2 hepatoma cells at concentrations up to 25 µg/mL after 4 h of stimulation.47 As no information regarding glioblastoma cells, astrocytes or microglia is given in the available literature, different concentrations of blank GMO cubosomes were tested with regard to their cytotoxic effects in A172 and LN229 cells, as well as in SVGA and HMC3 after 72 h of treatment (Figure 7A). Indeed, blank cubosomes exhibited the greatest cytotoxicity in HMC3 cells (IC50 = 32.5 µg/mL), while in LN229 (IC50 = 54.8 µg/mL), A172 (IC50 = 64.5 µg/mL) and SVGA (IC50 = 51.7 µg/mL) the same effect was only seen with higher concentrations (Figure 5A). This was further considered during the preparation of AT101-loaded cubosomes in order to reduce the cytotoxic effect of cubosomes themselves, but to aim therapeutically relevant concentrations of AT101. In the following, the cytotoxic effect of various concentrations of encapsulated AT101 compared with equal concentrations of free AT101 and blank cubosomes, respectively, was investigated in GBM cells (LN229, A172), as well as in normal brain cells (SVGA, HMC3) after 72 h of treatment to estimate the potentially harmful effects on healthy tissue (Figure 7B). Although treatment with 2.5 or 3.75 µM of free AT101 had no relevant effect on the viability of both GBM cell lines compared with DMSO-stimulated controls, cytotoxic efficacy was significantly improved after encapsulation of AT101 into GMO cubosomes (Figure 7B, left). In comparison, the same amounts of empty cubosomes were not toxic to LN229 and A172 cells. In contrast to this, stimulation of SVGA and HMC3 with encapsulated AT101 did not significantly change their viability compared with controls and free drug administration (Figure 7B, right). A slight cytotoxic effect was observed after the application of empty cubosomes in HMC3 corresponding to a 3.75 µM AT101 stimulation concentration. This is explained by a stronger response of microglia to the blank nanoparticles as shown in Figure 5A. However, addition of AT101 in such concentrations did not further decrease the n-fold viability of HMC3 (Figure 7B, right).
The improvement of the AT101 cytotoxicity after loading it into GMO cubosomes can be the result of several factors. Firstly, cubosomes are known to exist in stable colloidal water-based dispersions, preferentially exploited in biomedical formulations.2–4,34,49 These nanoparticles ensure the chemical and physical stability of drug molecules, that are often rapidly degraded in the body, particularly in the cell environment. Moreover, when considering the delivery of hydrophobic drugs, such as AT101, the benefit of using cubosomes is the improvement in the bioavailability of the drug due to its greater solubility in the lipid membrane of formed nanoparticles and finally their dispersion in water-based media. A similar approach to enhancing the solubility and, hence, the bioavailability of hydrophobic drugs by using monoolein- and phytantriol-based cubosomes was described by Ali et al.50,51 The improved cytotoxicity response to GMO-AT101 cubosomes can be further related to the pronounced internalization associated with endocytic pathways, as reported previously by Abdel-Bar et al (Figure 6).47 Moreover, AT101 molecules exhibit a high binding affinity to proteins resulting in a reduced AT101 activity.52,53 Another factor that improves the cytotoxicity of GMO-AT101 cubosomes, is that the cubosome-based carriers prevent AT101 from binding to the proteins in the culture medium. In addition, cubosomes are characterized by a good ability to penetrate into brain tissue as shown by Ahirrao et al, which predestines them as excellent tools, particularly for the treatment of glioblastoma multiforme in vivo.48 As cubosomes can be also further functionalized, similar to other nanoparticles used for drug delivery, their uptake into cancer cells can be enhanced. Such active targeting approaches can include, eg, modifications with specific cancer cell membrane lipids and proteins, where the cancer cells are powerful sources for providing intrinsic homotypic or heterotypic cell properties to the drug carriers.54,55 Another approach involves common modifications with specific ligands to protein receptors expressed on the surface of the cancer cells, typically folic acid ligands56 or the tumor-homing and tumor-penetrating cyclic iRGD peptide, also due to its ability to increases the tumor vascular permeability.57,58 Of course, the anti-cancer efficacy of the GMO-AT101 formulation requires further investigation in the GBM model in vivo.
Interestingly, nanoparticle approaches employing AT101 loaded into liposomes, micelles or polymeric capsules did not show an increased cytotoxicity of the encapsulated compared with free drug in prostate, ovarian or breast cancer cell lines.23,24,59,60 To our knowledge, no such studies focusing on GBM cells have yet been conducted. Exceptional tumor-growth control was established by multi-drug stimulation and functionalization of nanoparticles for targeted therapy.25,59-61 However, the potential cytotoxic effects of such nanoparticles on healthy tissue were not evaluated. With respect to our prepared nanoparticle formulation, the cytotoxicity of AT101 was augmented by encapsulation, while normal brain cells were not harmed to a significant extent. Thus, GMO-AT101 cubosomes represent a promising basic tool for alternative therapy attempts in GBMs.
The cytotoxic benefit of encapsulated AT101 in GBM cells is further underlined by the induction of marked cytoskeletal rearrangement of actin fibers by GMO-AT101 stimulation compared with application of the free drug (Figure 7C). Reorganization of the cytoskeleton is essential in the execution phase of apoptosis resulting in cell contraction, plasma membrane blebbing and other effects.62,63 Particularly the rearrangement and shortening of actin fibers is important for nuclear fragmentation and membrane blebbing.64–66 In detail, the actin-myosin system has been proposed to be the source of the contractile force that drives bleb formation. In this, the Rho effector protein ROCK I, which contributes to phosphorylation of myosin light-chains, myosin ATPase activity and coupling of actin-myosin filaments to the plasma membrane, is cleaved during apoptosis to generate a truncated active form. The activity of ROCK proteins is both necessary and sufficient for the formation of membrane blebs and for re-localization of fragmented DNA into blebs and apoptotic bodies.64 Phalloidin, a toxin from the death cap mushroom, was used since it binds to actin filaments, thereby visualizing actin remodeling.67 A network of actin fibers was still visible after 6 h of treatment with free AT101, particularly in LN229 cells, whereas GMO-AT101 administration led to a slight condensation of fibers at poles accompanied by reduced intensity of stained actin (Figure 7C, upper panel). Albeit a fiber network was less clearly visible in control-stimulated A172 cells, and, as above, the intensity of the phalloidin signal only decreased after GMO-AT101 stimulation (Figure 7C, lower panel). This rearrangement of actin fibers accompanies the destruction of the cell morphology during apoptosis and indicates the pronounced cytotoxic activity of AT101-loaded cubosomes. Thus, the reorganization of the cytoskeleton is one possible molecular mechanism of action for the greater anti-tumorigenic potential of AT101 cubosomes. A closer look at the transcriptional changes of β-actin after 24 h revealed significant downregulation in LN229 cells after treatment with both encapsulated and free AT101 (Figure 7C). Accordingly, the expression of ACTB was also significantly reduced in A172 after stimulation with GMO-AT101 cubosomes but not with free AT101. However, a slight downregulation of ACTB was also induced by blank cubosomes (Figure 7C). Thus, relevant rearrangement of the cytoskeleton due to the treatment of GBM cells with encapsulated AT101 might have only occurred at the protein level but was not associated with transcriptional changes of β-actin.
Conclusions
In this study, the AT101 drug, considered a promising drug in the treatment of glioblastoma multiforme, albeit with poor solubility in water-based media and low bioavailability, was successfully encapsulated into cubosomal nanoparticles prepared from glyceryl monooleate. The GMO-AT101 cubosomes were stable colloids with a high drug entrapment efficiency and a continuous sustained drug release. Efficient binding of AT101 to GMO cubosomes was verified by NMR diffusometry. The GMO-AT101 cubosomes were efficiently taken up by GBM cells and GBM 3D tumor spheroids. AT101 encapsulated in cubosomes had a stronger cytotoxic effect on GBM cells in vitro as evidenced by the induced rearrangement of cytoskeletal actin fibers in treated GBM cells, while normal brain cells were not significantly affected. GMO-AT101 cubosomes are a promising basic tool for alternative therapy attempts in GBM.
Abbreviations
aCSF, artificial cerebrospinal fluid; ACTB, β-actin; ANOVA, analysis of variance; BBB, blood–brain barrier; BSA, bovine serum albumin; CT, threshold cycle; DLC, drug loading capacity; DLS, dynamic light scattering; DMEM, Dulbecco’s modified Eagle’s medium; DMSO, dimethyl sulfoxide; EE, entrapment efficiency; ELS, electrophoretic light scattering; FBS, fetal bovine serum; GAPDH, glycerinaldehyde 3-phosphate dehydrogenase; GBM, glioblastoma multiforme; GMO, glyceryl monooleate; MWCO, molecular weight cut-off; NMR, nuclear magnetic resonance; NP, nanoparticle; PdI, polydispersity index; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; RNA, ribonucleic acid; RT, room temperature; SD, standard deviation; TEM, transmission electron microscopy; NMR, nuclear magnetic spectroscopy; TMZ, temozolomide.
Acknowledgments
The biological studies performed in this work were funded by the German Research Foundation (DFG) as part of the Research Training Group “Materials4Brain” (RTG2154; P8).
Work related to the preparation and physicochemical characterization of cubosomal nanoparticles was supported by the National Science Center, Poland, under grant SONATA BIS 6 “Cubosomes - liquid-crystal nanoparticles as a potential bioimaging systems” 2016/22/E/ST3/00458.
During the NMR diffusion experiments, KS and SJ were supported by the grant H2020-INFRAIA-2016–2017 under research grant EUSMI – European infrastructure for spectroscopy, scattering and imaging of soft-matter, contract number GA731019, funded under H2020-EU.1.4.1.2.RIA.
The authors would like to thank Dr. Łucja Przysiecka (NanoBioMedical Centre) for her advice on scientific and experimental matters during the research.
Disclosure
Dorota K Flak and Vivian Adamski shared first authorship. The authors report no conflicts of interest in this work.
References
1. Azmi IDM, Wibroe PP, Wu L-P, et al. A structurally diverse library of safe-by-design citrem-phospholipid lamellar and non-lamellar liquid crystalline nano-assemblies. Ther Deliv. 2016;239:1–9.
2. Barriga HMG, Holme MN, Stevens MM. Cubosomes: the Next Generation of Smart Lipid Nanoparticles? Angew Chem Int Ed Engl. 2018;58(10):2958–2978. doi:10.1002/anie.201804067
3. Guo C, Wang J, Cao F, Lee RJ, Zhai G. Lyotropic liquid crystal systems in drug delivery. Drug Discov Today. 2010;15(23):1032–1040. doi:10.1016/j.drudis.2010.09.006
4. Mulet X, Boyd BJ, Drummond CJ. Advances in drug delivery and medical imaging using colloidal lyotropic liquid crystalline dispersions. J Colloid Interface Sci. 2013;393:1–20. doi:10.1016/j.jcis.2012.10.014
5. Alavi M, Karimi N, Safaei M. Application of Various Types of Liposomes in Drug Delivery Systems. Adv Pharm Bull. 2017;7(1):3–9. doi:10.15171/apb.2017.002
6. Bulbake U, Doppalapudi S, Kommineni N, Khan W. Liposomal Formulations in Clinical Use: an Updated Review. Pharmaceutics. 2017;9(2):12.
7. Liu X, Situ A, Kang Y, et al. Irinotecan delivery by lipid-coated mesoporous silica nanoparticles shows improved efficacy and safety over liposomes for pancreatic cancer. ACS Nano. 2016;10(2):2702–2715.
8. Russell LM, Hultz M, Searson PC. Leakage kinetics of the liposomal chemotherapeutic agent Doxil: the role of dissolution, protonation, and passive transport, and implications for mechanism of action. J Control Release. 2018;269:171–176.
9. Davis ME. Glioblastoma: overview of Disease and Treatment. Clin J Oncol Nurs. 2016;20(5 Suppl):S2–S8.
10. Ozdemir-Kaynak E, Qutub AA, Yesil-Celiktas O. Advances in Glioblastoma Multiforme Treatment: new Models for Nanoparticle Therapy. Front Physiol. 2018;9:170. doi:10.3389/fphys.2018.00170
11. Jativa P, Cena V. Use of nanoparticles for glioblastoma treatment: a new approach. Nanomed. 2017;12(20):2533–2554. doi:10.2217/nnm-2017-0223
12. Lu Y, Li J, Dong CE, Huang J, Zhou H-B, Wang W. Recent advances in gossypol derivatives and analogs: a chemistry and biology view. Future Med Chem. 2017;9(11):1243–1275. doi:10.4155/fmc-2017-0046
13. Opydo-Chanek MG, Gonzalo O, Marzo I. Multifaceted anticancer activity of BH3 mimetics: current evidence and future prospects. Biochem Pharmacol. 2017;136:12–23. doi:10.1016/j.bcp.2017.03.006
14. Voss V, Senft C, Lang V, et al. The Pan-Bcl-2 Inhibitor (-)-gossypol triggers autophagic cell death in malignant glioma. Mol Cancer Res. 2010;8(7):1002–1016. doi:10.1158/1541-7786.MCR-09-0562
15. Warnsmann V, Meyer N, Hamann A, Kögel D, Osiewacz HD. A novel role of the mitochondrial permeability transition pore in (-)-gossypol-induced mitochondrial dysfunction. Mech Ageing Dev. 2018;170:45–58. doi:10.1016/j.mad.2017.06.004
16. Li F, Xia Y, Meiler J, Ferguson-Miller S. Characterization and modelling of the oligomeric state and ligand binding behavior of purified translocator protein 18kDa from Rhodobacter sphaeroides. Biochemistry. 2013;52(34):5884–5899. doi:10.1021/bi400431t
17. Mehner M, Kubelt C, Adamski V, Schmitt C, Synowitz M, Held-Feindt J. Combined treatment of AT101 and demethoxycurcumin yields an enhanced anti‑proliferative effect in human primary glioblastoma cells. J Cancer Res Clin Oncol. 2020;146(1):117–126. doi:10.1007/s00432-019-03107-7
18. Jarzabek MA, Amberger-Murphy V, Callanan JJ, et al. Interrogation of gossypol therapy in glioblastoma implementing cell line and patient-derived tumour models. Br J Cancer. 2014;111(12):2275. doi:10.1038/bjc.2014.529
19. Keshmiri-Neghab H, Goliaei B, Nikoofar A. Gossypol enhances radiation-induced autophagy in glioblastoma multiforme. Gen Physiol Biophys. 2014;33(4):433–442. doi:10.4149/gpb_2014017
20. Adamski V, Hempelmann A, Fluh C, et al. Dormant glioblastoma cells acquire stem cell characteristics and are differentially affected by Temozolomide and AT101 treatment. Oncotarget. 2017;8(64):108064–108078. doi:10.18632/oncotarget.22514
21. Adamski V, Schmitt C, Ceynowa F, et al. Effects of sequentially applied single and combined temozolomide, hydroxychloroquine and AT101 treatment in a long-term stimulation glioblastoma in vitro model. J Cancer Res Clin Oncol. 2018;144(8):1475–1485. doi:10.1007/s00432-018-2680-y
22. Bushunow P, Reidenberg MM, Wasenko J, et al. Gossypol Treatment of Recurrent Adult Malignant Gliomas. J Neurooncol. 1999;43(1):79–86. doi:10.1023/A:1006267902186
23. Zhai G, Wu J, Zhao X, et al. A Liposomal Delivery Vehicle for the Anticancer Agent Gossypol. Anticancer Res. 2008;28(5A):2801–2805.
24. Jin C-L, Chen M-L, Wang Y, et al. Preparation of novel (-)-gossypol nanoparticles and the effect on growth inhibition in human prostate cancer PC-3 cells in vitro. Exp Ther Med. 2015;9(3):675–678. doi:10.3892/etm.2015.2172
25. Heleg-Shabtai V, Aizen R, Sharon E, et al. Gossypol-capped mitoxantrone-loaded mesoporous SiO2 NPs for the cooperative controlled release of two anti-cancer drugs. ACS Appl Mater Interfaces. 2016;8(23):14414–14422. doi:10.1021/acsami.6b03865
26. Akhlaghi SP, Ribeiro IR, Boyd BJ, Loh W. Impact of preparation method and variables on the internal structure, morphology, and presence of liposomes in phytantriol-Pluronic® F127 cubosomes. Colloid Surf B Biointerfaces. 2016;145:845–853. doi:10.1016/j.colsurfb.2016.05.091
27. Stejskal EO, Tanner JE. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J Chem Phys. 1965;42(1):288–292. doi:10.1063/1.1695690
28. Schweighardt B, Shieh JTC, Atwood WJ. CD4/CXCR4-independent infection of human astrocytes by a T-tropic strain of HIV-1. J Neurovirol. 2001;7(2):155–162. doi:10.1080/13550280152058816
29. Henriksen S, Tylden GD, Dumoulin A, Sharma BN, Hirsch HH, Rinaldo C. The human fetal glial cell line SVG p12 contains infectious BK polyomavirus. J Virol. 2014;88(13):7556–7568. doi:10.1128/JVI.00696-14
30. Hattermann K, Held-Feindt J, Lucius R, et al. The Chemokine Receptor CXCR7 Is Highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer Res. 2010;70(8):3299–3308. doi:10.1158/0008-5472.CAN-09-3642
31. Qiu H, Caffrey M. The phase diagram of the monoolein/water system: metastability and equilibrium aspects. Biomaterials. 2000;21(3):223–234. doi:10.1016/S0142-9612(99)00126-X
32. Demurtas D, Guichard P, Martiel I, Mezzenga R, Hébert C, Sagalowicz L. Direct visualization of dispersed lipid bicontinuous cubic phases by cryo-electron tomography. Nat Commun. 2015;6(1):8915. doi:10.1038/ncomms9915
33. Wörle G, Drechsler M, Koch MHJ, Siekmann B, Westesen K, Bunjes H. Influence of composition and preparation parameters on the properties of aqueous monoolein dispersions. Int J Pharm. 2007;329(1):150–157. doi:10.1016/j.ijpharm.2006.08.023
34. Karami Z, Hamidi M. Cubosomes: remarkable drug delivery potential. Drug Discov Today. 2016;21(5):789–801. doi:10.1016/j.drudis.2016.01.004
35. Tilley AJ, Drummond CJ, Boyd BJ. Disposition and association of the steric stabilizer Pluronic-F127 in lyotropic liquid crystalline nanostructured particle dispersions. J Colloid Interface Sci. 2013;392:288–296. doi:10.1016/j.jcis.2012.09.051
36. Szlezak M, Nieciecka D, Joniec A, et al. Monoolein cubic phase gels and cubosomes doped with magnetic nanoparticles-hybrid materials for controlled drug release. ACS Appl Mater Interfaces. 2017;9(3):2796–2805. doi:10.1021/acsami.6b12889
37. Murgia S, Bonacchi S, Falchi AM, et al. Drug-Loaded Fluorescent Cubosomes: versatile Nanoparticles for Potential Theranostic Applications. Langmuir. 2013;29(22):6673–6679. doi:10.1021/la401047a
38. Nasr M, Ghorab MK, Abdelazem A. In vitro and in vivo evaluation of cubosomes containing 5-fluorouracil for liver targeting. Acta Pharm Sin B. 2015;5(1):79–88. doi:10.1016/j.apsb.2014.12.001
39. Zeng N, Gao X, Hu Q, et al. Lipid-based liquid crystalline nanoparticles as oral drug delivery vehicles for poorly water-soluble drugs: cellular interaction and in vivo absorption. Int J Nanomedicine. 2012;7:3703–3718. doi:10.2147/IJN.S32599
40. Pan X, Han K, Peng X, et al. Nanostructured Cubosomes as Advanced Drug Delivery System. Curr Pharm Des. 2013;19(35):6290–6297. doi:10.2174/1381612811319350006
41. Higuchi T. Mechanism of sustained-action medication. Theoretical analysis of rate of release of solid drugs dispersed in solid matrices. J Pharm Sci. 1963;52(12):1145–1149. doi:10.1002/jps.2600521210
42. Higuchi T. Rate of release of medicaments from ointment bases containing drugs in suspension. J Pharm Sci. 1961;50(10):874–875. doi:10.1002/jps.2600501018
43. Jia L, Coward LC, Kerstner-Wood CD, et al. Comparison of pharmacokinetic and metabolic profiling among gossypol, apogossypol and apogossypol hexaacetate. Cancer Chemother Pharmacol. 2008;61(1):63–73. doi:10.1007/s00280-007-0446-3
44. Boyd BJ, Whittaker DW, Khoo S-M, Davey G. Lyotropic liquid crystalline phases formed from glycerate surfactants as sustained release drug delivery systems.. Int J Pharm. 2006;309(1–2):218–226. doi:10.1016/j.ijpharm.2005.11.033
45. Clogston J, Caffrey M. Controlling release from the lipidic cubic phase. Amino acids, peptides, proteins and nucleic acids. J Control Release. 2005;107(1):97–111. doi:10.1016/j.jconrel.2005.05.015
46. Szutkowski K, Sikorska E, Bakanovych I, et al. Structural analysis and dynamic processes of the transmembrane segment inside different micellar environments-implications for the TM4 Fragment of the bilitranslocase protein. Int J Mol Sci. 2019;20(17):4172. doi:10.3390/ijms20174172
47. Abdel-Bar HM, El Basset Sanad RA. Endocytic pathways of optimized resveratrol cubosomes capturing into human hepatoma cells. Biomed Pharmacother. 2017;93:561–569. doi:10.1016/j.biopha.2017.06.093
48. Ahirrao M, Shrotriya S. In vitro and in vivo evaluation of cubosomal in situ nasal gel containing resveratrol for brain targeting. Drug Dev Ind Pharm. 2017;43(10):1686–1693. doi:10.1080/03639045.2017.1338721
49. Akbar S, Anwar A, Ayish A, et al. Phytantriol based smart nano-carriers for drug delivery applications. Eur J Pharm Sci. 2017;101:31–42. doi:10.1016/j.ejps.2017.01.035
50. Ali MA, Kataoka N, Ranneh A-H, et al. Enhancing the solubility and oral bioavailability of poorly water-soluble drugs using monoolein cubosomes. Chem Pharm Bull (Tokyo). 2017;65(1):42–48. doi:10.1248/cpb.c16-00513
51. Ali MA, Noguchi S, Iwao Y, Oka T, Itai S. Preparation and Characterization of SN-38-Encapsulated Phytantriol Cubosomes Containing α-Monoglyceride Additives. Chem Pharm Bull (Tokyo). 2016;64(6):577–584. doi:10.1248/cpb.c15-00984
52. Cater CM, Lyman CM. Reaction of gossypol with amino acids and other amino compounds. J Am Oil Chem Soc. 1969;46(12):649–653. doi:10.1007/BF02540621
53. Dabrowski K, Lee K-J, Rinchard J, Ciereszko A, Blom JH, Ottobre JS. Gossypol isomers bind specifically to blood plasma proteins and spermatozoa of rainbow trout fed diets containing cottonseed meal. Biochim Biophys Acta. 2001;1525(1):37–42. doi:10.1016/S0304-4165(00)00168-9
54. Li B, Wang F, Gui L, He Q, Yao Y, Chen H. The potential of biomimetic nanoparticles for tumor-targeted drug delivery. Nanomedicine. 2018;13(16):2099–2118. doi:10.2217/nnm-2018-0017
55. Sabu C, Rejo C, Kotta S, Pramod K. Bioinspired and biomimetic systems for advanced drug and gene delivery. J Control Release. 2018;287:142–155. doi:10.1016/j.jconrel.2018.08.033
56. Tian Y, Li J-C, Zhu J-X, et al. Folic Acid-Targeted Etoposide Cubosomes for Theranostic Application of Cancer Cell Imaging and Therapy. Med Sci Monit. 2017;23:2426–2435. doi:10.12659/MSM.904683
57. Simon-Gracia L, Hunt H, Scodeller P, et al. iRGD peptide conjugation potentiates intraperitoneal tumor delivery of paclitaxel with polymersomes. Biomaterials. 2018;104:247–257. doi:10.1016/j.biomaterials.2016.07.023
58. Sugahara KN, Teesaku T, Karmali PP, et al. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science. 2010;328(5981):1031. doi:10.1126/science.1183057
59. Cho H, Lai TC, Kwon GS. Poly(ethylene glycol)-block-poly(ε-caprolactone) micelles for combination drug delivery: evaluation of paclitaxel, cyclopamine and gossypol in intraperitoneal xenograft models of ovarian cancer. J Control Release. 2013;166(1):1–9. doi:10.1016/j.jconrel.2012.12.005
60. Liu H, Li K, Lan L, et al. Double-layered hyaluronic acid/stearic acid-modified polyethyleneimine nanoparticles encapsulating (-)-gossypol: a nanocarrier for chiral anticancer drugs. J Mat Chem B. 2014;2(32):5238–5248. doi:10.1039/C4TB00539B
61. Shen S, Wu Y, Li K, et al. Versatile hyaluronic acid modified AQ4N-Cu(II)-gossypol infinite coordination polymer nanoparticles: multiple tumor targeting, highly efficient synergistic chemotherapy, and real-time self-monitoring. Biomaterials. 2018;154:197–212. doi:10.1016/j.biomaterials.2017.11.001
62. Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26(4):239–257. doi:10.1038/bjc.1972.33
63. Laster SM, Mackenzie JJM. Bleb formation and F-actin distribution during mitosis and tumor necrosis factor-induced apoptosis. Microsc Res Tech. 1996;34(3):272–280. doi:10.1002/(SICI)1097-0029(19960615)34:3<272::AID-JEMT10>3.0.CO;2-J
64. Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol. 2001;3(4):339–345.
65. Seo MY, Rhee K. Caspase-mediated cleavage of the centrosomal proteins during apoptosis. Cell Death Dis. 2018;9(571). doi:10.1038/s41419-018-0632-8
66. Wickman GR, Julian L, Mardilovich K, et al. Blebs produced by actin-myosin contraction during apoptosis release damage-associated molecular pattern proteins before secondary necrosis occurs. Cell Death Differ. 2013;20(10):1293. doi:10.1038/cdd.2013.69
67. Lengsfeld AM, Löw I, Wieland T, et al. Interaction of phalloidin with actin. Proc Natl Acad Sci U S A. 1974;71(7):2803–2807. doi:10.1073/pnas.71.7.2803
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.