Back to Journals » Clinical Ophthalmology » Volume 15
Associations of ARMS2 and CFH Gene Polymorphisms with Neovascular Age-Related Macular Degeneration
Authors Supanji S , Romdhoniyyah DF , Sasongko MB , Agni AN, Wardhana FS, Widayanti TW, Prayogo ME, Perdamaian ABI, Dianratri A, Kawaichi M , Oka C
Received 20 December 2020
Accepted for publication 22 February 2021
Published 11 March 2021 Volume 2021:15 Pages 1101—1108
DOI https://doi.org/10.2147/OPTH.S298310
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Supanji Supanji,1– 4 Dewi Fathin Romdhoniyyah,1,2 Muhammad Bayu Sasongko,1,2,4 Angela Nurini Agni,1,2,4 Firman Setya Wardhana,1,2,4 Tri Wahyu Widayanti,1,2,4 Muhammad Eko Prayogo,1,2,4 Ayudha Bahana Ilham Perdamaian,1,2 Aninditta Dianratri,1,2 Masashi Kawaichi,5 Chio Oka5
1Department of Ophthalmology, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Yogyakarta, Indonesia; 2Department of Ophthalmology, Dr. Sardjito General Hospital, Yogyakarta, Indonesia; 3Ophthalmology Clinic, Military Air Force Central Hospital Dr. Suhardi Hardjolukito, Yogyakarta, Indonesia; 4Ophthalmology Clinic, Dr YAP Eye Hospital, Yogyakarta, Indonesia; 5Laboratory of Gene Function in Animals, Nara Institute of Science and Technology, Ikoma, Nara, Japan
Correspondence: Supanji Supanji
Department of Ophthalmology, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Jalan Farmako Sekip Utara, Yogyakarta, 55281, Indonesia
Tel +62 274 560300
Fax +62 274 581 876
Email [email protected]
Purpose: This study aimed to determine the association of ARMS2 A69S, ARMS2 del443ins54, and CFH Y402H polymorphisms with neovascular age-related macular degeneration (nAMD) for the first time in an Indonesian population.
Patients and Methods: Our case–control study involved 104 nAMD and 100 control subjects. AMD diagnosis was evaluated by retinal specialists based on color fundus photography and optical coherence tomography. The polymorphisms on CFH Y402H and ARMS2 A69S were analyzed by PCR-restriction fragment length polymorphism (PCR-RFLP), whereas ARMS2 del443ins54 was evaluated by PCR-based assay.
Results: Significant allelic associations with nAMD were detected on all polymorphisms (P< 0.05), with stronger association with the ARMS2 A69S (OR 3.13; 95% CI 2.08– 4.71; P< 0.001) and ARMS2 del443ins54 (OR 3.28; 95% CI 2.17– 4.95; P< 0.001) polymorphisms than with CFH Y402H (OR 2.08; 95% CI 1.08– 3.99; P=0.028). Genotype analysis showed a statistical difference between nAMD and the control group for all polymorphisms (P< 0.05). However, the association with nAMD was weaker for CFH Y402H (P=0.043) than for ARMS2 A69S and ARMS2 del443ins54 (P< 0.001). A significant interaction between ARMS2 A69S and hypertension was documented (OR 9.53; 95% CI 3.61– 25.1; P< 0.001).
Conclusion: Our findings indicate that ARMS2 A69S and ARMS2 del443ins54 polymorphisms are strongly associated with the risk of nAMD for the first time in an Indonesian population. The risk of nAMD increased when the presence of risk alleles from ARMS2 A69S was combined with the presence of hypertension.
Keywords: age-related macular degeneration, ARMS2, CFH, polymorphism
Introduction
Age-related macular degeneration (AMD) is a progressive degenerative disease affecting the macula and is the top five leading cause of irreversible blindness worldwide.1 It has been estimated that there are nearly 200 millions of individuals with AMD in 2020, and will be projected to rise to 288 millions in 2040.1
The prevalence of AMD increases exponentially with age.1 With ageing, a cascade of deterioration occurs in photoreceptors, retinal pigment epithelium (RPE) and Bruch’s membrane (BM) leaving permanent lesion observed clinically as geographic atrophy (dry AMD) or causing abnormal blood vessel originating from choroid to leak or to bleed at the macular area (neovascular AMD [nAMD]).2 These may ultimately cause irreversible visual impairment if left untreated. Interestingly, studies showed that not all aged individuals undergo the similar processes and develop AMD, suggesting a strong genetic-driven variation in the pathophysiology of this condition.3
There has been extensive literature reporting the genetic associations in AMD.4–6 Complement Factor H (CFH), Human high-temperature requirement serine protease A1 (HtrA1), and substitution from alanine to serine of amino acid 69 (A69S) in age-related maculopathy susceptibility 2 (ARMS2) at chromosome 10q26 are speculated to play key roles in cellular senescence, thus have been the most consistently associated with AMD in different populations.7–9 In previous studies, ARMS2 and HtrA1 were reported to have a strong linkage disequilibrium.10,11 Grassmann et al12 further asserted that the ARMS2 rs10490924 variant (not HtrA1 rs11200638) is more strongly associated with AMD than HtrA1 rs11200638. This finding was supported by Kanda et al,10 who identify that ARMS2 rs10490924 polymorphism alone can explain the association of the 200-kb region at chromosome 10q26 with AMD. Deletion/insertion consisting of a 443 bp deletion and an adjacent 54 bp insertion in the 3ʹ-untranslated region (3ʹ-UTR) of ARMS2 (del443ins54) and complement factor H Tyr402His (CFH Y402H) was also reported to be strongly associated with AMD.13–15 Deletion/insertion polymorphism in ARMS2 disrupts the stability of ARMS2 gene transcription products16 and induces HtrA1 transcription regulator activity.17
In Western populations, the associations of ARMS2 and CFH were documented in American, Dutch, Italian, Spanish, and Swiss populations.14,18–23 In Asian, similar associations were reported in Chinese, Japanese, and Indian populations.15,24–27 However, very limited evidence is available from Asian Malay population, which is also one of the biggest ethnic groups in Asia.
In this study, we aimed to investigate the associations of ARMS2 A69S, ARMS2 del443ins54, and CFH Y402H with AMD in Indonesian population, which constitutes the majority of Asian Malay ethnic group in the region.
Method
This was an age-matched case–control study of participants aged 45 years old or older. Cases were naïve nAMD patients in at least one eye attending retinal clinic at three tertiary hospitals in Yogyakarta: 1) Dr. Sardjito General Hospital; 2) Hardjolukito Military Air Force Central Hospital, and 3) Dr. YAP Eye Hospital with no previous history of AMD treatment, recruited consecutively from August 2016 to November 2018. The diagnosis of AMD was established from slit-lamp examination, fundus photograph and spectral-domain OCT, confirmed by a retinal specialist following the International Age-related Maculopathy (ARM) Epidemiological Study Group28 and AMD clinical classification criteria.29 We excluded cases with co-existing choroidal or other retinal inflammatory diseases. Controls were healthy individuals without AMD or other retinal lesions who underwent eye examination for senile cataract.
Each subject was fully informed about the purpose and the procedures of the study. Consent was obtained from all subjects in written form prior to participation. All study procedures adhered to the principles of the Declaration of Helsinki. The study was approved by the Institutional Review Board of Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada in August 2016.
Genotyping
The genomic DNA of each patient was extracted from venous blood placed into a tube containing EDTA as an anticoagulant. The blood samples were immediately processed utilizing a commercially available DNA extraction kit (GeneAid Genomic Human DNA Mini Kit [GB100/300], New Taipei City, Taiwan). DNA extraction and single nucleotide polymorphism (SNP) identification were conducted at the Integrated Research Laboratory, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada.
The specific variants for the ARMS2 genes were ARMS2 A69S rs10490924 and ARMS2 del443ins54 (c.*372_815del443ins54), whereas that for CFH Y402H was rs1061170. Polymerase chain reaction (PCR) was performed in a thermal cycler (ProFlex PCR System, Applied Biosystems) following the ready-to-use PCR kit protocol (KAPA Taq PCR Kit, Kapa Biosystems). The PCR cycling conditions were set as follows: 1 cycle (95 °C for 2 min), 30 cycles (95 °C for 30 s), 1 cycle (52 °C for 1 min for each gene), 1 cycle (72 °C for 1 min), and 1 cycle (72 °C for 5 min).
The primer sequences for the genes of interest are as follows: 1) ARMS2 A69S forward 5ʹ-TGTCACTGCATTCCCTCCTGTCAT-3ʹ and reverse 5ʹ-AAGCTTCTTACCCTGACTTCCAGC-3ʹ; 2) ARMS2 del443ins54 forward 5ʹ-TACCCAGGACCGATGGTAAC-3ʹ and reverse 5ʹ-GAGGAAGGCTGAATTGCCTA-3ʹ; and 3) CFH Y402H forward 5ʹ-CTTTAGTTCGTCTTCAGTTATAC-3ʹ and reverse 5ʹ-GTCATCTATGTTACTTAGAAAGT-3ʹ.
SNP identification involved PCR-based assay for ARMS2 del443ins54 and PCR-restriction fragment length polymorphism (PCR-RFLP) for ARMS2 A69S and CFH Y402H. Restriction digestion was performed at 37 °C for 18 h following the manufacturer’s protocol using PvuII restriction enzyme for ARMS2 A69S (Takara Bio, Japan) and Hsp92II for CFH Y402H (Promega). All amplified products were electrophoresed on 1.5% agarose gel containing FloroSafe DNA stain (1st Base Asia). Random sampling from each genotype in each SNP was conducted for genotype confirmation through Sanger DNA sequencing. Sequencing service was provided by 1st Base Asia, Singapore.
Statistical Analysis
Descriptive data were generated for all variables. Unpaired Student’s t-test for numerical variables or Chi-squared test and Fisher exact test for categorical variables was performed to compare baseline characteristics between nAMD and control groups. Two-sided p-values were reported. We tested for deviation from the Hardy–Weinberg equilibrium (HWE) in both groups through the chi-square test with the “genhwcci” command in Stata.
Associations between SNP and other risk factors for susceptibility to nAMD were assessed using logistic regression models measured by odds ratio (OR) and 95% confidence interval (CI). In the multivariable logistic regression model, the likelihood ratio test was performed to fit the model. We pooled one risk allele and two risk alleles as one category (risk allele) in the interaction analysis. Interaction analysis was performed by introducing the interaction term in the same regression model. All analyses were carried out using Stata (version 15.1, StataCorp, College Station, TX, USA).
Results
There were 116 cases [46 males (44.2%) and 58 females (55.8%)] and 100 controls [45 males (45.0%) and 55 females (55.0%)] included in the final analysis. Baseline characteristics of the participants are presented in Table 1. The mean age of cases was 66.3 ± 8.8 years while control was 67.9 ± 7.7 years. Cases showed very similar characteristics to control except that having higher BMI (23.7 vs 22.0; P=0.002) and were more likely to have hypertension (46.2% vs 18.0%; P<0.001) than controls.
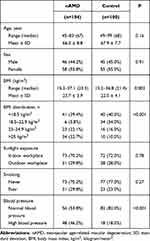 |
Table 1 Baseline Characteristics of Participants |
The allele/genotype distributions and odds ratio (OR) of each SNP are summarized in Table 2. Significant allelic associations with nAMD were detected on all SNPs (P<0.05). Compared to those having non-risk alleles, those with risk alleles of ARMS2 A69S, ARMS2 del443ins54, and CFH Y402H were more likely to have nAMD (OR 3.13; 95% Confidence Interval [CI] 2.08–4.71 for ARMS2 A69S, OR 3.28; 95% CI 2.17–4.95 for ARMS2 del443ins54, and OR 2.08; 95% CI 1.08–3.99 for CFH Y402H). Genotype analysis showed significant differences between the nAMD and control groups for all polymorphisms (Table 2). The associations of ARMS2 A69S and ARMS2 del443ins54 (P<0.001) with nAMD were stronger than that of CFH Y402H (P=0.043).
 |
Table 2 Case–Control Frequencies of Alleles and Genotypes of SNP on ARMS2 A69S, ARMS2 del443ins54 and CFH Y402H |
In Table 3, it is shown that homozygous risk allele carriers at the ARMS2 A69S polymorphism (OR 5.97; 95% CI 2.75–13.0) and ARMS2 del443ins54 (OR 7.99; 95% CI 3.45–18.6) were both strongly associated with nAMD. For CFH Y402H, individuals with one copy of the risk allele were more likely to have nAMD than control (OR 2.47; 95% CI 1.19–5.11). These associations remained significant even after controlling for age, gender, smoking, body mass index and blood pressure.
 |
Table 3 Distribution of Unadjusted and Adjusted Odds Ratio for Risk Genotypes in ARMS2 A69S, ARMS2 Del443ins54 and CFH Y402H |
In additional analyses, we documented significant interaction between ARMS2 A69S and hypertension. Table 4 shows that individuals who had ARMS2 A69S risk alleles and hypertension had significantly higher odds of nAMD than those with hypertension or ARMS2 A69S risk alleles only (OR 9.53; 95% CI 3.61–25.1; P<0.001).
 |
Table 4 Interaction Analysis of ARMS2 A69S and Hypertension |
Discussion
In this study population, we documented that gene polymorphisms of ARMS2 A69S and ARMS2 del443ins54 were strongly and independently associated with nAMD. In contrast, we also documented that the association of CFH Y402H with nAMD was weaker than that of ARMS2 A69S and ARMS2 del443ins54. We also documented a synergistic effect between ARMS2 A69S and hypertension meaning, that individuals with both ARMS2 A69S risk alleles and hypertension had a significantly higher risk of nAMD. Findings from our study reconfirm that ARMS2 genes are strongly associated with nAMD across different populations, at the same time suggest the existence of gene–hypertension interaction between this specific gene and hypertension.
We provided the first evidence of the associations of ARMS2 A69S, ARMS2 del443ins54, and CFH Y402H with nAMD in Indonesian population. There have been several studies from Asian population available for direct comparison.15,30–32 ARMS2 A69S gene polymorphisms have been consistently associated with nAMD in Malaysian,33 Chinese Singaporean,31 Thai,30 Chinese,34,35 Japanese,36,37 Korean,38 Indian,32 and European populations.39 It has also been reported that ARMS2 A69S has stronger associations with nAMD than CFH Y402H,40 which is comparable to our study findings. In addition to ARMS2 A69S, results from our study showed that ARMS2 del443ins54, also significantly associated with nAMD, which has been reported in Japanese, Caucasian, and Indian populations.13–15,41 In contrast to ARMS2, associations between CFH Y402H gene variants and nAMD have been less consistent.42 For example, CFH Y402H in Caucasian had a strong association with nAMD,4,14,43,44 but studies from Asian showed a conflicting result. Xu et al,34 Gotoh et al,45 Okamoto et al,46 Uka et al,47 and Chen et al48 showed a weak association of CFH Y402H with AMD while Lau et al49 showed a contradictory result.
The role of ARMS2 genes in nAMD has become a subject of interest for more than a decade.10 ARMS2 has been speculated to regulate the surface complement-mediated phagocytosis of cellular debris.50 Micklisch et al50 reported that decreases of the ARMS2 expression in AMD were associated with polymorphism of ARMS2 A69S and del443ins54. Decreases in ARMS2 protein would result in drusen accumulation due to impaired cellular debris clearance.50 Furthermore, a study by Yang and associates51 suggested that ARMS2 A69S risk allele may decrease antioxidant enzyme activity in end-stage AMD-specific induced pluripotent stem cells(iPSCs)-derived RPE model. RPE cells are exposed to intense photo-oxidative energy and excess oxygen, promoting reactive oxygen species (ROS). Decreases in antioxidant enzyme capacity lead to ROS accumulation, increasing oxidative damage contributed to AMD.
Some studies have suggested that inflammation may partly explain the link between AMD and ARMS2 polymorphisms.25,52 In iPSCs-derived RPE from AMD donor, Saini et al52 showed that ARMS2 risk allele increased the complement proteins and pro-inflammatory factors compared to iPSCs-RPE derived from healthy control. In addition, there was a study reporting that ARMS2 del443ins54 was correlated with an increase in the serum high sensitivity C-reactive protein (hs-CRP) levels of nAMD subjects in a Japanese study.25 High serum CRP is associated with the late stage of AMD in a systematic literature review and meta-analysis.53 Serum CRP represents systemic inflammatory activity and is a marker of chronic low-grade inflammation.53
The present study also documented gene–hypertension interactions of the ARMS2 A69S and hypertension. Hyman et al54 reported that nAMD and hypertensive disease may have a similar underlying systemic process, as nAMD is linked to high diastolic blood pressure (OR: 4.4; 95% CI: 1.4–14.2). The involvement of oxidative stress accumulation processes in both nAMD and hypertension might explain these associations.
The strengths of our study included age-matched cases and controls, detailed clinical and eye examinations by retinal specialist using advanced multimodal imaging to confirm the diagnosis of nAMD and the application of PCR that ensured the accuracy of genetic assessment. However, several limitations were also noted. First, we did not use indocyanine green angiography (ICGA) as the gold standard for nAMD diagnosis. Nevertheless, spectral-domain OCT had high sensitivity and specificity in distinguishing nAMD from polypoidal choroidal vasculopathy (PCV).55–57 Diagnosis of nAMD based on fundus photography and spectral-domain OCT had more than 90% agreement when compared to ICGA,58–60 thus reassuring the minimal bias in this study. Second, the hospital-based design of our study may have only captured the advanced profile of AMD patients, therefore limiting the representation of AMD in general population. Whether or not individuals with AMD from the general population have similar genetic associations remained questionable. Future population-based studies are warranted to address these questions.
In conclusion, our study highlighted a strong association of ARMS2 A69S and del443ins54 in people with nAMD in Yogyakarta, Indonesia. This is the first study on nAMD’s genetic risk factors and the first AMD research in Indonesia. Limited studies have been performed in Southeast Asia. Although our study found a weak relationship between the CFH Y402H polymorphism and nAMD risk, further studies are warranted to confirm the relationship of CFH Y402H and nAMD in Indonesian populations. Future work should have larger and more diverse sample sizes to allow subanalysis based on ethnic origin in Indonesia. Genetic information is important in the area of personalized medicine, and it may be useful as a baseline data to establish cohort studies of AMD clinical risk prediction scoring relevant to the Indonesian population.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
This study received funding from the Indonesian Government – Ministry of Research, Technology and Higher Education Research Grant and from Hibah Dana Masyarakat of Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada. The authors independently carried out this research without any interference from the funding bodies.
The authors report no conflicts of interest for this work.
References
1. Wong WL, Su X, Li X, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2(2):e106–e116. doi:10.1016/S2214-109X(13)70145-1
2. Yonekawa Y, Kim IK. Clinical characteristics and current treatment of age-related macular degeneration. Cold Spring Harb Perspect Med. 2015;5(1):a017178. doi:10.1101/cshperspect.a017178
3. Schwartz SG, Hampton BM, Kovach JL, Brantley MA. Genetics and age-related macular degeneration: a practical review for the clinician. Clin Ophthalmol. 2016;10:1229–1235. doi:10.2147/OPTH.S109723
4. Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi:10.1126/science.1109557
5. Fritsche LG, Igl W, Bailey JNC, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134–143. doi:10.1038/ng.3448
6. Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genom Hum Genet. 2014;15(1):151–171. doi:10.1146/annurev-genom-090413-025610
7. Parmeggiani F, Sorrentino FS, Romano MR, et al. Mechanism of inflammation in age-related macular degeneration: an up-to-date on genetic landmarks. Mediators Inflamm. 2013;2013:1–13. doi:10.1155/2013/435607
8. Bonyadi MHJ, Yaseri M, Soheilian M. Association of combined complement factor H Y402H and ARMS2/LOC387715 A69S polymorphisms with age-related macular degeneration: an updated meta-analysis. Ophthalmic Genet. 2020;1–7. doi:10.1080/13816810.2020.1765396
9. Bonyadi MHJ, Yaseri M, Nikkhah H, Bonyadi M, Soheilian M. Association of risk genotypes of ARMS2/LOC387715 A69S and CFH Y402H with age-related macular degeneration with and without reticular pseudodrusen: a meta-analysis. Acta Ophthalmol. 2018;96(2):e105–e110. doi:10.1111/aos.13494
10. Kanda A, Chen W, Othman M, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci U S A. 2007;104(41):16227–16232. doi:10.1073/pnas.0703933104
11. Wang G, Spencer KL, Court BL, et al. Localization of age-related macular degeneration-associated ARMS2 in cytosol, not mitochondria. Invest Ophthalmol Vis Sci. 2009;50(7):3084–3090. doi:10.1167/iovs.08-3240
12. Grassmann F, Heid IM, Weber BHF. Recombinant haplotypes narrow the ARMS2/HTRA1 association signal for age-related macular degeneration. Genetics. 2017;205(2):919–924. doi:10.1534/genetics.116.195966
13. Hadley D, Orlin A, Brown G, et al. Analysis of six genetic risk factors highly associated with AMD in the region surrounding ARMS2 and HTRA1 on chromosome 10, region q26. Invest Ophthalmol Vis Sci. 2010;51(4):2191–2196. doi:10.1167/iovs.09-3798
14. Ricci F, Zampatti S, D’Abbruzzi F, et al. Typing of ARMS2 and CFH in age-related macular degeneration: case-control study and assessment of frequency in the Italian population. Arch Ophthalmol. 2009;127(10):1368–1372. doi:10.1001/archophthalmol.2009.237
15. Gotoh N, Nakanishi H, Hayashi H, et al. ARMS2 (LOC387715) variants in Japanese patients with exudative age-related macular degeneration and polypoidal choroidal vasculopathy. Am J Ophthalmol. 2009;147(6):1037–1041.e2. doi:10.1016/j.ajo.2008.12.036
16. Fritsche LG, Loenhardt T, Janssen A, et al. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40(7):892–896. doi:10.1038/ng.170
17. Iejima D, Itabashi T, Kawamura Y, et al. HTRA1 (high temperature requirement a serine peptidase 1) gene is transcriptionally regulated by insertion/deletion nucleotides located at the 3′ end of the ARMS2 (age-related maculopathy susceptibility 2) gene in patients with age-related macular degeneration. J Biol Chem. 2015;290(5):2784–2797. doi:10.1074/jbc.M114.593384
18. Vavvas DG, Small KW, Awh CC, Zanke BW, Tibshirani RJ, Kustra R. CFH and ARMS2 genetic risk determines progression to neovascular age-related macular degeneration after antioxidant and zinc supplementation. Proc Natl Acad Sci USA. 2018;115(4):E696–E704. doi:10.1073/pnas.1718059115
19. Lechanteur YTE, van de Camp PL, Smailhodzic D, et al. Association of smoking and CFH and ARMS2 risk variants with younger age at onset of neovascular age-related macular degeneration. JAMA Ophthalmol. 2015;133(5):533–541. doi:10.1001/jamaophthalmol.2015.18
20. Smailhodzic D, Klaver CCW, Klevering BJ, et al. Risk alleles in CFH and ARMS2 are independently associated with systemic complement activation in age-related macular degeneration. Ophthalmology. 2012;119(2):339–346. doi:10.1016/j.ophtha.2011.07.056
21. Brión M, Sanchez‐Salorio M, Cortón M, et al. Genetic association study of age-related macular degeneration in the Spanish population. Acta Ophthalmol. 2011;89(1):e12–e22. doi:10.1111/j.1755-3768.2010.02040.x
22. Wang G, Spencer KL, Scott WK, et al. Analysis of the indel at the ARMS2 3′UTR in age-related macular degeneration. Hum Genet. 2010;127(5):595–602. doi:10.1007/s00439-010-0805-8
23. Zysset-Burri DC, Keller I, Berger LE, et al. Associations of the intestinal microbiome with the complement system in neovascular age-related macular degeneration. NPJ Genom Med. 2020;5(1):1–11. doi:10.1038/s41525-020-00141-0
24. Sundaresan P, Vashist P, Ravindran RD, et al. Polymorphisms in ARMS2/HTRA1 and complement genes and age-related macular degeneration in India: findings from the INDEYE study. Invest Ophthalmol Vis Sci. 2012;53(12):7492. doi:10.1167/iovs.12-10073
25. Yasuma TR, Nakamura M, Nishiguchi KM, et al. Elevated C-reactive protein levels and ARMS2/HTRA1 gene variants in subjects without age-related macular degeneration. Mol Vis. 2010;16:2923.
26. Hayashi H, Yamashiro K, Gotoh N, et al. CFH and ARMS2 variations in age-related macular degeneration, polypoidal choroidal vasculopathy, and retinal angiomatous proliferation. Invest Ophthalmol Vis Sci. 2010;51(11):5914–5919. doi:10.1167/iovs.10-5554
27. Zhuang W, Li H, Liu Y, et al. Association of specific genetic polymorphisms with age-related macular degeneration in a Northern Chinese population. Ophthalmic Genet. 2014;35(3):156–161. doi:10.3109/13816810.2014.921314
28. Bird AC, Bressler NM, Bressler SB, et al. An international classification and grading system for age-related maculopathy and age-related macular degeneration. Surv Ophthalmol. 1995;39(5):367–374. doi:10.1016/S0039-6257(05)80092-X
29. Ferris FL, Wilkinson CP, Bird A, et al. Clinical classification of age-related macular degeneration. Ophthalmology. 2013;120(4):844–851. doi:10.1016/j.ophtha.2012.10.036
30. Ruamviboonsuk P, Tadarati M, Singhanetr P, et al. Genome-wide association study of neovascular age-related macular degeneration in the Thai population. J Hum Genet. 2017;62(11):957–962. doi:10.1038/jhg.2017.72
31. Cheung CMG, Laude A, Yeo I, et al. Systemic, ocular and genetic risk factors for age-related macular degeneration and polypoidal choroidal vasculopathy in Singaporeans. Sci Rep. 2017;7. doi:10.1038/srep41386
32. Rajendran A, Dhoble P, Sundaresan P, et al. Genetic risk factors for late age-related macular degeneration in India. Br J Ophthalmol. 2018;102(9):1213–1217. doi:10.1136/bjophthalmol-2017-311384
33. Mohamad NA, Ramachandran V, Mohd Isa H, et al. Association of HTRA1 and ARMS2 gene polymorphisms with response to intravitreal ranibizumab among neovascular age-related macular degenerative subjects. Hum Genomics. 2019;13. doi:10.1186/s40246-019-0197-3
34. Xu Y, Guan N, Xu J, et al. Association of CFH, LOC387715, and HTRA1 polymorphisms with exudative age-related macular degeneration in a northern Chinese population. Mol Vis. 2008;14:1373.
35. Cheng Y, Huang L, Li X, Zhou P, Zeng W, Zhang C. Genetic and functional dissection of ARMS2 in age-related macular degeneration and polypoidal choroidal vasculopathy. Lee H-C, ed. PLoS One. 2013;8(1):e53665. doi:10.1371/journal.pone.0053665
36. Yanagisawa S, Kondo N, Miki A, et al. Difference between age-related macular degeneration and polypoidal choroidal vasculopathy in the hereditary contribution of the A69S variant of the age-related maculopathy susceptibility 2 gene (ARMS2). Mol Vis. 2011;17:3574–3582.
37. Aoki A, Tan X, Yamagishi R, et al. Risk factors for age-related macular degeneration in an elderly Japanese population: the Hatoyama Study. Invest Ophthalmol Vis Sci. 2015;56(4):2580–2585. doi:10.1167/iovs.14-16339
38. Lee SJ, Kim NR, Chin HS. LOC387715/HTRA1 polymorphisms, smoking and combined effects on exudative age-related macular degeneration in a Korean population. Clin Experiment Ophthalmol. 2010;38(7):698–704. doi:10.1111/j.1442-9071.2010.02316.x
39. Chakravarthy U, McKay GJ, de Jong PTVM, et al. ARMS2 increases the risk of early and late age-related macular degeneration in the European eye study. Ophthalmology. 2013;120(2):342–348. doi:10.1016/j.ophtha.2012.08.004
40. Jabbarpoor Bonyadi MH, Yaseri M, Nikkhah H, Bonyadi M, Nazari R, Soheilian M. Comparison of ARMS2/LOC387715 A69S and CFH Y402H risk effect in wet-type age-related macular degeneration: a meta-analysis. Int Ophthalmol. 2019;39(4):949–956. doi:10.1007/s10792-018-0853-y
41. Kaur I, Cantsilieris S, Katta S, et al. Association of the del443ins54 at the ARMS2 locus in Indian and Australian cohorts with age-related macular degeneration. Mol Vis. 2013;19:822–828.
42. Restrepo NA, Spencer KL, Goodloe R, et al. Genetic determinants of age-related macular degeneration in diverse populations from the PAGE study. Invest Ophthalmol Vis Sci. 2014;55(10):6839–6850. doi:10.1167/iovs.14-14246
43. Goverdhan SV, Hannan S, Newsom RB, Luff AJ, Griffiths H, Lotery AJ. An analysis of the CFH Y402H genotype in AMD patients and controls from the UK, and response to PDT treatment. Eye. 2007;22(6):849–854. doi:10.1038/sj.eye.6702830
44. Matušková V, Zeman T, Ewerlingová L, et al. An association of neovascular age-related macular degeneration with polymorphisms of CFH, ARMS2, HTRA1 and C3 genes in Czech population. Acta Ophthalmol. 2020;98:e691–e699. doi:10.1111/aos.14357
45. Gotoh N, Yamada R, Hiratani H, et al. No association between complement factor H gene polymorphism and exudative age-related macular degeneration in Japanese. Hum Genet. 2006;120(1):139–143. doi:10.1007/s00439-006-0187-0
46. Okamoto H, Umeda S, Obazawa M, et al. Complement factor H polymorphisms in Japanese population with age-related macular degeneration. Mol Vis. 2006;12:156–158.
47. Uka J, Tamura H, Kobayashi T, et al. No association of complement factor H gene polymorphism and age-related macular degeneration in the Japanese population. Retina. 2006;26(9):985–987. doi:10.1097/01.iae.0000244068.18520.3e
48. Chen LJ, Liu DTL, Tam POS, et al. Association of complement factor H polymorphisms with exudative age-related macular degeneration. Mol Vis. 2006;12:1536–1542.
49. Lau L-I, Chen S-J, Cheng C-Y, et al. Association of the Y402H polymorphism in complement factor H gene and neovascular age-related macular degeneration in Chinese patients. Invest Ophthalmol Vis Sci. 2006;47(8):3242. doi:10.1167/iovs.05-1532
50. Micklisch S, Lin Y, Jacob S, et al. Age-related macular degeneration associated polymorphism rs10490924 in ARMS2 results in deficiency of a complement activator. J Neuroinflammation. 2017;14. doi:10.1186/s12974-016-0776-3
51. Yang J, Li Y, Chan L, et al. Validation of genome-wide association study (GWAS)-identified disease risk alleles with patient-specific stem cell lines. Hum Mol Genet. 2014;23(13):3445–3455. doi:10.1093/hmg/ddu053
52. Saini JS, Corneo B, Miller JD, et al. Nicotinamide ameliorates disease phenotypes in a human iPSC model of age-related macular degeneration. Cell Stem Cell. 2017;20(5):635–647.e7. doi:10.1016/j.stem.2016.12.015
53. Feng C, Krogh Nielsen M, Sørensen TL, Subhi Y. Systemic levels of C-reactive protein in patients with age-related macular degeneration: a systematic review with meta-analyses. Mech Ageing Dev. 2020;191:111353. doi:10.1016/j.mad.2020.111353
54. Hyman L, Schachat AP, He Q, Leske M. Hypertension, cardiovascular disease, and age-related macular degeneration. Arch Ophthalmol. 2000;118(3):351–358. doi:10.1001/archopht.118.3.351
55. Chaikitmongkol V, Kong J, Khunsongkiet P, et al. Sensitivity and specificity of potential diagnostic features detected using fundus photography, optical coherence tomography, and fluorescein angiography for polypoidal choroidal vasculopathy. JAMA Ophthalmol. 2019;137(6):661–667. doi:10.1001/jamaophthalmol.2019.0565
56. Liu R, Li J, Li Z, et al. Distinguishing polypoidal choroidal vasculopathy from typical neovascular age-related macular degeneration based on spectral domain optical coherence tomography. Retina. 2016;36(4):778–786. doi:10.1097/IAE.0000000000000794
57. Cheung CMG, Lai TYY, Teo K, et al. Polypoidal choroidal vasculopathy: consensus nomenclature and non–indocyanine green angiograph diagnostic criteria from the Asia-pacific ocular imaging society PCV workgroup. Ophthalmology. 2020. doi:10.1016/j.ophtha.2020.08.006
58. Mowatt G, Hernández R, Castillo M, et al. Assessment of Diagnostic and Monitoring Studies. NIHR Journals Library; 2014. Available from: https://www.ncbi.nlm.nih.gov/books/NBK263940/.
59. Sandhu SS, Talks SJ. Correlation of optical coherence tomography, with or without additional colour fundus photography, with stereo fundus fluorescein angiography in diagnosing choroidal neovascular membranes. Br J Ophthalmol. 2005;89(8):967–970. doi:10.1136/bjo.2004.060863
60. Wilde C, Patel M, Lakshmanan A, Amankwah R, Dhar-Munshi S, Amoaku W. The diagnostic accuracy of spectral-domain optical coherence tomography for neovascular age-related macular degeneration: a comparison with fundus fluorescein angiography. Eye. 2015;29(5):602–610. doi:10.1038/eye.2015.44
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
