Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 15
Associations Between Genetic Polymorphisms Within Transporter Genes and Clinical Response to Methotrexate in Chinese Rheumatoid Arthritis Patients: A Pilot Study
Authors Cen H, Wen QW, Zhang HQ, Yu H, Zeng Z, Jin T, Wang TH, Qin W, Huang H, Wu XD
Received 26 November 2021
Accepted for publication 28 March 2022
Published 12 April 2022 Volume 2022:15 Pages 327—339
DOI https://doi.org/10.2147/PGPM.S350417
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Han Cen,1– 3 Qin-Wen Wen,4 Han-Qing Zhang,4 Hang Yu,4 Zhen Zeng,2 Ting Jin,2 Ting-Hui Wang,4 Wen Qin,4 Hua Huang,4 Xiu-Di Wu4
1Institute of Geriatrics, The Affiliated Hospital of Medical School, Ningbo University, Ningbo, Zhejiang, People’s Republic of China; 2Department of Preventive Medicine, Medical School of Ningbo University, Ningbo, Zhejiang, People’s Republic of China; 3Zhejiang Provincial Key Laboratory of Pathophysiology, School of Medicine, Ningbo University, Ningbo, Zhejiang, People’s Republic of China; 4Department of Rheumatology, Ningbo First Hospital, Ningbo Hospital of Zhejiang University, Ningbo, Zhejiang, People’s Republic of China
Correspondence: Xiu-Di Wu, Department of Rheumatology, Ningbo First Hospital, Ningbo Hospital of Zhejiang University, 59 Liuting Road, Ningbo, Zhejiang, 315010, People’s Republic of China, Tel +86 574 87085588, Fax +86 574 87291583, Email [email protected]
Purpose: To investigate the associations between genetic polymorphisms within transporter genes and clinical response to methotrexate (MTX) in Chinese rheumatoid arthritis (RA) patients.
Patients and Methods: A total of 100 RA patients receiving MTX were prospectively followed up for approximately 3 months to determine the clinical response based on several criteria, including European League Against Rheumatism (EULAR) good and moderate response, disease activity score in 28 joint counts – erythrocyte sedimentation rate (DAS28-ESR) low disease activity (LDA), change in DAS28-ESR (ΔDAS28-ESR) and ΔDAS28-ESR > 0.6. Fifty-four single nucleotide polymorphisms (SNPs) within seven transporter genes, including SLC19A1, ABCB1, ABCC1∼ 4 and ABCG2, were genotyped.
Results: Multivariable analysis revealed that SLC19A1 rs12659 and rs3788200, ABCC2 rs3740066, rs4148396 and rs717620 were significantly associated with EULAR good and moderate response, and ABCC2 rs3740066 and rs717620 were significantly associated with DAS28-ESR LDA, and ABCB1 rs1128503, rs4148737 and ABCC3 rs2277624, rs4148416 were significantly associated with ΔDAS28-ESR. Moreover, 12 genetic polymorphisms were found to be significantly associated with ΔDAS28-ESR > 0.6. With adjustment for corresponding confounders, SLC19A1 TGAA haplotype consisting of rs1051266, rs1131596, rs12659 and rs3788200 was significantly associated with EULAR good and moderate response and ΔDAS28-ESR > 0.6 compared with the most common haplotype CAGG. The ABCC2 haplotype TTT composed of rs717620, rs4148396 and rs3740066 was significantly associated with EULAR good and moderate response and ΔDAS28-ESR > 0.6 compared with the most common haplotype CCC.
Conclusion: Our results highlight the potential of genetic polymorphisms within transporter genes, particularly SLC19A1 and ABCC2, as predictors of clinical response to MTX in Chinese RA patients.
Keywords: rheumatoid arthritis, methotrexate, transporter, single-nucleotide polymorphism, clinical response
Introduction
Rheumatoid arthritis (RA) is a debilitating systemic autoimmune disease affecting about 5 per 1000 adults worldwide, and irreversible joint damage and disability would ensue if without timely diagnosis and treatment.1 Over the past decades, the RA treatment landscape has dramatically changed, attributed to the application of treat-to-target approach and the increasing availability of biological disease-modifying anti-rheumatic drugs (bDMARDs).2 In spite of this, methotrexate (MTX) is still recommended as the initial therapeutic drug for most newly diagnosed RA patients owing to its good efficacy, long-term safety and low price.3–5 A considerable proportion of RA patients, however, do not respond satisfactorily to MTX,6,7 often necessitating a switch to or addition of another DMARD according to the treat-to-target principle. Of note, since early sufficient response to MTX determines the long-term prognosis of RA,8,9 it is of great clinical significance to investigate predictors of clinical response to MTX in RA patients.
Currently, although the precise mechanism of action of MTX has not been fully elucidated, accumulating evidence indicates that MTX entering cells by transporters undergoes serial polyglutamation to form MTX-polyglutamates (MTX-PGs), which then exert anti-inflammatory and immunosuppressive response through multiple mechanisms such as the inhibition of purine and pyrimidine synthesis, transmethylation reactions and the promotion of adenosine release. Finally, the long-chain MTX-PGs are converted into short-chain MTX and ultimately to MTX, which could be transported out of cells via ATP binding cassette (ABC) transporters.10 Notably, available evidence indicates that solute carrier family 19 member 1 (SLC19A1) is the main transporter mediating the influx of MTX, while the efflux of MTX mainly involves ABCB1, ABCC1~4 and ABCG2.11,12 These transporters are expressed in multiple tissues and remarkably determine the pharmacokinetics of MTX, including absorption, distribution and elimination. Over the past decades, there is growing interest in pharmacogenetic studies owing to the success of high throughput genotyping technology development and the stable quality of genetic polymorphisms, and much more efforts have been made to dissect the role of single-nucleotide polymorphisms (SNPs) within genes encoding proteins implicated in pharmacokinetics and pharmacodynamics in the inter-individual variability of therapeutic drug response.13 In the context of pharmacogenetic studies of MTX in RA, based on the considerable effects of transporters on pharmacokinetics, there is indeed a great deal of studies investigating the associations of genetic polymorphisms within aforementioned transporter genes with clinical response to MTX in RA patients with inconsistent results.11,14 However, the majority of these studies focused on a few SNPs within some transporters, and most were performed in Caucasians. In view of the genetic heterogeneity among different ethnic populations, we determined to systematically examine the associations of genetic polymorphisms within transporter genes (SLC19A1, ABCB1, ABCC1~4 and ABCG2) with clinical response to MTX in Chinese RA patients.
Methods
Study Design
A total of 100 adult RA patients (age ≥18 years) with active disease, who received MTX monotherapy and were prospectively followed up for approximately 3 months, were recruited from the Department of Rheumatology, Ningbo First Hospital between August 2015 and January 2021. All these RA patients fulfilled the 1987 American College of Rheumatology revised criteria15 or the 2010 American College of Rheumatology/European League Against Rheumatism (EULAR) criteria for the classification of RA.16 Only DMARD-naive RA patients or RA patients who had discontinued the use of DMARD for more than six months were eligible. The dose of MTX was increased to the maximum based on efficacy and toxicity considerations. Folic acid was given to all patients with RA. Other concomitant DMARDs were not allowed, while the low-dose glucocorticoid (GC) and nonsteroidal anti-inflammatory drugs (NSAIDs) were permitted. This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Ningbo University (Ethics ID: SX2019139). Informed consent was obtained from all participants.
Data Collection
The baseline information of the following variables was collected: age, gender, smoking and drinking status, disease duration, rheumatoid factor (RF) and anti-cyclic citrullinated peptide (CCP) antibody status, tender joint count (TJC) and swollen joint count (SJC) in 28 joints, erythrocyte sedimentation rate (ESR), MTX dosage, concomitant GC use and prednisolone equivalent dosage, and concomitant NSAIDs use. Moreover, the values of the components of disease activity score in 28 joint counts – ESR (DAS28-ESR),17 namely TJC, SJC and ESR, were collected again at the end of approximate 3-month follow-up.
Clinical Response Assessment
The clinical response to MTX was evaluated based on multiple response criteria, including European League Against Rheumatism (EULAR) response criteria (RA patients who achieved good and moderate response were classified as responders and the remainders achieving poor response were classified as non-responders),18 DAS28-ESR low disease activity (LDA) (DAS28-ESR ≤ 3.2), change in DAS28-ESR (ΔDAS28-ESR) (the difference in DAS28-ESR from baseline to 3 months) and ΔDAS28-ESR > 0.6. When patients received additional DMARDs during the follow-up period, those patients were also classified as non-responders.
SNP Selection
Genetic polymorphisms within genes encoding transporters (SLC19A1, ABCB1, ABCC1~4 and ABCG2) were systematically selected based on literature report and genomic locations. In brief, the list of SNPs reported in the literature and the list of SNPs located in potentially functional regions, including exons, 5’-untranslated regions (5’-UTRs) and 3’-UTRs, with minor allele frequency (MAF) of more than 0.1 in Han Chinese, Beijing were merged. Finally, a total of 57 SNPs were chosen, and details about the selected SNPs are summarized in Table S1. Among these selected SNPs, three (SLC19A1 rs1051296, rs1051298 and rs7499) were not included for the final genotyping experiment due to technical issues; eventually, a total of 54 genetic polymorphisms were genotyped.
DNA Extraction and Genotyping
About 2 mL EDTA anti-coagulated venous blood sample was collected from each participant, and genomic DNA was extracted from peripheral blood lymphocytes using a full automatic nucleotide acid extraction instrument based on the standard procedures of the corresponding commercial DNA extraction kit (NP968-S system, Tianlong, China). The concentration and purity of DNA samples were determined utilizing a NanoDrop 2000 UV spectrophotometer. The genetic polymorphisms were detected by Shanghai Biowing Applied Biotechnology Co., Ltd. (www.biowing.com.cn) applying the method based on multiplex PCR with next-generation sequencing.19 The SNPs of seven randomly selected patients were genotyped twice, and the genotyping results are 100% concordant.
Statistical Analysis
Mean (standard deviation, SD) or median (interquartile range, IQR) was employed to describe quantitative variable based on its distribution, and the normality of the distribution was assessed by histograms and Q-Q plots. Qualitative variable was expressed as absolute number and percentage (%). For normally distributed data, two groups were compared using unpaired t test, while Mann–Whitney U-test was applied when data were non-normally distributed. Chi-square test or Fisher’s exact test was applied for categorical variables. The chi-square goodness-of-fit test was applied to test Hardy–Weinberg equilibrium (HWE). For patients who withdrew during the follow-up, the last observation carried forward (LOCF) method was applied Multivariable log binomial regression model was applied to estimate the relative risk (RR) and 95% confidence interval (CI) for the association between transporter gene polymorphism and clinical response to MTX assessed according to dichotomous classification criteria (EULAR response, DAS28-ESR LDA and REM, and ΔDAS28-ESR >0.6) with adjustment for potential confounding factors. Multivariable linear regression model was applied to estimate the beta (β) coefficient and 95% CI for the association between transporter gene polymorphism and ΔDAS28-ESR with adjustment for potential confounding factors. Haplotype analysis was performed for genetic polymorphisms within the same transporter gene shown to be significantly associated with clinical response to MTX based on the SNPStats software.20 Utilizing the most frequent haplotype as a reference, only relatively common haplotype (frequency greater than 5%) was examined for its association with clinical response to MTX with adjustment for potential confounders. All the above statistical analyses were performed using Stata version 15.0 for Windows (StataCorp, College Station, TX, USA), and a two-tailed P value less than 0.05 was considered statistically significant.
Results
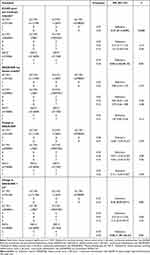
The baseline characteristics of RA patients included in our present study are shown in Table 1. In total, 100 RA patients aged 22~78 years (mean age of 52.23 ± 12.71 years) were enrolled, and the majority were female (81.00%) and DMARD-naïve (98.00%). A total of seven genetic polymorphisms were excluded from further data analysis due to the following reasons: 5 (SLC19A1 rs8199; ABCC1 rs129081, rs212090 and rs3784862; ABCC3 rs9895420) with genotyping call rate less than 95%, 1 with genotype distribution deviating from HWE (ABCG2 rs2231137) (Table S2), and one being triallelic SNP (ABCB1 rs2032582). Therefore, a total of 47 genetic polymorphisms within transporter genes were analyzed for their associations with clinical response to MTX.
 |
Table 1 Baseline Characteristics of Rheumatoid Arthritis Patients Included in the Present Study |
Associations of Genetic Polymorphisms Within Transporter Genes with EULAR Good and Moderate Response
When the baseline characteristics were compared between responders and non-responders classified according to whether achieving ELUAR good and moderate response, significant difference was found for the variables including smoking, drinking, TJC, ESR, DAS28-ESR and concomitant NSAIDs use (Table S3). Since TJC and ESR are components of DAS28-ESR, only DAS28-ESR in addition to smoking, drinking and concomitant NSAIDs use were adjusted in the multivariable analysis, and the detailed results could be found in Table S4. As shown in Table 2, after multivariable analysis, the major alleles of SLC19A1 rs12659 (G/G + A/G versus A/A, RR = 1.42, 95% CI = 1.02–1.97, P = 0.04), rs3788200 (G/G + A/G versus A/A, RR = 1.45, 95% CI = 1.04–2.01, P = 0.03) were significantly associated with EULAR good and moderate response under dominant models, and the major alleles of ABCC2 rs4148396 (C/C versus C/T + T/T, RR = 0.79, 95% CI = 0.65–0.96, P = 0.02), rs717620 (C/C versus C/T + T/T, RR = 0.77, 95% CI = 0.63–0.94, P = 0.01) were significantly associated with EULAR good and moderate response under recessive models. Moreover, a significant association of the major allele of ABCC2 rs3740066 with EULAR good and moderate response was found under both the dominant model (C/C + C/T versus T/T, RR = 0.88, 95% CI = 0.80–0.98, P = 0.02) and the recessive model (C/C versus C/T + T/T, RR = 0.80, 95% CI = 0.65–0.98, P = 0.03).
Associations of Genetic Polymorphisms Within Transporter Genes with DAS28-ESR LDA
When the baseline characteristics were compared between responders and non-responders classified according to whether achieving DAS28-ESR LDA, only ESR and DAS28-ESR were found to be significantly different (Table S5). Since ESR is one component of DAS28-ESR, only baseline DAS28-ESR was adjusted in the multivariable analysis, with the detailed results shown in Table S6. As displayed in Table 3, only the major alleles of ABCC2 rs3740066 (C/C versus C/T + T/T, RR = 0.67, 95% CI = 0.47–0.94, P = 0.02) and rs717620 (C/C versus C/T + T/T, RR = 0.66, 95% CI = 0.48–0.92, P = 0.02) were found to be significantly associated with DAS28-ESR LDA under recessive models after multivariable analysis.
Associations of Genetic Polymorphisms Within Transporter Genes with ΔDAS28-ESR
When the ΔDAS28-ESR was compared across different groups of baseline categorical variables and the correlation of ΔDAS28-ESR with baseline continuous variables was checked, significant evidence was found for the variables including TJC, SJC, ESR and DAS28-ESR (Table S7). As explained previously, only baseline DAS28-ESR was adjusted in the multivariable analysis, with the detailed results presented in Table S8. As shown in Table 4, RA patients carrying major alleles of ABCB1 rs1128503 (β = 0.89, 95% CI = 0.28–1.50, P = 0.005), ABCB1 rs4148737 (β = 0.90, 95% CI = 0.32–1.49, P = 0.003) and ABCC3 rs2277624 (β = 0.75, 95% CI = 0.04–1.46, P = 0.04) achieved more ΔDAS28-ESR compared with minor allele homozygotes, while the major allele homozygotes of ABCC3 rs4148416 (β = 0.52, 95% CI = 0.03–1.01, P = 0.04) achieved more ΔDAS28-ESR compared with minor allele carriers.
Associations of Genetic Polymorphisms Within Transporter Genes with ΔDAS28-ESR > 0.6
In a bivariate association analysis between baseline characteristics and clinical response to methotrexate assessed according to ΔDAS28-ESR > 0.6, significant evidence was found for the following variables: disease duration, drinking, TJC, ESR, DAS28-ESR and concomitant NSAIDs use (Table S9). As mentioned previously, DAS28-ESR in addition to disease duration, drinking and concomitant NSAIDs use were adjusted in the multivariable analysis, with the detailed results shown in Table S10. As shown in Table 5, 12 genetic polymorphisms within transporter genes were found to be significantly associated with ΔDAS28-ESR > 0.6. The major alleles of SLC19A1 rs12659 (G/G + A/G versus A/A, RR = 1.44, 95% CI = 1.07–1.96, P = 0.02), rs3788200 (G/G + A/G versus A/A, RR = 1.47, 95% CI = 1.08–1.99, P = 0.01) and ABCG2 rs4367138 (A/A + A/G versus G/G, RR = 0.74, 95% CI = 0.58–0.95, P = 0.02) were found to be significantly associated with ΔDAS28-ESR > 0.6 under dominant models, while the major alleles of SLC19A1 rs79091853 (C/C versus C/T + T/T, RR = 0.81, 95% CI = 0.68–0.95, P = 0.01), ABCB1 rs3842 (T/T versus C/T + C/C, RR = 1.23, 95% CI = 1.01–1.49, P = 0.04), ABCC1 rs3743527 (C/C versus C/T + T/T, RR = 1.22, 95% CI = 1.02–1.46, P = 0.03), ABCC2 rs3740066 (C/C versus C/T + T/T, RR = 0.81, 95% CI = 0.67–0.99, P = 0.04), rs4148396 (C/C versus C/T + T/T, RR = 0.81, 95% CI = 0.68–0.98, P = 0.03) and rs717620 (C/C versus C/T + T/T, RR = 0.79, 95% CI = 0.65–0.95, P = 0.01) were found to be significantly associated with ΔDAS28-ESR > 0.6 under recessive models. In addition, the major alleles of SLC19A1 rs1051266 (C/C + C/T versus T/T, RR = 1.45, 95% CI = 1.04–2.02, P = 0.03; C/C versus C/T + T/T, RR = 1.23, 95% CI = 1.02–1.48, P = 0.03), rs1131596 (A/A +A/G versus G/G, RR = 1.45, 95% CI = 1.04–2.02, P = 0.03; A/A versus A/G + G/G, RR = 1.23, 95% CI = 1.02–1.48, P = 0.03) and rs2838956 (A/A +A/G versus G/G, RR = 1.39, 95% CI = 1.01–1.90, P = 0.04; A/A versus A/G + G/G, RR = 1.23, 95% CI = 1.03–1.48, P = 0.02) were found to be significantly associated with ΔDAS28-ESR > 0.6 under either dominant or recessive models.
Associations of Haplotypes with Clinical Response to MTX
Based on the findings that all genetic polymorphisms within SLC19A1 and three genetic polymorphisms within ABCC2 (rs3740066, rs4148396 and rs717620) were significantly associated with clinical response to MTX, we also determined whether these genetic polymorphisms were in high linkage disequilibrium (LD) and then performed haplotype association analysis. As shown in Figure S1, two LD blocks were defined within SLC19A1, with rs1051266, rs1131596, rs12659 and rs3788200 constituting one block and the rest genetic polymorphisms (rs2838956, rs7867 and rs79091853) forming another block. Moreover, genetic polymorphisms within ABCC2 were found to be significantly associated with clinical response to MTX in high LD (Figure S2). With adjustment for corresponding confounders, TGAA haplotype consisting of SLC19A1 rs1051266, rs1131596, rs12659 and rs3788200 was found to be significantly associated with EULAR good and moderate response (OR = 0.37, 95% CI = 0.14–0.98, P = 0.048) and ΔDAS28-ESR > 0.6 (OR = 0.23, 95% CI = 0.07–0.71, P = 0.01) compared with the most common haplotype CAGG (Table 6). With regard to ABCC2 haplotype, the haplotype TTT composed of rs3740066, rs4148396 and rs717620 was found to be significantly associated with EULAR good and moderate response (OR = 8.83, 95% CI = 1.62–48.15, P = 0.01) and ΔDAS28-ESR > 0.6 (OR = 14.96, 95% CI = 1.79–125.24, P = 0.01) compared with the most common haplotype CCC (Table 6).
Discussion
In view of the significant effects of transporters on pharmacokinetics of MTX, this study was carried out to comprehensively evaluate the associations of genetic polymorphisms within transporter genes with clinical response to MTX based on multiple response criteria in Chinese RA patients. By adjusting for potential confounding factors, multiple significant signals were found in single marker analysis as well as haplotype analysis, indicating that genetic polymorphisms within transporter genes, especially SLC19A1 and ABCC2, should be incorporated into the future prediction model for clinical response to MTX in Chinese RA patients.
The SLC transporter superfamily consists of more than 300 membrane-bound proteins that mediate the translocation of substrates across biological membranes.21 Among them, SLC19A1, also known as reduced folate carrier 1 (RFC1), is mainly implicated in the uptake of MTX into target cells, and the roles of genetic polymorphisms within SLC19A1 in clinical response to MTX have been extensively examined with inconsistent results. A recent meta-analysis found that SLC19A1 rs105126622 and rs283895623 were significantly associated with MTX efficacy in RA patients. To date, the SLC19A1 rs1051266 is the most extensively studied variant, since the allele substitution of this SNP results in amino acid change and substantial impact on the red blood cell (RBC) MTX-PGs levels.24 While SLC19A1 rs2838956 is an intronic SNP, it has been postulated that this SNP might potentially influence RNA splicing and thereby affect the structure and function of SLC19A1.25 In our present study, the major alleles of SLC19A1 rs1051266 and rs2838956 were found to be significantly associated with ΔDAS28-ESR > 0.6 under either dominant model or recessive model, and borderline significant evidence was found for the association of the major allele of SLC19A1 rs1051266 with EULAR good and moderate response under dominant model (Table S4). Apart from the two aforementioned SNPs, significant evidence was also detected for two synonymous variants, namely rs12659 and rs79091853, one intronic variant, rs3788200, and one 5’-UTR variant, rs1131596. Specifically, RA patients carrying the major alleles of SLC19A1 rs12659 and rs3788200 were more likely to achieve EULAR good and moderate response and ΔDAS28-ESR > 0.6 compared with patients homozygous to the minor alleles, yet no studies on the relationships between these two SNPs and clinical response to MTX have been reported. It is worth mentioning that the genotype distribution of SLC19A1 rs12659 was found to be different between MTX-sensitive and MTX-resistant human cancer cell lines,26 possibly accounting for the observed association of SLC19A1 rs12659 with clinical response to MTX. Intriguingly, SLC19A1 rs1131596 was predicted to be located in the putative activator protein-1 (AP1) transcription factor recognition region, and G allele of this variant was found to be associated with reduced protein expression in contrast to A allele.27 Consistently, we found that RA patients carrying the major allele A allele of SLC19A1 rs1131596 were more likely to obtain ΔDAS28-ESR > 0.6. Additionally, the major allele A was associated with increased likelihood of achieving EULAR good and moderate response under the dominant model with borderline significance (Table S4). As for SLC19A1 rs3788200 and rs79091853, although the biological significance of these two variants remains unknown, the intronic SNP might be involved in alternative splicing, while the synonymous SNP might influence the secondary structure of the mRNA and further regulate protein expression. However, it is also possible that these observed associations might result from their strong LD with other variants possessing significant biological effects. In addition to the single marker analysis, the haplotype analysis was also carried out, and patients carrying the haplotype TGAA comprising rs1051266, rs1131596, rs12659 and rs3788200 were found to be more likely to achieve EULAR good and moderate response and ΔDAS28-ESR > 0.6 in contrast to the most common haplotype CAGG, suggesting that alleles in the form of combination might be more predictive. Collectively, these findings highlighted the impact of SLC19A1 genetic polymorphisms and haplotype on clinical response to MTX in Chinese RA patients.
As the largest transmembrane transporter family, the ABC transporter superfamily consists of 48 members classified into seven subfamilies, from ABC-A to G.28 These transporters are ubiquitously expressed in multiple tissues and organs such as small intestinal, liver and kidney, thus playing an essential role in absorption, distribution, metabolism and excretion of oral therapeutic drugs. Of note, the major ABC transporters mediating the efflux of MTX include ABCB1, ABCC1~4 and ABCG2,11,12 and the impact of genetic polymorphisms within genes encoding aforementioned ABC transporters on clinical response to MTX has also become a focus of increasing interest during the past decade. As for ABCB1, rs1045642 (C3435T) is the most widely investigated variant. Consistent with one recent meta-analysis showing that ABCB1 rs1045642 was associated with MTX efficacy in Caucasians but not in non-Caucasians,29 we did not find significant evidence for this SNP. Similarly, one recent study performed in Chinese patients with RA applying the same response criterion (ΔDAS28-ESR > 0.6) also did not find significant association between ABCB1 rs1045642 and clinical response to MTX.30 Apart from rs1045642, a few studies have assessed the effect of ABCB1 rs1128503 on MTX efficacy with non-significant results.11,31,32 Nevertheless, we found that RA patients carrying the major allele A of ABCB1 rs1128503 were more likely to achieve more DAS28-ESR reduction compared with minor allele homozygotes. The discrepancy might be due to difference in response criteria, duration of MTX therapy and consideration of confounding factors. Intriguingly, our finding was consistent with the results in patients with osteosarcoma, in which the major allele A of ABCB1 rs1128503 was significantly associated with better response to chemotherapy including MTX.33,34 As one synonymous variant in exon 13, the allele substitution of ABCB1 rs1128503 does not lead to an amino acid alteration but might exert effect on ABCB1 expression via codon usage.33 Moreover, we found that ABCB1 rs3842 was significantly associated with ΔDAS28-ESR > 0.6 and rs4148737 was significantly associated with ΔDAS28-ESR, although on studies on these associations have been reported. Of note, ABCB1 rs3842 was previously reported to be associated with active anti-retroviral treatment response in human immunodeficiency syndrome virus (HIV)/acquired immunodeficiency syndrome (AIDS) patients.35 As one 3’-UTR SNP, the allele substitution of ABCB1 rs3842 might disrupt or create microRNA (miRNA) binding site, therefore influencing protein expression. As for ABCB1 rs4148737, consistent with our finding that RA patients carrying the major allele A were more likely to achieve more reduction in DAS28-ESR, the major allele A of this variant was previously documented to be linked to a better treatment outcome in patients with osteosarcoma33 and Ewing Sarcoma.36 Although the biological function of ABCB1 rs4148737 remains unknown, as one intronic variant, it might lead to alternative splicing and the altered protein expression. Regarding ABCC1, only one study performed in Caucasian patients with RA reported that rs246240 and rs3784864 were significantly associated clinical response to MTX.11 However, we did not successfully replicate these findings, and this might be due to the difference in clinical characteristics, length of follow-up and genetic heterogeneity. Indeed, a total of 15 variants were selected including the two above-mentioned variants. While three variants were excluded due to genotyping call rate less than 95%, 12 variants were eligible for analysis. In our present study, one 3’-UTR variant, namely rs3743527, was detected to be associated with ΔDAS28-ESR > 0.6, and RA patients homozygous for the major allele C were more likely to achieve reduction in DAS28-ESR greater than 0.6 compared with minor allele carries. One previous study found that this SNP was significantly associated with chemotherapy‐related toxicities in Chinese acute myeloid leukemia (AML) patients.37 The possible underlying mechanism responsible for the observed association originated from ABCC1 rs3743527 might be the alteration of binding ability with miRNA due to its location in 3’-UTR. To date, the impact of genetic polymorphisms within ABCC2 on clinical response to MTX has only been evaluated in a few studies.11,31,38,39 One cross-sectional study performed in New Zealand did not find significant associations of ABCC2 rs2273697 and rs4148396 with DAS28 LDA in RA patients who had been treated by MTX for a median of 3 years before study entry.38 One retrospective study carried out in Japanese revealed that there was no significant difference in mean DAS28 after MTX monotherapy treatment among RA patients with different genotypes of rs2273697 and rs3740066.31 Besides, another retrospective cohort study performed in Portuguese did not detect significant evidence for associations of ABCC2 rs4148396 and rs717620 with clinical response to DAS28 LDA in RA patients treated by for 6 months.11 However, we found that three out of four selected variants within ABCC2, namely rs3740066, rs4148396 and rs717620, were significantly associated with clinical response to MTX. Factors possible accounting for the controversy include different study designs, durations, and regimens of MTX treatment and response criteria. Remarkably, one promoter variant in 5’-UTR, ABCC2 rs717620, has been recently elucidated to be significantly associated with clinical response to low-dose MTX in patients with psoriasis. In accordance with our finding here, patients carrying the minor allele T were more likely to be responders,39 and this might be explained by this allele associated with reduced activity and an increased bioavailability of methotrexate.40,41 As for ABCC2 rs3740066, one study conducted in Han Chinese epilepsy patients with generalized seizure found that patients carrying the major allele C of this variant were more likely to be resistant to valproic acid (VPA), the first-line antiepileptic drug used to control seizure in epilepsy patients.42 Moreover, advanced nonsmall cell lung cancer (NSCLC) patients homozygous for the minor allele T of this SNP were more likely to achieve higher response rates and progression-free survival.43 In agreement with these studies, we also found that patients carrying the minor allele T were more likely to achieve better response to MTX. Regarding ABCC2 rs4148396, the minor allele T was found to confer increased risk of developing intolerance to antiretroviral therapy in Brazilian HIV-1 positive individuals,44 possibly due to T allele leading to reduced expression or activity of ABCC2 and increased intracellular drug concentrations. Consistently, we found that patients carrying the minor allele T of this variant were more likely to obtain EULAR good and moderate response and ΔDAS28-ESR > 0.6. Of note, similar to SLC19A1, the haplotype TTT consisting of the minor allele of rs3740066, rs4148396 and rs717620 were found to be more likely to achieve EULAR good and moderate response and ΔDAS28-ESR > 0.6 in contrast to the most common haplotype CCC, suggesting that alleles in the form of combination might be more predictive of clinical response. With regard to ABCC3, we found that two synonymous variants, rs2277624 and rs4148416, were significantly associated with change in DAS28-ESR, yet no studies on their relationships have been reported. The minor allele T of ABCC3 rs2277624 was previously revealed to be associated improved survival of small-cell lung cancer (SCLC) patients with borderline significance.45 We found that the major alleles carriers of ABCC3 rs2277624 were more likely to achieve reduction in DAS28-ESR compared with minor allele homozygous. Consistent with previous studies demonstrating individuals homozygous for the major allele C of ABCC3 rs4148416 having improved survival rate and better response to chemotherapy (including MTX) of osteosarcoma patients,33,34,46 we found that patients with RA homozygous for the major allele were more likely to achieve reduction in DAS28-ESR. Although the biological significance of these two missense variants remains unknown, the allele substitutions might exert effect on ABCC3 expression via codon usage.33 To date, there has been no studies on the relationships between genetic polymorphisms within ABCC4 and clinical response to MTX. In our present study, a total of 14 variants with MAF greater than 0.1 located in potentially functional regions were systematically selected. However, non-significant evidence was found for any variant even though multiple response criteria were employed, suggesting that ABCC4 genetic polymorphisms might not contribute to inter-individual variability in MTX efficacy. Regarding ABCG2, to date, only two studies have examined the relationship between one missense SNP, namely rs2231142, and MTX efficacy with non-significant results.31,38 In accordance with these studies, we did not find significant evidence for this variant. Among the remaining three SNPs, only one intronic variant, rs4367138, was found to be associated with ΔDAS28-ESR > 0.6, and RA patients carrying the major allele A of this variant were less likely to achieve ΔDAS28-ESR > 0.6. Although this intronic variant has not reported in literature, we postulated that the allele substitution may influence alternative splicing and further regulate the expression of encoded protein.
The main strength of our study lies in the comprehensively selected SNPs and the multiple response criteria applied. Nonetheless, several potential limitations should be noted. First, the sample size of our present study was relatively small, thus statistical power was relatively limited to detect variants with weak effect sizes. However, we should also bear in mind that only genetic polymorphisms with relatively stronger effect sizes hold the potential to be applied to future clinical practice. Second, our findings for several polymorphisms showing significant evidence might have limited clinical implication presently. Moreover, most of these significant associations would become non-significant after multiple corrections, whereas this could be due to our relatively small sample size. If our results could be replicated in independent studies with larger numbers of RA patients, these genetic polymorphisms within transporter genes should be incorporated into the future prediction model of clinical response to MTX in Chinese RA patients, thereby guiding individualized treatment of MTX in daily clinical practice. Finally, the biological significance of several SNPs found to be associated with clinical response to MTX needs further clarification.
Conclusion
Our findings herein highlight the potential of genetic polymorphisms within transporter genes, especially SLC19A1 and ABCC2, as predictors of clinical response to MTX in Chinese RA patients.
Acknowledgments
This work was supported by Zhejiang Provincial Natural Science Foundation of China (Grant No. LY20H260001), Medical and Health Planned Science and Technology Project of Zhejiang Province (Grant No. 2019KY570), Nature Science Foundation of Ningbo city (Grant No. 202003N4112).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016;388(10055):2023–2038. doi:10.1016/S0140-6736(16)30173-8
2. Aletaha D, Smolen JS. Diagnosis and management of rheumatoid arthritis: a review. JAMA. 2018;320(13):1360–1372. doi:10.1001/jama.2018.13103
3. Fraenkel L, Bathon JM, England BR, et al. 2021 American college of rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Rheumatol. 2021;73(7):1108–1123. doi:10.1002/art.41752
4. Smolen JS, Landewé RBM, Bijlsma JWJ, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis. 2020;79(6):685–699. doi:10.1136/annrheumdis-2019-216655
5. Lau CS, Chia F, Dans L, et al. 2018 update of the APLAR recommendations for treatment of rheumatoid arthritis. Int J Rheum Dis. 2019;22(3):357–375. doi:10.1111/1756-185X.13513
6. Chatzidionysiou K, Sfikakis PP. Low rates of remission with methotrexate monotherapy in rheumatoid arthritis: review of randomised controlled trials could point towards a paradigm shift. RMD Open. 2019;5(2):e000993. doi:10.1136/rmdopen-2019-000993
7. Hazlewood GS, Barnabe C, Tomlinson G, et al. Methotrexate monotherapy and methotrexate combination therapy with traditional and biologic disease modifying antirheumatic drugs for rheumatoid arthritis: abridged Cochrane systematic review and network meta-analysis. BMJ. 2016;353:i1777. doi:10.1136/bmj.i1777
8. Krause C, Herborn G, Braun J, et al. Response to methotrexate predicts long-term mortality of patients with rheumatoid arthritis independent of the degree of response: results of a re-evaluation 30 years after baseline. Clin Exp Rheumatol. 2017;35(3):384–389.
9. Krause D, Gabriel B, Herborn G, et al. Response to methotrexate predicts long-term patient-related outcomes in rheumatoid arthritis. Clin Rheumatol. 2016;35:1123–1127.
10. Cronstein BN, Aune TM. Methotrexate and its mechanisms of action in inflammatory arthritis. Nat Rev Rheumatol. 2020;16(3):145–154. doi:10.1038/s41584-020-0373-9
11. Lima A, Bernardes M, Azevedo R, et al. Pharmacogenomics of methotrexate membrane transport pathway: Can clinical response to methotrexate in rheumatoid arthritis be predicted? Int J Mol Sci. 2015;16(12):13760–13780. doi:10.3390/ijms160613760
12. Mikkelsen TS, Thorn CF, Yang JJ, et al. PharmGKB summary: methotrexate pathway. Pharmacogenet Genomics. 2011;21(10):679–686. doi:10.1097/FPC.0b013e328343dd93
13. Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. Lancet. 2019;394(10197):521–532. doi:10.1016/S0140-6736(19)31276-0
14. Gao J, Wang C, Wei W. The effects of drug transporters on the efficacy of methotrexate in the treatment of rheumatoid arthritis. Life Sci. 2021;268:118907. doi:10.1016/j.lfs.2020.118907
15. Arnett FC, Edworthy SM, Bloch DA, et al. The American rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31(3):315–324. doi:10.1002/art.1780310302
16. Aletaha D, Neogi T, Silman AJ, et al. 2010 Rheumatoid arthritis classification criteria: an American college of rheumatology/European league against rheumatism collaborative initiative. Arthritis Rheum. 2010;62(9):2569–2581. doi:10.1002/art.27584
17. Prevoo ML, van ‘t Hof MA, Kuper HH, et al. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38(1):44–48. doi:10.1002/art.1780380107
18. van Gestel AM, Prevoo ML, van ‘t Hof MA, et al. Development and validation of the European league against rheumatism response criteria for rheumatoid arthritis. Comparison with the preliminary American college of rheumatology and the World Health Organization/International league against rheumatism criteria. Arthritis Rheum. 1996;39(1):34–40. doi:10.1002/art.1780390105
19. Chen K, Zhou YX, Li K, et al. A novel three-round multiplex PCR for SNP genotyping with next generation sequencing. Anal Bioanal Chem. 2016;408(16):4371–4377. doi:10.1007/s00216-016-9536-6
20. Solé X, Guinó E, Valls J, et al. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi:10.1093/bioinformatics/btl268
21. Lin L, Yee SW, Kim RB, et al. SLC transporters as therapeutic targets: emerging opportunities. Nat Rev Drug Discov. 2015;14(8):543–560. doi:10.1038/nrd4626
22. Naushad SM, Alrokayan SA, Almajhdi FN, et al. Influence of RFC1 c.80A>G polymorphism on methotrexate-mediated toxicity and therapeutic efficacy in rheumatoid arthritis: a meta-analysis. Ann Pharmacother. 2021;55(12):1429–1438. doi:10.1177/10600280211002053
23. Qiu Q, Huang J, Shu X, et al. Polymorphisms and pharmacogenomics for the clinical efficacy of methotrexate in patients with rheumatoid arthritis: a systematic review and meta-analysis. Sci Rep. 2017;7(1):44015. doi:10.1038/srep44015
24. Dervieux T, Kremer J, Lein DO, et al. Contribution of common polymorphisms in reduced folate carrier and gamma-glutamylhydrolase to methotrexate polyglutamate levels in patients with rheumatoid arthritis. Pharmacogenetics. 2004;14(11):733–739. doi:10.1097/00008571-200411000-00004
25. Lima A, Bernardes M, Azevedo R, et al. SLC19A1, SLC46A1 and SLCO1B1 polymorphisms as predictors of methotrexate-related toxicity in Portuguese rheumatoid arthritis patients. Toxicol Sci. 2014;142(1):196–209. doi:10.1093/toxsci/kfu162
26. Yoon SA, Choi JR, Kim JO, et al. Influence of reduced folate carrier and dihydrofolate reductase genes on methotrexate-induced cytotoxicity. Cancer Res Treat. 2010;42(3):163–171. doi:10.4143/crt.2010.42.3.163
27. Chatzikyriakidou A, Georgiou I, Voulgari PV, et al. Transcription regulatory polymorphism −43T>C in the 5′-flanking region of SLC19A1 gene could affect rheumatoid arthritis patient response to methotrexate therapy. Rheumatol Int. 2007;27(11):1057–1061. doi:10.1007/s00296-007-0339-0
28. Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11(7):1156–1166. doi:10.1101/gr.184901
29. Chen Y, Zou K, Sun J, et al. Are gene polymorphisms related to treatment outcomes of methotrexate in patients with rheumatoid arthritis? A systematic review and meta-analysis. Pharmacogenomics. 2017;18(2):175–195. doi:10.2217/pgs-2016-0158
30. Lv S, Fan H, Yang H, et al. Membrane-spanning protein genetic polymorphisms related to methotrexate therapeutic outcomes in a Chinese rheumatoid arthritis population. J Clin Pharmacol. 2019;59(11):1471–1476. doi:10.1002/jcph.1446
31. Kato T, Hamada A, Mori S, et al. Genetic polymorphisms in metabolic and cellular transport pathway of methotrexate impact clinical outcome of methotrexate monotherapy in Japanese patients with rheumatoid arthritis. Drug Metab Pharmacokinet. 2012;27(2):192–199. doi:10.2133/dmpk.DMPK-11-RG-066
32. Sandhu A, Ahmad S, Kaur J, et al. Do SNPs in folate pharmacokinetic pathway alter levels of intracellular methotrexate polyglutamates and affect response? A prospective study in Indian patients. Clin Rheumatol. 2018;37(12):3221–3228. doi:10.1007/s10067-018-4206-z
33. Caronia D, Patiño-Garcia A, Peréz-Martínez A, et al. Effect of ABCB1 and ABCC3 polymorphisms on osteosarcoma survival after chemotherapy: a pharmacogenetic study. PLoS One. 2011;6(10):e26091. doi:10.1371/journal.pone.0026091
34. Yang J, Wang ZG, Cai HQ, et al. Effect of variation of ABCB1 and ABCC3 genotypes on the survival of bone tumor cases after chemotherapy. Asian Pac J Cancer Prev. 2013;14(8):4595–4598. doi:10.7314/APJCP.2013.14.8.4595
35. Wart M, Ren Y, Smith P, et al. ABCB1 4036A>G and 1236C>T polymorphisms affect plasma efavirenz levels in South African HIV/AIDS patients. Front Genet. 2012;3:236. doi:10.3389/fgene.2012.00236
36. Ruiz-Pinto S, Pita G, Patiño-García A, et al. Identification of genetic variants in pharmacokinetic genes associated with Ewing Sarcoma treatment outcome. Ann Oncol. 2016;27(9):1788–1793. doi:10.1093/annonc/mdw234
37. Cao HX, Miao CF, Yan L, et al. Polymorphisms at microRNA binding sites of Ara-C and anthracyclines-metabolic pathway genes are associated with outcome of acute myeloid leukemia patients. J Transl Med. 2017;15(1):235. doi:10.1186/s12967-017-1339-9
38. Stamp LK, Chapman PT, O’Donnell JL, et al. Polymorphisms within the folate pathway predict folate concentrations but are not associated with disease activity in rheumatoid arthritis patients on methotrexate. Pharmacogenet Genomics. 2010;20(6):367–376. doi:10.1097/FPC.0b013e3283398a71
39. Grželj J, Mlinarič-Raščan I, Marko PB, et al. Polymorphisms in GNMT and DNMT3b are associated with methotrexate treatment outcome in plaque psoriasis. Biomed Pharmacother. 2021;138:111456. doi:10.1016/j.biopha.2021.111456
40. Haenisch S, Zimmermann U, Dazert E, et al. Influence of polymorphisms of ABCB1 and ABCC2 on mRNA and protein expression in normal and cancerous kidney cortex. Pharmacogenomics J. 2007;7(1):56–65. doi:10.1038/sj.tpj.6500403
41. Rau T, Erney B, Göres R, et al. High-dose methotrexate in pediatric acute lymphoblastic leukemia: impact of ABCC2 polymorphisms on plasma concentrations. Clin Pharmacol Ther. 2006;80(5):468–476. doi:10.1016/j.clpt.2006.08.012
42. Chen J, Su Q, Qin J, et al. Correlation of MCT1 and ABCC2 gene polymorphisms with valproic acid resistance in patients with epilepsy on valproic acid monotherapy. Drug Metab Pharmacokinet. 2019;34(3):165–171. doi:10.1016/j.dmpk.2018.01.006
43. Han JY, Lim HS, Yoo YK, et al. Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer. 2007;110(1):138–147. doi:10.1002/cncr.22760
44. Arruda MB, Campagnari F, de Almeida TB, et al. Single nucleotide polymorphisms in cellular drug transporters are associated with intolerance to antiretroviral therapy in Brazilian HIV-1 positive individuals. PLoS One. 2016;11(9):e0163170. doi:10.1371/journal.pone.0163170
45. Sun Z, Chen J, Aakre J, et al. Genetic variation in glutathione metabolism and DNA repair genes predicts survival of small-cell lung cancer patients. Ann Oncol. 2010;21(10):2011–2016. doi:10.1093/annonc/mdq212
46. Liu S, Yi Z, Ling M, et al. Predictive potential of ABCB1, ABCC3, and GSTP1 gene polymorphisms on osteosarcoma survival after chemotherapy. Tumour Biol. 2014;35:9897–9904.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.