Back to Journals » Cancer Management and Research » Volume 10
Association of two FOXP3 polymorphisms with breast cancer susceptibility in Chinese Han women
Authors Tian T, Wang M, Zheng Y , Yang T, Zhu W, Li H, Lin S , Liu K, Xu P , Deng Y , Zhou L , Dai Z
Received 29 November 2017
Accepted for publication 13 February 2018
Published 26 April 2018 Volume 2018:10 Pages 867—872
DOI https://doi.org/10.2147/CMAR.S158433
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
Tian Tian,1,* Meng Wang,1,* Yi Zheng,1,* Tielin Yang,2 Wenge Zhu,3 Hongtao Li,4 Shuai Lin,1 Kang Liu,1 Peng Xu,1 Yujiao Deng,1 Linghui Zhou,1 Zhijun Dai1
1Department of Oncology, The Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, People’s Republic of China; 2School of Life Science and Technology, Xi’an Jiaotong University, Xi’an, People’s Republic of China; 3Department of Biochemistry and Molecular Medicine, The George Washington University Medical School, Washington, DC, USA; 4Department of Breast Head and Neck Surgery, Affiliated Tumor Hospital of Xinjiang Medical University, Urumchi, People’s Republic of China
*These authors contributed equally to this work
Background: Forkhead box P3 (FOXP3) is a key gene in the immune system which also plays a role in tumor development. This study aims to explore the association of two FOXP3 polymorphisms (rs3761548 and rs3761549) with susceptibility to breast cancer (BC).
Method: A case–control study was conducted, involving 560 patients and 583 healthy individuals from the Chinese Han population. The genotypes of FOXP3 polymorphisms were detected using the Sequenom MassARRAY method. The association between FOXP3 polymorphisms and BC risk was evaluated using a χ2 test with an odds ratio (OR) and 95% confidence intervals (95% CIs) under six genetic models. False-positive report probability was utilized to examine whether the significant findings were noteworthy.
Results: We observed that rs3761548 was associated with a higher BC risk in heterozygous, dominant, overdominant, and allele genetic models (CA vs CC: OR =1.32, P=0.031; CA/AA vs CC: OR =1.32, P=0.023; CA vs CC/AA: OR =1.29, P=0.042; A vs C: OR =1.26, P=0.029), whereas no significant association was found between rs3761549 and BC risk. In addition, CA, CA/AA genotype, and A allele of rs3761548 were related to larger tumor size, and the A allele was also correlated with a positive status of Her-2 in BC patients.
Conclusion: Our study suggests that FOXP3 polymorphism rs3761548 is associated with BC susceptibility in the Chinese and may be involved in tumor progression. Future studies are needed to confirm the results in a larger population with more races.
Keywords: forkhead box P3, polymorphism, breast cancer, risk
Introduction
Breast cancer (BC) is the most common type of cancer and the leading cause of cancer death among females globally.1 Hereditary factors are considered to play a key role in the genesis of breast cancer. Among all breast carcinomas, about 5%–10% are inherited, arising from genetic variants in susceptible genes.2,3 Single-nucleotide polymorphisms (SNPs) are the most frequent variation that occur in a single nucleotide at a specific position in the genome. Numerous SNPs have been identified through sequencing, and many of them in critical genes such as BRCA1/2, TP53, TNF, and VEGF were demonstrated to be associated with cancer susceptibility.4–6
FOXP3 is a protein-coding gene located at human chromosome Xp11.23. Its product, forkhead box P3 (FoxP3), also known as scurfin, is specifically expressed in CD4+CD25+ regulatory T cells (Tregs) and functions mainly as a key regulator for the development and function of Tregs.7,8 As a member of the FOX family, FoxP3 also regulates transcription and DNA repair and is involved in cell growth and differentiation as well as embryogenesis.9,10 In addition to its critical function in immune response, FOXP3 plays an important role in cancer development, although it is still a controversy whether it is an oncogene or tumor suppressor gene.11,12
Nevertheless, several FOXP3 SNPs have been reported to be associated with susceptibility to multiple cancers including lung cancer, hepatocellular carcinoma (HCC), and colorectal cancer.13–15 Among these SNPs, rs3761548 and rs3761549 were the most common polymorphisms. Studies revealed that A allele and AA/AC genotype of rs3761548 significantly increased the risk of thyroid cancer, colorectal cancer, and non-small-cell lung cancer.13,15,16 T allele of rs3761549 was found to be associated with susceptibility to lung cancer, while C allele was related with higher risk of HCC. In addition, TT/CT genotype was linked to an increased incidence of tumor recurrence in HCC.17,18 Here, we conducted a case–control study to explore the association of the two polymorphisms with BC risk in a Chinese Han population.
Methods
Study subjects
Cases were selected from female BC patients treated at the Department of Oncology, the Second Affiliated Hospital of Xi’an Jiaotong University (Xi’an, People’s Republic of China). Healthy women who had a checkup at the hospital during the same period of time were recruited as controls. The criteria for enrollment were the same as our previous study.19 Cases were confirmed by pathology, and controls were matched according to age and menopausal status. None of the cases had received chemotherapy or radiotherapy before surgery. Patients with other types of cancer were excluded. Finally, 560 patients and 583 healthy individuals were enrolled in our study. All of the subjects were from the Chinese Han population, had a mean age of 49 years, and were not related to each other.
Ethics statement
This study was approved by the Ethics Committee of Xi’an Jiaotong University. The purpose of this study was well informed to the subjects and written informed consent was obtained from each of them. Then, personal and clinical information of the subjects was collected from their medical records.
Genotyping assay
Peripheral blood of each subject was collected in tubes containing EDTA and stored at −80°C. Then, DNA was extracted from whole blood using the Universal Genomic DNA Extraction Kit (TaKaRa, Kyoto, Japan) according to the manufacturer’s protocol, and the quantity of extracted DNA was assessed utilizing the UV/Vis spectrophotometer (DU530, Beckman Instruments, Brea, CA, USA). Two tag-SNPs (rs3761548 and rs3761549) were selected from the HapMap database, with minor allele frequency (MAF) >0.01 in Chinese Han population. Sequenom MassARRAY Assay Design Software (version 3.0, Agena Bioscience, San Diego, CA, USA) was used to design multiplexed SNP MassEXTEND assay. And SequenomMassARRAY RS1000 was used to detect SNP genotyping.20 Primers of the two SNPs are listed in Table 1. Data were analyzed with Sequenom Typer Software (version 4.0, Sequenom, San Diego, CA, USA).
  | Table 1 Primers used in this study Abbreviations: SNP, single-nucleotide polymorphism; PCRP, primer for polymerase chain reaction; UEP_SEQ, primer for single nucleotide extension. |
Statistical analysis
SPSS software (version 22.0, IBM Corporation, Armonk, NY, USA) was used for statistical analyses of data. The differences of basic parameters between the two groups were examined with Student t-test or χ2 test. For each SNP, Hardy–Weinberg equilibrium as well as the differences in allele frequencies between patients and healthy controls were examined by χ2 test. The association of SNPs with BC risk was evaluated by odds ratios (ORs) with 95% confidence intervals (CIs) for five different genetic models (“A” indicates the major allele and “a” indicates the minor allele): codominant model including homozygote (aa vs AA) and heterozygote (Aa vs AA), dominant model (Aa/aa vs AA), recessive model (aa vs AA/Aa), overdominant model (Aa vs AA/aa), and allele model (a vs A). P-values were adjusted by logistic regression analysis. All the tests were two-tailed, and P-value <0.05 was considered statistically significant. We further calculated false-positive report probability (FPRP) to examine whether the significant findings were noteworthy. The prior probability of 0.1 was set in our study, and 0.5 was determined as a cut-off value for FPRP.21,22
Results
The basic characteristics of study subjects
No significant difference was observed between the two groups in regards to age, menopausal status, and procreative times. However, the mean value of body mass index (BMI) in the patient group was significantly lower compared with that in control group (P=0.038). As obesity is also associated with BC risk,23 BMI may be a confounding factor for the analysis. Therefore, the results were adjusted for BMI. The basic parameters of cases and controls are presented in Table 2.
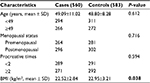  | Table 2 The comparison of basic characteristics between cases and controls Note: P-value <0.05 was shown in bold Abbreviation: BMI, body mass index. |
Association of FOXP3 SNPs with BC susceptibility
For both of the FOXP3 polymorphisms, the genotype distribution in controls complied with Hardy–Weinberg equilibrium (P=0.895 and 0.934 respectively). In the overall analysis, FOXP3-rs3761548 was associated with a higher BC risk in heterozygous, dominant, overdominant, and allele genetic models (CA vs CC: OR =1.32, 95% CI =1.03–1.69, P=0.031; CA/AA vs CC: OR =1.32, 95% CI =1.04–1.69, P=0.023; CA vs CC/AA: OR =1.29, 95% CI =1.01–1.66, P=0.042; A vs C: OR =1.26, 95% CI =1.02–1.54, P=0.029). FPRP analysis showed that the associations were noteworthy. However, FOXP3-rs3761549 was not related to BC susceptibility (Table 3). We then carried out stratified analysis based on age and menopausal status in five genetic models. However, no relationship was observed between the two SNPs and BC susceptibility in either of the subgroups (Table S1).
Relationship between FOXP3 SNPs and clinical features of BC
We also explored the relationship between the two FOXP3 SNPs and BC clinical features, including tumor size, metastasis of lymph node, status of hormone receptor (estrogen receptor and progesterone receptor) as well as Her-2, and the expression level of Ki-67. We observed that CA, CA/AA genotype, and A allele of rs3761548 were related with a larger size of tumor (CA vs CC: OR =1.47, 95% CI =1.01–2.15; CA/AA vs CC: OR =1.54, 95% CI =1.07–2.23; A vs C: OR =1.48, 95% CI =1.08–2.03). In addition, the A allele was correlated with a positive status of Her-2 (A vs C: OR =1.36, 95% CI =1.01–1.85) (Table 4). As for rs3761549, there was still no significant association with BC clinicopathological features existing in any of the genetic models (Table S2).
Discussion
FOXP3 is a key gene in the immune system. It encodes a protein for a transcriptional regulator which belongs to the forkhead/winged-helix family. Defects in this gene can cause immunodeficiency syndrome.8,24 But the role of FOXP3 in tumorigenesis has long been controversial. FOXP3 was found to represses some oncogenes such as MYC, HER2, and SATB1. Its expression was downregulated in several tumors including breast, prostate, and ovarian tumors.11,12,25 However, it was reported that most human tumors were infiltrated by Tregs with high FoxP3 labeling, and an excess of Treg activity can prevent the immune system from destroying cancer cells.26,27
No matter what the exact role of this gene in tumor is, the genetic variation of FOXP3 can indeed affect cancer susceptibility.9,26 The two common polymorphisms rs3761549 (C>T) and rs3761548 (C>A) were located in the promoter region of the FOXP3 gene, which is considered to affect FoxP3 production and activity.14 They were investigated in several tumors previously and were demonstrated to be associated with cancer susceptibility.13,15–18 However, the conclusions of the relationship between these two SNPs and BC susceptibility were controversial. Lopes et al28 observed that the homozygote of rs3761548 was associated with triple-negative breast cancer risk in Brazilians (OR =3.78; 95% CI =1.02–14.06). Jahan et al29 evaluated both rs3761548 and rs3761549 in an Indian population but failed to find any correlation with BC risk. However, the AA genotype of rs3761548 was significantly associated with advanced tumor stage (T3-T4) (OR =3.90; p=0.03),29 whereas Zheng et al’s30 and Raskin et al’s31 study reported that neither rs3761548 nor rs3761549 were associated with BC. Among all these researchers, only Zheng et al30 studied the Chinese population. However, their study evaluated just two genetic models (homozygotes and heterozygotes) and did not explore the relationship of these two SNPs with clinical parameters of BC.
Our study focused on a Chinese Han population and found that the CA, CA/AA genotype, and the A allele of rs3761548 can increase the risk of BC. Moreover, the A allele of rs3761548 was related with a larger size of tumor (≥2 cm). It was also correlated with a Her-2 positive status, indicating that patients with A allele of rs3761548 are more likely to have overexpression of HER2. It means that the polymorphism of this locus may be a potential biomarker for tumor subtype classification and help to guide treatment. The results were in accord with some of previous studies, suggesting that the A allele of rs3761548 is a risk factor for BC and may be involved in tumor progression. The mechanism of action is not clear, which is a potential subject for future studies. Maybe the critical location of this SNP could affect the production and activity of FoxP3, thus influencing tumorigenesis though FoxP3’s function.
Some limitations could not be ignored in the study. First, selection bias is inevitable as this is a hospital-based, single-center study. Second, our sample size was insufficient to support stratified analysis for tumor subtypes. Finally, we did not analyze the impact of other risk factors such as lifestyle, radiation exposure to the chest, family history, and other benign breast lesions because of lacking relative data. Hence, population-based studies are required in the future to improve the accuracy of evaluation and to explore gene–environment interactions as well.
Conclusion
To sum up, the present study suggests that the FOXP3 polymorphism rs3761548 is associated with BC susceptibility in the Chinese and may be involved in tumor progression. Future studies in a large population with more races are needed to confirm the results. There is also a need to explore gene–environment interactions and possible mechanisms of action for this SNP.
Acknowledgment
This study was supported by National Natural Science Foundation, People’s Republic of China (No. 81471670); the Key research and development plan, Shaanxi Province, People’s Republic of China (2017ZDXM-SF-066); and Science and Technology Branch Project of Xinjiang Uygur Autonomous Region, People’s Republic of China (2017E0262). The sponsors had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Tan DS, Marchio C, Reis-Filho JS. Hereditary breast cancer: from molecular pathology to tailored therapies. J Clin Pathol. 2008;61(10):1073–1082. | ||
Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M. Genetic susceptibility to breast cancer. Mol Oncol. 2010;4(3):174–191. | ||
Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409(6822):928–933. | ||
Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9(2):95–107. | ||
Loktionov A. Common gene polymorphisms, cancer progression and prognosis. Cancer Lett. 2004;208(1):1–33. | ||
Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–1061. | ||
Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. | ||
Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328(2):198–206. | ||
Brunkow ME, Jeffery EW, Hjerrild KA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. | ||
Triulzi T, Tagliabue E, Balsari A, Casalini P. FOXP3 expression in tumor cells and implications for cancer progression. J Cell Physiol. 2013;228(1):30–35. | ||
Douglass S, Ali S, Meeson AP, Browell D, Kirby JA. The role of FOXP3 in the development and metastatic spread of breast cancer. Cancer Metastasis Rev. 2012;31(3–4):843–854. | ||
He YQ, Bo Q, Yong W, Qiu ZX, Li YL, Li WM. FoxP3 genetic variants and risk of non-small cell lung cancer in the Chinese Han population. Gene. 2013;531(2):422–425. | ||
Jiang LL, Ruan LW. Association between FOXP3 promoter polymorphisms and cancer risk: a meta-analysis. Oncol Lett. 2014;8(6):2795–2799. | ||
Chen L, Yu Q, Liu B, Zhu L. Association of FoxP3 rs3761548 polymorphism with susceptibility to colorectal cancer in the Chinese population. Med Oncol. 2014;31(12):374. | ||
Jiang W, Zheng L, Xu L, et al. Association between FOXP3 gene polymorphisms and risk of differentiated thyroid cancer in Chinese Han population. J Clin Lab Anal. 2017;31(5). | ||
Chen Y, Zhang H, Liao W, et al. FOXP3 gene polymorphism is associated with hepatitis B-related hepatocellular carcinoma in China. J Exp Clin Cancer Res. 2013;32:39. | ||
Fazelzadeh Haghighi M, Ali Ghayumi M, Behzadnia F, Erfani N. Investigation of FOXP3 genetic variations at positions -2383 C/T and IVS9+459 T/C in southern Iranian patients with lung carcinoma. Iran J Basic Med Sci. 2015;18(5):465–471. | ||
Dai ZJ, Liu XH, Kang HF, et al. Genetic variation in metastasis-associated in colon cancer-1 and the risk of breast cancer among the Chinese Han population: a STROBE-compliant observational study. Medicine (Baltimore). 2016;95(6):e2801. | ||
Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;Unit 2.12. | ||
Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434–442. | ||
He J, Wang F, Zhu J, et al. Association of potentially functional variants in the XPG gene with neuroblastoma risk in a Chinese population. J Cell Mol Med. 2016;20(8):1481–1490. | ||
Picon-Ruiz M, Morata-Tarifa C, Valle-Goffin JJ, Friedman ER, Slingerland JM. Obesity and adverse breast cancer risk and outcome: mechanistic insights and strategies for intervention. CA Cancer J Clin. 2017;67(5):378–397. | ||
Bennett CL, Christie J, Ramsdell F, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27(1):20–21. | ||
Tan B, Anaka M, Deb S, et al. FOXP3 over-expression inhibits melanoma tumorigenesis via effects on proliferation and apoptosis. Oncotarget. 2014;5(1):264–276. | ||
Martin F, Ladoire S, Mignot G, Apetoh L, Ghiringhelli F. Human FOXP3 and cancer. Oncogene. 2010;29(29):4121–4129. | ||
Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. | ||
Lopes LF, Guembarovski RL, Guembarovski AL, et al. FOXP3 transcription factor: a candidate marker for susceptibility and prognosis in triple negative breast cancer. Biomed Res Int. 2014;2014:341654. | ||
Jahan P, Ramachander VR, Maruthi G, Nalini S, Latha KP, Murthy TS. Foxp3 promoter polymorphism (rs3761548) in breast cancer progression: a study from India. Tumour Biol. 2014;35(4):3785–3791. | ||
Zheng J, Deng J, Jiang L, et al. Heterozygous genetic variations of FOXP3 in Xp11.23 elevate breast cancer risk in Chinese population via skewed X-chromosome inactivation. Hum Mutat. 2013;34(4):619–628. | ||
Raskin L, Rennert G, Gruber SB. FOXP3 germline polymorphisms are not associated with risk of breast cancer. Cancer Genet Cytogenet. 2009;190(1):40–42. |
Supplementary materials
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




