Back to Journals » Infection and Drug Resistance » Volume 10
Association between the IL-6 gene polymorphism and tuberculosis risk: a meta-analysis
Authors Wang H, Pang C, Zeng N, Wan C, Shen Y, Wen F
Received 19 June 2017
Accepted for publication 6 October 2017
Published 27 November 2017 Volume 2017:10 Pages 445—454
DOI https://doi.org/10.2147/IDR.S144296
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Hao Wang,1,* Caishuang Pang,2,* Ni Zeng,1 Chun Wan,1 Yongchun Shen,1 Fuqiang Wen1
1Department of Respiratory and Critical Care Medicine, West China Hospital of Sichuan University, and Division of Pulmonary Diseases, State Key Laboratory of Biotherapy of China, Chengdu, China; 2Chongqing Cancer Institute & Hospital & Cancer Center, Chongqing, China
*These authors contributed equally to this work
Background: The gene polymorphism of interleukin-6 (IL-6) has been shown to be implicated in tuberculosis susceptibility in many studies, but with conflicting results. This study aimed to provide more accurate estimation of the relationship between IL-6 gene polymorphism and tuberculosis risk through a meta-analysis
Method: A literature search was performed in PubMed, EMBASE, and other databases. Data were retrieved, and pooled odds ratio (OR) with 95% CI were calculated. Statistical analyses were performed by using STATA 12.0.
Results: Twelve publications with 2635 cases and 3049 controls were included. The pooled analysis demonstrated significant evidence of association between IL-6 (-174G/C) and low risk of tuberculosis in dominant model (CC+GC vs GG: OR =0.693, 95% CI 0.581–0.826, p<0.001). Subgroup analysis got similar results for IL-6 (-174G/C) in Asians and Latinos, but the significance did not exist in Caucasians. IL-6 (-572C/G) polymorphism was also associated with low risk of tuberculosis in dominant model (CC+GC vs GG: OR =0.719, 95% CI 0.577–0.896, p=0.003). No publication bias was detected in either of the polymorphisms.
Conclusion: In summary, IL-6 -572 C/G polymorphism may be associated with a decreased risk of tuberculosis, and C allele is the protective factor against tuberculosis for IL-6 -174G/C among Asians and Latinos, but not in Caucasian population.
Keywords: interleukin-6, rs1800795, rs1800796, TB
Introduction
Tuberculosis remains the main cause of mortality and morbidity worldwide, especially in developing countries.1,2 The precise etiology and pathogenesis of tuberculosis is still inconclusive. However, among those infected with Mycobacterium tuberculosis, only 10% develop active tuberculosis, and most of infected individuals remain latently infected, implying that each stage of the host response to M. tuberculosis may be under genetic control.3 Growing number of reports have demonstrated that various host genetic factors are critical in susceptibility to tuberculosis.4,5
Cytokines play a significant role in host susceptibility and development of tuberculosis.6 For instance, the polymorphism of interleukin (IL)-10 and interferon (IFN)-γ were reported to be associated with pleural tuberculosis.7 Similarly, Hussain et al reported that suppressed T-cell-derived IFN-γ but greater monocyte-derived IL-6 and IL-10 production might be associated with pulmonary tuberculosis susceptibility.8 Lopez-Maderuelo et al found that IFN-γ (+874) A allele significantly increased the risk of tuberculosis.9 IL-6 is a pro-inflammatory cytokine secreted by monocytes, endothelial cells and fibroblasts, which is able to stimulate B and T lymphocytes, and regulate immune response to tuberculosis.10 It is found to be essential for generating protective Th1 immune responses which is necessary for the resistance to M. tuberculosis infection.11,12 Previously, several studies have shown that the IL-6 polymorphism is significantly associated with a number of diseases, such as sepsis, coronary heart disease, and systemic lupus erythematosus, in which it plays an important role in the etiology.13–15 With regard to tuberculosis, two positions of IL-6 gene polymorphism, -174G/C (rs1800795) and -572C/G (rs1800796), were reported to be related to tuberculosis risk in different populations.16–18 However, the results of these studies were variable and inconclusive. Since meta-analysis is a powerful tool for analyzing cumulative data from individual studies, the sample sizes of which are small and the statistical power low,19,20 two recently published meta-analysis reviews investigated the relationship between -174G/C polymorphism and risk of tuberculosis based on the results of studies performed before 2012.21,22 However, 5 years have passed, thus a meta-analysis based on the most current published independent studies was performed in the current review, which may provide a comprehensive assessment of the associations of both IL-6 (-174G/C) and IL-6 (-572C/G) gene polymorphisms with the risk of tuberculosis.
Materials and methods
Search strategy
Databases, including PubMed, EMBASE, Wanfang, VIP, and CNKI, were systematically searched for publications regarding the association between the IL-6 gene polymorphism and tuberculosis risk up to October 2016. The search terms were set as follows: (“Interleukin-6” or “IL-6” or “rs1800795” or “rs1800796”) and (“polymorphism” or “variant” or “mutation” or “SNP”) and “tuberculosis.” Articles were also identified using the related articles function in PubMed. References within the identified articles or review articles were also searched manually.
Quality assessment
Quality of the included studies was evaluated with a standard method recommended by Thakkinstian et al.23 The assessment was carried out according to the following variables: representativeness of cases and controls, credibility of controls, specimens of cases determining genotypes, Hardy–Weinberg equilibrium (HWE) in controls, and total sample size. A quality score was ultimately obtained by summing each component giving a range from 0 (the lowest) to 15 (the highest), the higher the publication scored, the better quality it is, and vice versa, and studies with scores ≥10 were classified as high-quality studies, whereas studies with scores <10 were considered as low-quality studies.
Inclusion and exclusion criteria
Articles were included in this meta-analysis if they met the following criteria: 1) they were case–control clinical studies published in English or Chinese; 2) they evaluated the IL-6 gene polymorphism and tuberculosis risk; 3) they comprised sufficient data (genotype frequency for cases and controls) to estimate an odds ratio (OR) with its 95% confidence interval (95% CI). The exclusion criteria included the following: 1) they were abstracts and reviews; 2) there were studies without sufficient data reported. When publications involved the same or overlapping data sets, only the study with the largest number of participants was included.
Data extraction
Data of all eligible publications were extracted according to the inclusion and exclusion criteria by two independent investigators (HW and CP). A consensus was reached in case of disagreement. The following characteristics were collected from each study: first author, year of publication, study design, country, ethnicity, sample size, alleles, and IL-6 genotype distributions in both cases and controls. If necessary information could not be obtained from the original article, we contacted the corresponding author via the email listed.
Statistical analysis
With the purpose of assessing the associations between the IL-6 gene polymorphisms and tuberculosis risk, pooled OR with corresponding 95% CI was calculated based on allele contrast (C vs G), co-dominant (CC vs GG), dominant (CC+GC vs GG), and recessive (CC vs GC+GG) models. A fixed-effects or random-effects model was used for pooling data according to the results of heterogeneity examination, which was checked by a χ2-based Q statistic test. When p<0.05, OR was calculated by the random-effects model, otherwise the fixed-effects model was applied. The significance of the pooled OR was determined by a z-test, and p<0.05 was considered statistically significant.
Subgroup analysis was also conducted to determine whether there was an association between the IL-6 gene polymorphism and tuberculosis risk in different ethnicities. Sensitivity analysis was performed by individually removing the included studies to assess the stability of results. Publication bias was analyzed by Begg’s funnel plots and Egger’s test.24 HWE was tested by Pearson’s χ2 test (p<0.05 means deviated from HWE). All statistical tests were performed by STATA 12.0 software (StataCorp, College Station, TX, USA) two-sided, and p<0.05 was considered statistically significant.
Results
Study characteristics of included studies
A total of 12 publications with 2635 cases and 3049 controls were justified eligible according to inclusion criteria.7,17,18,25–33 Figure 1 outlines the selection and inclusion process of publications.
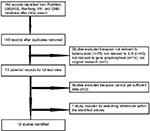  | Figure 1 The flow diagram of included and excluded studies. |
Among the included publications, eight evaluated the association between the IL-6 (-174G/C) polymorphism and tuberculosis risk, with 959 cases and 1608 controls;7,18,25–30 three focused on IL-6 (-572C/G) polymorphism, including 686 cases and 725 controls.17,32,33 One article assessed both IL-6 (-174G/C) and (-572C/G) with 495 cases and 358 cases,18 and it was treated as two independent studies in the present meta-analysis. Consequently, 13 studies were available for analyzing the association between IL-6 gene polymorphism and tuberculosis risk, nine (8 publications) of them were conducted in Asians,17,18,25,27,29,30,32,33 while two were in Caucasians,26,28 and two were in Latinos.7,31 Ten studies (9 publications) recruited pulmonary tuberculosis patient,17,18,25,27–32 three investigated other types of tuberculosis such as tuberculous pleurisy, renal tuberculosis, tuberculous lymphadenitis, and central nervous system tuberculosis.7,26,33 The genotype distributions in the controls of four studies were not consistent with HWE.7,25,17,30 In the 13 studies (12 publications), 10 are with quality score ≥10, suggesting the reliability of our results. The general characteristics of each study included in this meta-analysis are summarized in Table 1. Genotype distributions and HWE examination results are listed in Table 2.
  | Table 2 Distributions of IL-6 genotype and allele among tuberculosis patients and controls Abbreviations: HWE, Hardy–Weinberg equilibrium; IL, interleukin. |
Quantitative data synthesis
IL-6 (-174G/C) polymorphism and tuberculosis risk
We analyzed the heterogeneity of four genetic models: allele contrast (C vs G), co-dominant model (CC vs GG), dominant (CC+GC vs GG), and recessive (CC vs GC+GG) models. The values of χ2 were 7.98, 8.09, 11.26, and 9.71, respectively (p=0.281, 0.324, 0.188, and 0.206, respectively). Thus, all data were synthesized using the fixed-effects model. Overall, OR was 0.693 (95% CI 0.581–0.826) and the test for overall effect z value was 4.09 (p<0.001) for CC+GC vs GG (Figure 2). We also analyzed the other three models: C vs G model: OR =0.714 (95% CI 0.619–0.825), z=4.59, p<0.002; CC vs GG model: OR =0.506 (95% CI 0.349–0.734), z=3.59, p<0.001; CC vs GC+GG model: OR =0.570 (95% CI 0.397–0.819), z=3.04, p=0.002, as listed in Table 3.
Additionally, subgroup analysis was carried out by ethnicity for IL-6 (-174G/C). IL-6 (-174G/C) polymorphism was not significantly associated with tuberculosis in Caucasian patients under the four genetic models. However, in Asian and Latino patients, the results indicated that there were significant associations between IL-6 (-174G/C) polymorphism and tuberculosis under all genetic models, similar to overall text effect (Figure 3). A summary of results from these comparisons is listed in Table 4.
IL-6 (-572C/G) polymorphism and tuberculosis risk
Based on the examination results of heterogeneity (χ2=7.03, p=0.071), a fixed-effects model was applied to pool the OR under dominant model (CC+GC vs GG), and the OR was 0.719 (95% CI 0.577–0.896), with a z value of 2.93 (p=0.003), the forest plot is shown in Figure 4. The results are summarized in Table 3.
  | Figure 4 Meta-analysis for the association between the IL-6 -572C/G polymorphism and tuberculosis risk (CC+GC vs GG). Abbreviations: IL, interleukin; OR, odds ratio; CI, confidence interval. |
Sensitivity analysis and publication bias
In order to evaluate the influence of single studies on overall estimates, sensitivity analyses were carried out for four genetic models by removing each study in turn. No obvious changes were found, which suggested that our results were stable in these models.
Begg’s funnel plots and Egger’s test were performed to evaluate the publication bias qualitatively and quantitatively. Figure 5A shows the funnel plots for IL-6 (-174G/C). The shape of the funnel plots seemed symmetrical, and Egger’s test did not find obvious publication bias (CC+GC vs GG, p=0.714). Figure 5B shows the funnel plots of IL-6 (-572C/G); similarly, Egger’s test for all models also suggested the absence of publication bias (CC+GC vs GG, p=0.423).
Discussion
To date, numerous studies have evaluated the effects of gene polymorphism on tuberculosis risk and confirmed that host susceptibility to tuberculosis was associated with genetic factors.34,35 With regard to IL-6, the results were paradoxical and some studies failed to identify the association. Recently, several systematic reviews focused on the relationship between -174G/C or -572C/G polymorphism and risk of tuberculosis;21,22,36 however, the enrolled studies and subjects were limited. In order to increase the statistical power and generate an absolute conclusion, we performed this meta-analysis to integrate the results from parallel studies regarding the association between IL-6 gene polymorphism (-174G/C and -572C/G) and tuberculosis risk.
In this meta-analysis, we evaluated the association between IL-6 (-174G/C and -572C/G) polymorphisms and tuberculosis risk under the allele comparison, co-dominant, recessive, and dominant models, respectively. There was an association between IL-6 (-174G/C) and low tuberculosis risk under allele contrast (for C vs G: OR =0.714, 95% CI 0.619–0.825, p<0.001) and dominant models (for CC+GC vs GG: OR =0.506, 95% CI 0.349–0.734, p<0.001), and also in recessive and co-dominant model (CC vs GC+GG: OR =0.570, 95% CI 0.397–0.819, p=0.002; CC vs GG: OR =0.506, 95% CI 0.349–0.734, p<0.001), which indicated that the C allele in IL-6 (-174G/C) may be related to a lower risk of tuberculosis. These were consistent with previous reviews that summarized the correlation of -174G/C polymorphism and risk of tuberculosis based on the results of the seven studies performed before 2012.21,22,36 However, in the current review, we enrolled two more studies conducted in 2015 and 2016, respectively, and our findings are the powerful update of these reviews. Besides, we also found a significant association between IL-6 (-572C/G) and low tuberculosis risk under dominant model (for CC+GC vs GG: OR =0.719, 95% CI 0.577–0.896, p=0.003) and co-dominant model (for CC vs GG: OR =0.713, 95% CI 0.517–0.984, p=0.039), which suggested that the CC+GC carriers have a 29% decreased risk of tuberculosis compared with those with GG. Recently, Sun et al reported similar findings that -572C/G polymorphism was related to the decreased risk of tuberculosis (CC vs GC: OR =0.71, 95% CI 0.55–0.90);36 compared with this review, we enrolled two more studies in the current study, and we got some new points through the co-dominant model (CC vs GG). Nevertheless, what should be addressed is that the role of IL-6 in tuberculosis is complicated. It has been reported that IL-6 played a critical role in protection against murine M. tuberculosis infection, while it has also been suggested to downregulate macrophage microbicidal activity and promote the growth of Mycobacterium avium in vitro.37,38 Additionally, environmental factors may interact with gene polymorphism and influence tuberculosis susceptibility. How to adjust the environmental effects on tuberculosis risk is still being investigated. More studies should be performed to investigate the mechanism how IL-6 polymorphism regulates IL-6 production and plays a role in tuberculosis susceptibility.
Ethnicity is one of the considerable factors for tuberculosis risk with controversial conclusions in previous studies.16,39 Therefore, we conducted subgroup analyses based on ethnicity in IL-6 (-174G/C), but not in IL-6 (-572C/G), as all studies investigating IL-6 (-572C/G) in this meta-analysis were performed in Asian population. The significant associations between IL-6 (-174G/C) polymorphism and tuberculosis risk were found in Asians and Latinos, but no obvious association was observed in Caucasian population, suggesting that ancestral genetic factors in different ethnic populations may have an impact on tuberculosis risk.
Heterogeneity and publication bias should be taken into consideration while interpreting the results of a meta-analysis. The heterogeneity in our meta-analysis existed in IL-6 (-572C/G), but not in overall or subgroup analyses of IL-6 (-174G/C). Several factors may be used to explain the heterogeneity, such as genetic background of cases and controls, diverse genotype distributions of IL-6 in different ethnicities which suggest that they are always subjected to natural selection.40 Besides, the uneven selection criteria for cases and controls in different original studies have also contributed to this heterogeneity. According to our inclusion and exclusion criteria, only four studies involved the association between IL-6 (-572C/G) and tuberculosis risk, all of which were conducted in Asian population. Thus, the reason for the heterogeneity may be attributed to the moderate number of studies and the absence of studies conducted in populations other than Asians. Referring to -174G/C polymorphism, our meta-analysis enrolled studies performed in Asians (five studies), Caucasians (two studies), and Latinos (two studies), which might make our conclusion inapplicable for the other populations. In addition, only two studies were conducted in Caucasians or Latinos, respectively. Therefore, more studies will be needed in the future to evaluate the relationship between IL-6 polymorphism and tuberculosis risk in different population, especially in Africans which were not enrolled in the current review. In this meta-analysis, publication bias was analyzed by Begg’s funnel plots and Egger’s test, and no significant publication bias was detected, suggesting that our results are reliable. However, more studies should be performed to verify the results of our meta-analysis.
Limitations should be pointed out in our meta-analysis. First, after strict search strategy and study selection, only limited numbers of publications were included. There are possibilities that certain articles were missed in this study, due to database or language limitation, which may bias the results. In the present meta-analysis, we included only publications in English or Chinese. Although we did not detect any publication bias, it is possible that including relevant studies in other languages or unpublished studies which had null outcomes may alter our results.
Second, due to the limited number of studies included, we did not exclude studies in which the genotype distributions in controls were not consistent with HWE. Large-scale and well-designed case–control studies should be done to estimate the accurate association between the IL-6 and tuberculosis risk in the years to come.
Lastly, the studies included in the meta-analysis were conducted in Caucasian, Asian, and Latino patients, which may not represent the associations in ethnicities worldwide. Meanwhile, due to the limitation of original information, the influence of gene–gene interactions or environmental factors was not considered in this meta-analysis. Last but not least, the current genotyping method may cause errors, which should be paid attention to in clinical explanation.
However, it is worthwhile to mention several highlights of this study. First, this was the first meta-analysis to assess the relationship between the IL-6 (-572C/G) polymorphism and tuberculosis risk. Second, heterogeneity, stability of results, and publication bias in this meta-analysis were all well investigated, indicating a correct methodology. Also, strict inclusion and exclusion criteria were followed to limit the potential bias among studies.
Taken together, our results suggested that in Asian and Latino population, IL-6 -572C/G polymorphism decreased the risk of tuberculosis, and C allele was a protective factor against tuberculosis for IL-6 -174 G/C polymorphism, which may not be in Caucasian population. In future, additional case–control studies should be performed to validate our findings.
Acknowledgments
This work was supported by a grant from the National Scientific and Technological Major Project of China (2012ZX10004-901) and grants 81230001, 81470236 from the National Natural Science Foundation of China. We are indebted to the authors of the primary studies included in this meta-analysis; without their contributions, this work would not have been possible.
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Dara M, Acosta CD, Melchers NV, et al. Tuberculosis control in prisons: current situation and research gaps. Int J Infect Dis. 2015;32:111–117. | ||
WHO. Global tuberculosis report 2016; 2016. Available from: http://www.who.int/tb/publications/global_report/en/. Accessed September 28, 2017. | ||
O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. The immune response in tuberculosis. Ann Rev Immunol. 2013;31:475–527. | ||
Bellamy R. Susceptibility to mycobacterial infections: the importance of host genetics. Genes Immun. 2003;4(1):4–11. | ||
Moller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb). 2010;90(2):71–83. | ||
Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. Innate immune recognition of Mycobacterium tuberculosis. Clin Dev Immunol. 2011;2011:405310. | ||
Henao MI, Montes C, Paris SC, Garcia LF. Cytokine gene polymorphisms in Colombian patients with different clinical presentations of tuberculosis. Tuberculosis (Edinb). 2006;86(1):11–19. | ||
Hussain R, Kaleem A, Shahid F, et al. Cytokine profiles using whole-blood assays can discriminate between tuberculosis patients and healthy endemic controls in a BCG-vaccinated population. J Immunol Methods. 2002;264(1–2):95–108. | ||
Lopez-Maderuelo D, Arnalich F, Serantes R, et al. Interferon-gamma and interleukin-10 gene polymorphisms in pulmonary tuberculosis. Am J Respir Crit Care Med. 2003;167(7):970–975. | ||
Martinez AN, Mehra S, Kaushal D. Role of interleukin 6 in innate immunity to Mycobacterium tuberculosis infection. J Infect Dis. 2013;207(8):1253–1261. | ||
Gouzy A, Nigou J, Gilleron M, Neyrolles O, Tailleux L, Gordon SV. Tuberculosis 2012: biology, pathogenesis and intervention strategies; an update from the city of light. Res Microbiol. 2013;164(3):270–280. | ||
Scola L, Crivello A, Marino V, et al. IL-10 and TNF-alpha polymorphisms in a sample of Sicilian patients affected by tuberculosis: implication for ageing and life span expectancy. Mech Ageing Dev. 2003;124(4):569–572. | ||
Gao JW, Zhang AQ, Pan W, et al. Association between IL-6-174G/C polymorphism and the risk of sepsis and mortality: a systematic review and meta-analysis. PLoS One. 2015;10(3):e0118843. | ||
Song C, Liu B, Yang D, et al. Association between Interleukin-6 gene -572G>C polymorphism and coronary heart disease. Cell Biochem Biophys. 2015;71(1):359–365. | ||
Yang Z, Liang Y, Qin B, Zhong R. A meta-analysis of the association of IL-6 -174 G/C and -572 G/C polymorphisms with systemic lupus erythematosus risk. Rheumatol Int. 2014;34(2):199–205. | ||
Larcombe LA, Orr PH, Lodge AM, et al. Functional gene polymorphisms in Canadian aboriginal populations with high rates of tuberculosis. J Infect Dis. 2008;198(8):1175–1179. | ||
Sun YH, Liu XX, Guo M, Feng FM. 白细胞介素-6基因多态性与肺结核易感性的关系研究 [A case control study on interleukin-6 genetic polymorphism and susceptibility to pulmonary tuberculosis]. Modem Prev Med. 2008;35(19):3796–3802. Chinese. | ||
Zhang G, Zhou B, Wang W, et al. A functional single-nucleotide polymorphism in the promoter of the gene encoding interleukin 6 is associated with susceptibility to tuberculosis. J Infect Dis. 2012; 205(11):1697–1704. | ||
Tang M, Slud EV, Pfeiffer RM. Goodness of fit tests for linear mixed models. J Multivar Anal. 2014;130:176–193. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Ke Z, Yuan L, Ma J, Zhang X, Guo Y, Xiong H. IL-10 polymorphisms and tuberculosis susceptibility: an updated meta-analysis. Yonsei Med J. 2015;56(5):1274–1287. | ||
Mao X, Ke Z, Liu S, et al. IL-1beta+3953C/T, -511T/C and IL-6 -174C/G polymorphisms in association with tuberculosis susceptibility: a meta-analysis. Gene. 2015;573(1):75–83. | ||
Thakkinstian A, McEvoy M, Minelli C, et al. Systematic review and meta-analysis of the association between beta2-adrenoceptor polymorphisms and asthma: a HuGE review. Am J Epidemiol. 2005;162(3):201–211. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Amirzargar AA, Rezaei N, Jabbari H, et al. Cytokine single nucleotide polymorphisms in Iranian patients with pulmonary tuberculosis. Eur Cytokine Netw. 2006;17(2):84–89. | ||
Oral HB, Budak F, Uzaslan EK, et al. Interleukin-10 (IL-10) gene polymorphism as a potential host susceptibility factor in tuberculosis. Cytokine. 2006;35(3–4):143–147. | ||
Selvaraj P, Alagarasu K, Harishankar M, Vidyarani M, Nisha Rajeswari D, Narayanan PR. Cytokine gene polymorphisms and cytokine levels in pulmonary tuberculosis. Cytokine. 2008;43(1):26–33. | ||
Trajkov D, Trajchevska M, Arsov T, et al. Association of 22 cytokine gene polymorphisms with tuberculosis in Macedonians. Indian J Tuberc. 2009;56(3):117–131. | ||
Ansari A, Hasan Z, Dawood G, Hussain R. Differential combination of cytokine and interferon- gamma +874 T/A polymorphisms determines disease severity in pulmonary tuberculosis. PLoS One. 2011;6(11):e27848. | ||
Hu Y, Wu L, Li D, Zhao Q, Jiang W, Xu B. Association between cytokine gene polymorphisms and tuberculosis in a Chinese population in Shanghai: a case-control study. BMC Immunol. 2015;16:8. | ||
Milano M, Moraes MO, Rodenbusch R, et al. Single nucleotide polymorphisms in IL17A and IL6 are associated with decreased risk for pulmonary tuberculosis in Southern Brazilian population. PLoS One. 2016;11(2):e0147814. | ||
Feng FM, Liu XX, Sun YH, et al. Independent and joint effects of the IL-6 and IL-10 gene polymorphisms in pulmonary tuberculosis among the Chinese Han population. Genet Mol Res. 2014;13(3):7766–7772. | ||
Shen C, Qi H, Sun L, et al. A 3′UTR polymorphism of IL-6R is associated with Chinese pediatric tuberculosis. BioMed Res Int. 2014;2014:1–7. | ||
Azad AK, Sadee W, Schlesinger LS. Innate immune gene polymorphisms in tuberculosis. Infect Immun. 2012;80(10):3343–3359. | ||
Wang T, Shen Y, Chen L, et al. The -159C/T polymorphism in the CD14 gene and tuberculosis risk: a meta-analysis. Int J Med Sci. 2013;10(11):1524–1529. | ||
Sun Y, Wang M. Association between IL-6 gene polymorphisms and susceptibility of tuberculosis: evidence based on a meta-analysis. Int J Clin Exp Med. 2017;10(3):4297–4304. | ||
Ladel CH, Blum C, Dreher A, Reifenberg K, Kopf M, Kaufmann SH. Lethal tuberculosis in interleukin-6-deficient mutant mice. Infect Immun. 1997;65(11):4843–4849. | ||
Shiratsuchi H, Johnson JL, Ellner JJ. Bidirectional effects of cytokines on the growth of Mycobacterium avium within human monocytes. J Immunol. 1991;146(9):3165–3170. | ||
Delgado JC, Baena A, Thim S, Goldfeld AE. Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis. 2002;186(10):1463–1468. | ||
Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4(2):99–111. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






