Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 13
Association Between lncRNA HULC rs7763881 Polymorphism and Gastric Cancer Risk
Authors Hong JH , Jin EH , Chang IA, Kang H, Lee SI , Sung JK
Received 23 January 2020
Accepted for publication 31 March 2020
Published 9 April 2020 Volume 2020:13 Pages 121—126
DOI https://doi.org/10.2147/PGPM.S247082
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Jang Hee Hong,1,2,* Eun-Heui Jin,3,* In Ae Chang,1 Hyojin Kang,1 Sang-Il Lee,4 Jae Kyu Sung5
1Clinical Trials Center, Chungnam National University Hospital, Daejeon, Republic of Korea; 2Department of Pharmacology, Chungnam National University College of Medicine, Daejeon, Republic of Korea; 3Research Institute for Medical Sciences, Chungnam National University College of Medicine, Daejeon, Republic of Korea; 4Department of Surgery, Chungnam National University Hospital, Chungnam National University College of Medicine, Daejeon, Republic of Korea; 5Department of Internal Medicine, Chungnam National University Hospital, Chungnam National University College of Medicine, Daejeon, Republic of Korea
*These authors contributed equally to this work
Correspondence: Jae Kyu Sung; Sang-Il Lee
Department of Internal Medicine, Chungnam National University Hospital, Chungnam National University College of Medicine, Daejeon, 35015, Republic of Korea; Department of Surgery, Chungnam National University Hospital, Chungnam National University College of Medicine, Daejeon 35015, Republic of Korea
Tel +82-42-280-7186
; +82-42-280-7180
Fax +82-42-254-4553
Email [email protected]; [email protected]
Purpose: Gastric cancer (GC) is one of the most common cancers in the world. Recently, several studies have suggested that single-nucleotide polymorphisms (SNPs) of long noncoding RNA (lncRNA) are associated with GC risk. However, the association of the lncRNA highly upregulated in liver cancer (HULC) SNP with GC risk is not yet known. The aims of this study were to evaluate the association between HULC rs7763881 SNP and the risk of GC and GC subgroups via a case–control study.
Patients and Methods: rs7763881 was genotyped using TaqMan genotyping assay with 459 GC patients and 379 controls.
Results: A significant association between HULC rs7763881 SNP and GC risk was not found. However, after adjustment for age and gender, the rs7763881 recessive model (CC) showed a significant association with an increased GC risk in the undifferentiated (odds ratio (OR) = 1.85, 95% confidence interval (CI) = 1.17– 2.94, P = 0.009), diffuse-type GC (OR = 1.72, 95% CI = 1.05– 2.82, P = 0.033), LNM-positive (OR = 2.02, 95% CI = 1.24– 3.27, P = 0.004), T3/T4 (OR = 1.75, 95% CI = 1.05– 2.91, P = 0.032), and tumor stage III (OR = 2.01, 95% CI = 1.17– 3.45, P = 0.011) subgroups when compared to the rs7763881 combined genotypes (AA+AC). Furthermore, after adjusting for age and gender, the rs7763881 additive model (CC) indicated a significantly higher GC risk than rs7763881 AA genotype in the undifferentiated (OR = 1.96, 95% CI = 1.15– 3.32, P = 0.013), diffuse-type GC (OR = 2.08, 95% CI = 1.23– 3.52, P = 0.004), and LNM-positive (OR = 2.00, 95% CI = 1.14– 3.49, P = 0.016) subgroups.
Conclusion: Our findings suggest that the HULC rs7763881 SNP is associated with increased susceptibility to GC. However, further studies are required to validate our results in large populations as well as different ethnic groups.
Keywords: lncRNA, HULC, gastric cancer, polymorphism
Introduction
Gastric cancer is one of the most common cancers worldwide and the fourth most common cancer with high mortality in South Korea in 2015. Gastric cancer (GC) incidence remains high in Asian countries despite its global decrease over the past few years.1,2
Long noncoding RNAs (lncRNAs) are defined as non-coding transcripts with lengths > 200 nucleotides.3 In recent years, a number of studies have demonstrated that non-coding RNAs, such as lncRNAs, are implicated in cancer development through regulation of cancer-related gene expression, thus acting as oncogenes and tumor suppressors in several cancers.4 LncRNAs including H19 imprinted maternally expressed transcript (H19),5,6 HOX transcript antisense RNA (HOTAIR),7,8 colon cancer-associated transcript 2(CCAT2),9,10 Pvt1 oncogene (PVT1),11,12 and metastasis-associated lung adenocarcinoma transcript 1(MALAT1)13,14 have been reported as oncogenes that are upregulated in gastric cancer. However, lncRNAs including maternally expressed 3 (MEG 3)15,16 and growth arrest-specific 5 (GAS5)17 have demonstrated as tumor suppressors that were downregulated in gastric cancer. LncRNA highly up- have been demonstrated as tumor suppressors that are downregulated in gastric cancer. The lncRNA highly upregulated liver cancer (HULC) gene, located on chromosome 6p24.3, has a length of 1638 bp and comprises two exons and one intron. HULC has been reported as an oncogene that is upregulated in several cancers, including hepatocellular carcinoma (HCC),18 gastric cancer,19 pancreatic cancer,20 osteosarcoma,21 glioma22 and ovarian cancer.23 Additionally, recent studies have reported that genetic variations in HULC are associated with susceptibility to various cancers. For instance, in Chinese populations, Liu et al reported the association between HULC rs7763881 SNP and decreased HCC risk,24 and Kang et al reported the association between HULC rs7763881 SNP and decreased esophageal squamous cell carcinoma (ESCC) risk.25 In Egyptian populations, Shaker et al reported the association between HULC rs7763881 SNP and decreased colorectal cancer (CRC) risk,27 and Motawi et al reported the association between HULC rs7763881 SNP and decreased HCC risk.26 However, the association between HULC rs7763881 SNP and GC risk is not yet reported.
Based on previous study, we hypothesized HULC rs7763881 SNP may contribute to susceptibility to GC. Therefore, we investigated the association between HULC rs7763881 SNP and GC risk in a Korean population. We further evaluated the correlation of HULC rs7763881 SNP with clinical features, including age, gender, tumor differentiation, histological type, LNM, T classification, and tumor stage.
Patientss and Methods
Study Subjects
This study was approved and reviewed by the Ethics Committee of the institutional review board of Chungnam National University Hospital, and in compliance with the Declaration of Helsinki. A total of 459 GC subjects and 379 control subjects were enrolled. The blood samples used in this study were provided by the Chungnam National Hospital Biobank, a member of the National Biobank of Korea, which is supported and audited by the Ministry of Health and Welfare of Korea. All individuals enrolled in this study provided written informed consent for blood collection and use. GC patients were recruited from the outpatient clinic at the Chungnam National University Hospital and classified according to Lauren’s classification.28 The subjects for the control group were randomly selected among healthy volunteers visiting the Chungnam National University Hospital medical center for their annual physical examinations; only individuals who had no history of cancer were included.
DNA Isolation and Genotyping
Genomic DNA was isolated from peripheral blood samples of all subjects using the QIAamp DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer’s instructions. HULC rs7763881 SNP was genotyped by the Applied Biosystems TaqMan SNP Genotyping Assay using predesigned primer/probe sets (C_29335152_10). PCR was performed using the StepOnePlus Real-time PCR System (Applied Biosystems) according to the following conditions: one cycle at 95 °C for 10 min; 45cycles at 92 °C for 15 s and 60 °C for 90 s.
Statistical Analysis
Hardy Weinberg equilibrium (HWE) for each SNP in the control groups was estimated using the chi-square test. Differences in age and gender between the GC and control groups were calculated using the two-sided Pearson chi-square test and the Mann–Whitney U-test. Five genetic models, including codominant (CC or AC vs AA), dominant (AC+CC vs AA), recessive (CC vs AA+AC), additive CC vs AA), and allelic (C vs A) models, were used to analyze the associations. A binary logistic regression was used to estimate the GC risk according to odds ratios (ORs) and 95% confidence intervals (CIs). The association analysis was adjusted by age and sex, which were included in the model as covariates. All statistical analyses were performed using the SPSS (SPSS Inc., Chicago, IL, USA), version 20.0 for Windows. P < 0.05 was considered statistically significant.
Results
Characteristics of Study Subjects
The characteristics of 459 GC and 379 control subjects are summarized in Table 1. There were statistically significant differences in the age and gender distribution between the two groups (P < 0.001 and P < 0.001, respectively). The mean age was 65.2±10.6 years for GC patients and 56.1±10.9 years for the controls. The percentage of GC male subjects (70.2%) was higher than that of females (29.8%), whereas the number of female control subjects (68.1%) was higher than that of males (31.9%). The GC group consisted of 48.4% well-differentiated GC, 56.2% intestinal-type, 51.0% T1, 62.3% lymph node metastasis, and 59.9% stage I tumor cases.
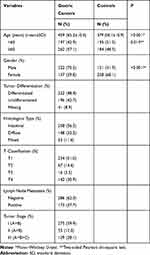 |
Table 1 Demographic and Clinical Features of the Gastric Cancer Group and the Control Group |
Association of SNP and GC Risk
We genotyped rs7763881 SNP in HULC, which has been previously associated with cancer risk. The genotype distribution of rs7763881 SNP, both among the GC group and the control group, was in accordance with HWE (P = 0.998 and P = 0.373, respectively). We applied five genetic models to assess the possible association between the rs7763881 SNP and GC risk. However, no significant association between rs7763881 SNP and GC risk was observed (Table 2).
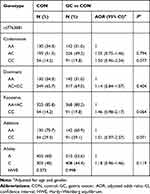 |
Table 2 Genotype and Allele Frequencies for HULC rs7763881 Polymorphism Among Subjects and Their Association with GC Risk |
Stratified Analysis for rs7763881 SNP
Further, we performed stratified analyses to evaluate the possible association between rs7763881 SNP and GC risk based on various disease characteristics, including age, gender, tumor differentiation, histological type, LNM, T classification, and tumor stage. The results are shown in Table 3. After adjusting age and gender, the rs7763881 recessive model (CC) showed a significant association with an increased GC risk in the undifferentiated (OR = 1.85, 95% CI = 1.17–2.94, P = 0.009), diffuse-type GC (OR = 1.72, 95% CI = 1.05–2.82, P = 0.033), LNM-positive (OR = 2.02, 95% CI = 1.24–3.27, P = 0.004), T3/T4 (OR = 1.75, 95% CI = 1.05–2.91, P = 0.032), and tumor stage III (OR = 2.01, 95% CI = 1.17–3.45, P = 0.011) subgroups, when compared to the rs7763881 combined genotypes (AA+AC). Furthermore, the rs7763881 additive model (CC) indicated a significantly higher GC risk than rs7763881 AA genotype in the undifferentiated (OR = 1.96, 95% CI = 1.15–3.32, P = 0.013), diffuse-type GC (OR = 2.08, 95% CI = 1.23–3.52, P = 0.004), and LNM-positive (OR = 2.00, 95% CI = 1.14–3.49, P = 0.016) subgroups (Table 3).
 |
Table 3 Stratified Analysis for HULC rs7763881 Polymorphism in GC Patients and Controls |
Discussion
In recent years, a number of studies have reported the association of lncRNA HULC polymorphisms with decreased risk of HCC, ESCC, and CRC.24–27 In this case-control study, we first investigated the association between HULC rs7763881 SNP and GC risk but did not observe a significant association between them. However, stratified analysis by age, gender, tumor differentiation, histological type, LNM, T classification, and tumor stage revealed statistically significant association between HULC rs7763881 CC genotype and increased GC risk of the undifferentiated, diffuse-type, LNM-positive, T3/T4, and tumor stage III groups. Till date, contradictory to our results, previous studies have reported the relationship between rs7763881 AC or AC+CC genotype with the decreased risk of several cancers, including HBV carried HCC, ESCC, and CRC.24–27 Through stratification analysis. Of Chinese populations, Kang et al showed that rs7763881 AC genotype is significantly associated with decreased ESCC risk in male patients compared to rs7763881 AA genotype. Moreover, stratification analysis of Egyptian populations by Shaker et al suggested that rs7763881 AC genotype is significantly associated with decreased CRC risk in younger patients compared to rs7763881 AA genotype.25,27 However, we did not observe the association of rs7763881 genotypes with GC risk in stratified subgroups by age and gender. To evaluate the possible clinical applications of lncRNA as a GC diagnosis biomarker, recent studies have assessed the correlation between lncRNA expression levels and clinical features such as tumor differentiation, histological type, LNM, T classification, and tumor stage in GC and reported their association. Moreover, it has recently been reported that lncRNAs play a pivotal role in development and progression of human cancer. In GC, overexpression of lncRNAs is correlated with clinical features of GC patients such as tumor differentiation, LNM, and tumor stage. Li et al showed an association between enhanced expression of H19 and LN number (N2-3) and tumor stage (III–IV) of GC.5 Zhang et al and Liu et al reported the relationship between HOTAIR overexpression and poor differentiation, LNM, and advanced tumor stage (III–IV).8,29 Sun et al showed an association between decreased expression of MEG3 and GAS5 and TNM stage and LNM of GC.15,17 Zhao et al revealed the relationship between increased expression of HULC and LNM present and GC tumor stage (III–IV).19 Ding et al observed a positive correlation between increased expression of PVT1 and LNM of GC.30 Additionally, Hong et al showed an association between lncRNA PRNCR1 SNPs and risk of GC in LNM-positive and tumor stage III subgroups.31 In our data, although we did not investigate the correlation between HULC rs7763881 CC genotype and HULC gene expression, we found that HULC rs7763881 CC genotype in the dominant and additive genetic model was associated with a higher risk for GC of undifferentiated, diffuse-type, LNM-positive, T3/T4, and tumor stage III groups.
There were a few limitations of this case-control study. First, the sample size is relatively small for a stratified analysis, leading to the reduction of the statistical power. Second, we failed to investigate the association between the genetic factors and other clinical features such as Helicobacter pylori infection, smoking, drinking, and diet, owing to lack of these data from the GC and control groups. Fourth, large prospective studies are needed to validate our results in different ethnic groups.
Conclusions
In conclusion, based on previous studies and our results, we suggest that HULC rs7763881 CC genotype may contribute to GC development by affecting HULC as an oncogene. Further studies are required to validate our findings in large populations and different ethnic groups to clarify whether HULC rs7763881 CC increases GC risk by altering HULC gene expression.
Abbreviations
AOR, adjusted odds ratio; CI, confidence interval; CON, control; CCAT2, colon cancer-associated transcript 2; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; GAS5, growth arrest-specific 5; GC, Gastric cancer; H19, imprinted maternally expressed transcript; HCC, hepatocellular carcinoma; HOTAIR, HOX transcript antisense RNA; HULC, highly upregulated in liver cancer; HWE, Hardy Weinberg equilibrium; LncRNAs, long non-coding RNAs; LNM, lymph node metastasis; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MEG 3, maternally expressed 3; PVT1, Pvt1 oncogene; SD, standard deviation; SNPs, single-nucleotide polymorphisms.
Acknowledgments
This work was supported by research fund of Chungnam National University, by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (NRF-2016R1D1A3B03936067), and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1D1A1B04033515, NRF-2018R1D1A1B07050285, and NRF-2019R1F1A1063469).
Disclosure
The authors have declared that no competing interest exists in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Jung KW, Won YJ, Kong HJ, Lee ES; Community of Population-Based Regional Cancer Registries. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2015. Cancer Res Treat. 2018;50(2):303–316. doi:10.4143/crt.2018.143
3. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
4. Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109(7):2093–2100. doi:10.1111/cas.13642
5. Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. doi:10.18632/oncotarget.1913
6. Chen JS, Wang YF, Zhang XQ, et al. H19 serves as a diagnostic biomarker and up-regulation of H19 expression contributes to poor prognosis in patients with gastric cancer. Neoplasma. 2016;63(2):223–230. doi:10.4149/207_150821N454
7. Lee NK, Lee JH, Park CH, et al. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451(2):171–178. doi:10.1016/j.bbrc.2014.07.067
8. Zhang ZZ, Shen ZY, Shen YY, et al. HOTAIR long noncoding RNA promotes gastric cancer metastasis through suppression of Poly r(C)-Binding Protein (PCBP) 1. Mol Cancer Ther. 2015;14(5):1162–1170. doi:10.1158/1535-7163.MCT-14-0695
9. Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ, Hu JH. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2015;8(1):779–785.
10. Wu S-W, Hao Y-P, Qiu J-H, Zhang D-B, Yu C-G, Li W-H. High expression of long non-coding RNA CCAT2 indicates poor prognosis of gastric cancer and promotes cell proliferation and invasion. Minerva Med. 2017;108(4):317–323. doi:10.23736/S0026-4806.17.04703-6
11. Kong R, Zhang EB, Yin DD, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82. doi:10.1186/s12943-015-0355-8
12. Yuan CL, Li H, Zhu L, Liu Z, Zhou J, Shu Y. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma. 2016;63(3):442–449. doi:10.4149/314_150825N45
13. Li J, Gao J, Tian W, Li Y, Zhang J. Long non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297. Cancer Cell Int. 2017;17:44. doi:10.1186/s12935-017-0408-8
14. Li Y, Wu Z, Yuan J, et al. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31–44. doi:10.1016/j.canlet.2017.02.035
15. Sun M, Xia R, Jin F, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35(2):1065–1073. doi:10.1007/s13277-013-1142-z
16. Guo W, Dong Z, Liu S, et al. Promoter hypermethylation-mediated downregulation of miR-770 and its host gene MEG3, a long non-coding RNA, in the development of gastric cardia adenocarcinoma. Mol Carcinog. 2017;56(8):1924–1934. doi:10.1002/mc.22650
17. Sun M, Jin FY, Xia R, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. doi:10.1186/1471-2407-14-319
18. Yang X, Xie X, Xiao YF, et al. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360(2):119–124. doi:10.1016/j.canlet.2015.02.035
19. Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31(1):358–364. doi:10.3892/or.2013.2850
20. Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31(12):346. doi:10.1007/s12032-014-0346-4
21. Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8(3):2994–3000.
22. Zhu Y, Zhang X, Qi L, et al. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget. 2016;7(12):14429–14440. doi:10.18632/oncotarget.7418
23. Chen S, Wu DD, Sang XB, et al. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 2017;8(10):e3118. doi:10.1038/cddis.2017.486
24. Liu Y, Pan S, Liu L, et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2012;7(4):e35145. doi:10.1371/journal.pone.0035145
25. Kang M, Sang Y, Gu H, et al. Long noncoding RNAs POLR2E rs3787016 C/T and HULC rs7763881 A/C polymorphisms are associated with decreased risk of esophageal cancer. Tumour Biol. 2015;36(8):6401–6408. doi:10.1007/s13277-015-3328-z
26. Motawi TMK, El-Maraghy SA, Sabry D, Mehana NA. The expression of long non coding RNA genes is associated with expression with polymorphisms of HULC rs7763881 and MALAT1 rs619586 in hepatocellular carcinoma and HBV Egyptian patients. J Cell Biochem. 2019;120(9):14645–14656. doi:10.1002/jcb.28726
27. Shaker OG, Senousy MA, Elbaz EM. Association of rs6983267 at 8q24, HULC rs7763881 polymorphisms and serum lncRNAs CCAT2 and HULC with colorectal cancer in Egyptian patients. Sci Rep. 2017;7(1):16246. doi:10.1038/s41598-017-16500-4
28. Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi:10.1111/apm.1965.64.1.31
29. Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi:10.1186/1476-4598-13-92
30. Ding J, Li D, Gong M, et al. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther. 2014;7:1625–1630. doi:10.2147/OTT.S68854
31. Hong JH, Jin EH, Kang H, Chang IA, Lee SI, Sung JK. Correlations between genetic polymorphisms in long non-coding RNA PRNCR1 and gastric cancer risk in a Korean population. Int J Mol Sci. 2019;20(13):3355. doi:10.3390/ijms20133355
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
