Back to Journals » International Journal of Nanomedicine » Volume 11
Arginine–glycine–aspartic acid–polyethylene glycol–polyamidoamine dendrimer conjugate improves liver-cell aggregation and function in 3-D spheroid culture
Authors Chen Z, Lian F, Wang X, Chen Y, Tang N
Received 23 May 2016
Accepted for publication 11 July 2016
Published 31 August 2016 Volume 2016:11 Pages 4247—4259
DOI https://doi.org/10.2147/IJN.S113407
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Linlin Sun
Zhanfei Chen,1,* Fen Lian,1,* Xiaoqian Wang,1 Yanling Chen,1,2 Nanhong Tang1,2
1Fujian Institute of Hepatobiliary Surgery, Fujian Medical University Union Hospital, 2Key Laboratory of Ministry of Education for Gastrointestinal Cancer, Research Center for Molecular Medicine, Fujian Medical University, Fuzhou, People’s Republic of China
*These authors contributed equally to this work
Abstract: The polyamidoamine (PAMAM) dendrimer, a type of macromolecule material, has been used in spheroidal cell culture and drug delivery in recent years. However, PAMAM is not involved in the study of hepatic cell-spheroid culture or its biological activity, particularly in detoxification function. Here, we constructed a PAMAM-dendrimer conjugate decorated by an integrin ligand: arginine–glycine–aspartic acid (RGD) peptide. Our studies demonstrate that RGD–polyethylene glycol (PEG)–PAMAM conjugates can promote singly floating hepatic cells to aggregate together in a sphere-like growth with a weak reactive oxygen species. Moreover, RGD-PEG-PAMAM conjugates can activate the AKT–MAPK pathway in hepatic cells to promote cell proliferation and improve basic function and ammonia metabolism. Together, our data support the hepatocyte sphere treated by RGD-PEG-PAMAM conjugates as a potential source of hepatic cells for a biological artificial liver system.
Keywords: dendrimer, arginine–glycine–aspartic acid (RGD), liver cell, spheroid culture, ammonia metabolism
Corrigendum for this paper has been published.
Introduction
The role of bioartificial liver in the treatment of liver-function failure has been widely recognized. Current research focuses on the long-term reproduction and normal function of hepatic cells in in vitro culture.1 Primary hepatocytes cultured in monolayer culture can meet the requirements of the general experiment. However, conducting passage culture and mass reproduction is difficult, because cell biological activity is maintained for a short time and primary hepatocytes are terminal cells. To maintain the activity and function of cells as far as possible, many research groups are committed to improving the methods of cell culture. Commonly used methods are coculture with other cells,2 microencapsulated culture,3 spheroidal aggregate culture,4 and bioreactor culture.5 Spheroidal aggregate culture makes hepatic cells aggregate into a sphere, in which the contact area is the largest. This phenomenon leads to the formation of a cube morphology and cytoskeleton structure similar to in vivo and simulates the microenvironment in vivo.6 This type of culture method is mainly used when combining biological materials. For example, polyurethane foam is used to culture rat primary hepatocyte spheres7 and HepG2 cell spheres.4 However, cells in the center of aggregation are in a poor-nutrition and hypoxic environment. In addition, these cells age and die easily, so the diameter of formed spherical aggregates must be controlled. In recent years, to solve this problem, some scholars have tried to use cell-linker molecules8 or tried to make a sandwich culture of hepatocytes by arginine–glycine–aspartic acid (RGD) adhesion peptide and galactose ligand together covalently bounding to polyethylene terephthalate membrane.9,10
Polyamidoamine (PAMAM) dendrimers were the first complete dendrimer family to be synthesized, characterized, and commercialized.11 In addition to its use in the chemical industry, PAMAM and its nanocomposites have made important progress in biomedical applications, including drug-controlled release,12 drug delivery,13–15 a magnetic resonance imaging agent,16,17 and dental material,18 due to good biocompatibility, no immunogenicity, and the easy introduction of various chemical groups at terminal and center positions. In recent years, as a novel type of biological material, PAMAM has been used in the culture of human mesenchymal stem cells19 and NIH3T3.20 However, PAMAM is not involved in the study of hepatic cell-sphere culture or biological activity, particularly in detoxification. Hepatic cells usually express integrin, so this study aimed to construct a PAMAM dendrimer decorated with an integrin ligand RGD. Through a series of research programs, we successfully constructed RGD–polyethylene glycol (PEG)–PAMAM conjugates, which are used in hepatic cell-sphere culture. Results showed that the conjugates can improve the aggregation of hepatocytes and metabolic function of ammonia with a weak reactive oxygen species (ROS).
Materials and methods
Chemicals and cell culture
Generation 3 PAMAM dendrimers (G3-PAMAM) were purchased from Sigma-Aldrich (St Louis, MO, USA). LY294002 (an inhibitor of the PI3K–AKT signaling pathway) was from Cell Signaling Technology Inc (Danvers, MA, USA). The human hepatoblastoma cell line HepG2 (HB-8065; American Type Culture Collection, Manassas, VA, USA), hepatoma cell line Huh7 (JCRB0403), and embryonic kidney cell line 293A (R705-07; Thermo Fisher Scientific, Waltham, MA, USA) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific) supplemented with 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific) in a 37°C humidified atmosphere containing 5% CO2.
Synthesis of RGD-PEG-PAMAM conjugate
RGD-PEG-PAMAM conjugates were synthesized by Dangang Biotechnology Inc (Hangzhou, People’s Republic of China). Briefly, 9-fluorenylmethyloxycarbonyl (Fmoc)-PEG2,000-2Cl(TRT)-resin was selected as a raw material, and the Fmoc of the resin was removed by piperidine. The first amino acid Fmoc-Lys(dde)-OH was added to the PEG2,000-2-Cl-(TRT) resin using the condensation agent O-(benzotriazol-1-yl)-N, N, N′, N′-tetramethyluronium tetrafluoroborate (TBTU) and ethyldiisopropylamine (DIEA). After the reaction was completed, these steps were repeated to link the remaining amino acid until the last amino acid cysteine. The Fmoc in the resin was removed by piperidine and added a mixture of N, N-dimethylformamide (DMF), pyridine, and acetic anhydride. When the reaction ended, the DDE groups were removed by using hydrazine and DMF mixed solution, and fluorescein isothiocyanate (FITC), DIEA, and DMF mixed solution were added for reacting overnight. After that, the resin was dried by methanol. After removal of the resin for 2 hours with the full-protection lysis solution, the filtrate was collected and distilled by rotation. The product was collected and an equal amount of G3-PAMAM added for 1–2 hours’ reaction in DMF, TBTU, and DIEA solution. At the end of the reaction, the white solids were precipitated by adding saturated citric acid aqueous solution, filtrated, and pump-dried. Then, the side-chain protection bases of the product were removed by using trifluoroacetic acid. The solids were precipitated by adding ether and pump-dried for mass spectrometry detection and purification. The final product was yellow powder. The control product of PEG-PAMAM was synthesized by using PAMAM directly linking Fmoc-PEG2,000-2Cl(TRT)-resin. To determine the modification level of RGD peptide on PAMAM scaffolds, the RGD-PEG-PAMAM conjugates (~10 mg) were dissolved in 1 mL dimethyl sulfoxide, and then detected by 1H nuclear magnetic resonance-imaging spectrometry (400 MHz Avance III Ascend; Bruker Corporation, Billerica, MA, USA). To determine nanoparticle size and ζ-potential, the RGD-PEG-PAMAM conjugates (1 mg) were dissolved in 1 mL DMEM medium with free serum, and then detected with an analyzer (Nanotrac Wave II; Nikkiso, Tokyo, Japan).
Cytotoxicity assay
HepG2, Huh7, and 293A cells were suspended in media containing 10% FBS at a density of 1.6×106 cells/mL. To make the spheroid formation, 0.1 mL (1.6×105) cell solution and 0.3 mL media were plated on 24-well ultralow-attachment plates (ULAPs, Corning, NY, USA), RGD-PEG-PAMAM (1 mg/mL in free serum media) was then added to a final concentration of 25, 50, 100, 200, 400, or 800 μg/mL. Then, the ULAPs were rotated at 80 rpm for 15 minutes on an orbital shaker every 30 minutes for three times. On the next day, the supernatant of wells in ULAPs was removed as far as possible (eliminating the fluorescence background), and the new culture solution was added. After 24 hours’ culture, cellular spheroids were collected by centrifugation at 1,000 rpm for 5 minutes. Cell viability (%) was analyzed using a Cell Counting Kit 8 (CCK-8; Beyotime, Shanghai, People’s Republic of China). CCK-8 solution (10 μL) was added to each well and incubated at 37°C for 2 hours. Absorbance at a wavelength of 450 nm was read on the μQuant Universal Microplate Spectrophotometer (Bio-Tek Instruments, Winooski, VT, USA). Cell viability was calculated using the equation:
|
|
where [OD]test, [OD]control, and [OD]blank represented the absorbance values of the wells with RGD-PEG-PAMAM, without RGD-PEG-PAMAM, and without RGD-PEG-PAMAM and cells, respectively. For each sample, the absorbance was the average value measured from five wells in parallel.
RGD-PEG-PAMAM conjugate binding to cells in 2-D culture and incorporation in 3-D spheroid culture
RGD-PEG-PAMAM conjugates were dissolved in DMEM with free serum at a concentration of 1 mg/mL. Huh7 cells were seeded at 3×105 cells/well on a six-well plate (Nunc; Thermo Fisher Scientific) with a sterile coverslip (24×24 mm) and allowed to attach to glass surfaces for 12 hours. The supernatants were then removed and replaced with medium containing RGD-PEG-PAMAM at 100 μg/mL under 10% FBS. To study the incorporation of RGD-PAMAM in 3-D spheroids, Huh7 cells were suspended in media containing 10% FBS at a density of 1.6×106 cells/mL. RGD-PEG-PAMAM conjugates were then added to a concentration of 100 μg/mL. The cells (1.6×105) were plated on ULAPs for spheroid formation, as described in the “Cytotoxicity assay” section. After 24 hours’ culture, the supernatants of these samples were removed and gently washed with warm Dulbecco’s phosphate-buffered saline (PBS) three times and stained with DAPI (4′,6-diamidino-2-phenylindole) for 10 minutes. The samples were imaged under a Zeiss LSM 780 Confocal Microscope (Carl Zeiss Meditec AG, Jena, Germany).
Measurement of intracellular ROS
Intracellular ROS concentrations were analyzed with an ROS-assay kit (Beyotime) using 2′,7′-dichlorodihydrofluorescin diacetate (DCFH-DA). Briefly, Huh7 and 293A cells were seeded in 96-well luminescence test plates (JetBio, Guangzhou, People’s Republic of China) at a density of 4×104 cells/mL. After 24 hours of cell attachment, plates were washed with 100 μL/well PBS and the cells treated with G3-PAMAM, increasing concentrations of PEG-PAMAM and RGD-PEG-PAMAM prepared in 10% FBS containing media. Loxapine (0.5 mg/mL) was used as positive control to validate the protocol. All incubations were performed at 37°C in a 5% CO2 humidified incubator. Three replicate wells were used for each control and test concentrations per 96-well microplate. After the specified incubation time (1, 2, 4, and 6 hours), the plates were washed with 100 μL/well PBS, and then 100 μL/well of 10 μM DCFH-DA was added to each well. The plates were incubated at 37°C for 30 minutes. The cells were washed with PBS three times. Fluorescence was measured with a versatile fluorescent light analyzer (Orion II Microplate Luminometer; Berthold Detection Systems GmbH, Germany) at an excitation/emission wavelength of 488/525 nm. The mean fluorescence values of DCFH-DA-loaded cells were corrected by subtracting the autofluorescence background. In order to exclude the effect of the FITC fluorescence of RGD-PEG-PAMAM material, the RGD-PEG-PAMAM-treatment group was specially set up with a control group without DCFHDA-loading. Quantification of intracellular ROS was calculated as:
|
|
Cell-proliferation assays
The HepG2, Huh7, and 293A cells were plated on ULAPs for spheroid formation as described in the “Cytotoxicity assay” section. RGD-PEG-PAMAM conjugates were then added to a concentration of 100 μg/mL. At 1, 2, 3, 5, and 7 days of culture, cellular spheroids cultured under different conditions were centrifuged and collected. After the supernatants in each tube were collected (for determination), 20 μL CellTiter 96® Aqueous One Solution (Promega Corporation, Fitchburg, WI, USA) was diluted five times with fresh medium and added to each tube. The fully mixed cell solutions were then transferred into the 96-well plates, and then kept at 37°C in the dark for 2 hours. The absorbance of the supernatant was measured at 490 nm using the Bio-Tek μQuant Universal Microplate Spectrophotometer. The control groups included the cells treated with PEG-PAMAM and the cells treated with nothing (blank). For each sample, absorbance was the average value measured from five wells in parallel. Meanwhile, at 1, 2, and 3 days of culture, the effects of different concentrations of RGD-PEG-PAMAM on the formation of the spheres were observed. Phase-contrast and fluorescent images were then recorded using an Eclipse TE2000-U fluorescence microscope (Nikon, Tokyo, Japan).
Albumin secretion and urea production
The albumin secretion of samples collected as described in the “Cell-proliferation assays” section on days 1, 2, 3, 5, and 7 of cultivation was measured using a human albumin ELISA kit (RayBiotech, Norcross, GA, USA). Urea production in Huh7 cells was measured as reported previously21 with modifications. Briefly, the Huh7 cells were plated on ULAPs (with 100 μg/mL RGD-PEG-PAMAM) for spheroid formation as described in the “Cytotoxicity assay” section. On the next day, when the spheroid formation had completed, the supernatant was replaced by 400 μL standard buffer with 5, 10, 20 mM NH4Cl. After being incubated for 12 hours, 50 μL supernatant was used for the detection assay. Sulfuric acid (25 μL, 1:3, 1.8305 g/mL)/phosphoric acid (1.874 g/mL) was added and urea production determined by adding 25 μL α-isonitrosopropiophenone (9% in absolute ethanol), incubating at 100°C in the dark for 15 minutes, and measuring absorbance at 490 nm. Meanwhile, the standard curve and regression equation for detection were produced using a series of urea standards with different concentrations, which were used to calculate the urea concentration in the samples.
Immunoblotting
Cell protein was extracted by conventional methods. Each treatment was repeated in triplicate. A total of 40 μg of protein was subjected to 12% sodium dodecyl sulfate polyacrylamide-gel electrophoresis and electrophoretic transfer to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA). Protein blots were incubated separately with a panel of specific antibodies from Santa Cruz Biotechnology Inc (Dallas, TX, USA), which included anti-FAK (1:1,000, sc-271195), p-FAK (1:1,000, sc-374668), anti-ERK1/2 (1:1,000, sc-135900), anti-Arg1 (1:1,000, sc-20150), anti-OTC (1:1,000, sc-102051) and anti-β-actin (1:4,000, sc-47778), and antibodies from Cell Signaling Technology, which included anti-p-ERK1/2 (1:1,000, 137F5), anti-AKT (1:1,000, 40D4), anti-p-AKT (1:1,000, 587F11) overnight at 4°C and then incubated with different horseradish peroxidase-conjugated secondary antibodies at room temperature for 1 hour. Visualization of the immunoreactive proteins was undertaken with a chemiluminescence kit (BeyoECL Plus; Beyotime). Intensities of band signals were quantified using the densitometric software Quantity One (Bio-Rad, Hercules, CA, USA), and relative intensity to internal control (β-actin) was calculated. All measurements were repeated in triplicate.
Statistical analysis
Results are expressed as mean ± standard deviation. Data were analyzed by Student’s t-test for pairwise comparisons and analysis of variance with Bonferroni posttest for multiple comparisons, using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA). Statistical significance was set at P<0.05.
Results
Construction of RGD-polypeptide dendrimer (RGD-PEG-PAMAM)
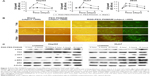
During cell adhesion and growth, as the ligand of some integrins, RGD depends on the conformation of amino acid residues; one of the indicators for measuring its biological activity is the distance between guanidine and carboxyl on both sides of the RGD polypeptide. The fourth amino acid linked to the RGD sequence has significant impact on the activity of RGD. Studies have shown that if the fourth amino acid is phenylalanine (F), then RGD has the highest activity.22 From this, we link the RGDF polypeptide with the spherical polymer G3-PAMAM, which has 32 amino terminals, by using PEG2,000 as a bridging molecule. For smooth linkage, two protective amino acids – cysteine and glycine (CG) – are added in the front of RGDF, and the lysine (K) for fluorescence labeling is added behind RGDF. The product CGRGDFK-PEG2,000-PAMAM (abbreviated RGD-PEG-PAMAM) is formed finally. According to the design program (Figure 1A), we have successfully synthesized a PAMAM conjugate that carries the end of the RGD polypeptide after a series of experiments. The nuclear magnetic resonance spectra showed that RGD-PEG-APMAM had several peaks in the range of 1.6–2.2 and 3.4–4.9 ppm (Figure 1B). In addition, the examination showed that average particle size and ζ-potential of RGD-PEG-APMAM conjugates were 7.45 nm and 104 mV, respectively.
RGD-PEG-PAMAM conjugate cytotoxicity, cell-binding property in 2-D culture, and incorporation in multicellular spheroids
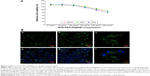
As RGD-PEG-PAMAM is used in cell culture, its biological compatibility and toxicity to cells must be considered. We added different concentrations of RGD-PEG-PAMAM (final concentration 25–800 μg/mL) into three types of culture solutions (HepG2, Huh7, and 293A) for 24 hours to test its impact on the activity of cells (cytotoxicity test). From the trend, in the range of final concentration 25–100 μg/mL, the activities of three kinds of cells were 129.28%–136.5%; in the range of final concentration 200–800 μg/mL, the activities gradually decreased from 115.04% to 82.16% (Figure 2A). A final concentration of 100 μg/mL was the relative maximum concentration, which had the smallest impact on cells. Therefore, 100 μg/mL was selected to be the final concentration to conduct relative research.
To investigate the binding property, the RGD-PEG-PAMAM conjugates were incubated with Huh7 cells in 2-D culture and subjected to microscopic investigation. As in Figure 2B (a and b), RGD-PEG-PAMAM can be seen along the edge of the spreading cells, showing the outlines of cell membranes. The fluorescent curves also seemed inhomogeneous, indicating the discontinuous distribution of the conjugates on the cell membrane. The result demonstrated that RGD-PEG-PAMAM was able to bind with Huh7 cells. To examine RGD-PEG-PAMAM conjugate incorporation into 3-D spheroids, the material was added into culture when Huh7 cells were induced to aggregate. The confocal analysis showed that most of the Huh7 cells were spherical after forming multicellular aggregates. The fluorescent stains indicated that the location of RGD-PEG-PAMAM conjugates was also observed in round curves in correspondence with the outer surface of cells (Figure 2B [c–f]). However, it is hard to determine whether any material had been internalized by the Huh7 cells.
Effects of RGD-PEG-PAMAM on intracellular ROS
Oxidative stress has been established as one of the key factors determining the toxicity of several nanomaterials, including PAMAM.23–26 Because of its importance, the potential of RGD-PEG-PAMAM to elicit such a response should be investigated. The intracellular ROS study was performed at different time points (1, 2, 4, and 6 hours) and with exposure of 2 μM G3-PAMAM and different concentrations of PEG-PAMAM and RGD-PEG-PAMAM. Intracellular ROS production in the Huh7 cells upon the case of a 2-hour exposure to G3-PAMAM, PEG-PAMAM, and RGD-PEG-PAMAM was visualized using fluorescence microscopy. As shown in Figure 3A, compared with the negative control, the cells treated by positive control (loxapine) produced strong fluorescence, but the cells treated by PAMAM or PEG-PAMAM produced weak fluorescence. Meanwhile, in the presence of RGD-PEG-PAMAM, the cells produced brighter fluorescence than with PAMAM or PEG-PAMAM treatment. This may have been due to the FITC-labeled RGD in the RGD-PEG-PAMAM material. This showed that G3-PAMAM, PEG-PAMAM, or RGD-PEG-PAMAM treatment may enable cells to produce less ROS. Next, we further detected the fluorescence quantification using a plate reader, which provides an average of the statistically variable response of Huh7 and 293A cells. The concentration-dependent increase in ROS production for four different time points for the three dendrimer generations was monitored, and the results are shown in Figure 3B. In addition to the Huh7 cells exposed to three concentrations of RGD-PEG-PAMAM for 6 hours as well as the 293A cells exposed to 250 μg/mL RGD-PEG-PAMAM for 4 and 6 hours, the other detection points were not significantly different between the experimental groups.
Effects of RGD-PEG-PAMAM on the proliferation of cell spheres
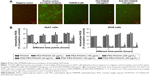
To observe the impact of RGD-PEG-PAMAM on the proliferation of spherical cells, we added RGD-PEG-PAMAM (final concentration 100 μg/mL) into three kinds of culture systems (HepG2, Huh7, and 293A) for 7 days to observe the culture process by using an MTS Cell Proliferation Colorimetric Assay Kit. The result showed that 1) with RGD-PEG-PAMAM, the proliferation of two kinds of hepatic cells reached a peak at day 3 and then decreased; proliferation was not obvious in the control groups, showing a gradual declining trend over time; 2) the OD values of three kinds of cells at 2, 3, 5, 7, and other time points were higher than those in the control groups (P<0.05); and 3) the proliferation trends of nonhepatic 293A cells with or without RGD-PEG-PAMAM were the same. Proliferation with RGD-PEG-PAMAM at all time points was slightly better than that without RGD-PEG-PAMAM. This result indicates that RGD-PEG-PAMAM is more suitable for the culture of hepatic cells, which may be based on the different expression of RGD receptors on the cell surface (Figure 4A).
RGD-PEG-PAMAM was added to the culture solution of Huh7 hepatic cells (final concentration 100 μg/mL) for 5 days to observe the growth of cells and study the impact of RGD-PEG-PAMAM on the formation of cell spheres. The result showed that in 1–3 days of culture, the promotion effect of RGD-PEG-PAMAM on the formation of Huh7 cell spheres was the most significant, and the formed spheres had the largest diameter. The spheres were not significantly changed on days 4 and 5 compared with those on day 3. RGD-PEG-PAMAM in a final concentration of 100 μg/mL was used to culture hepatic cells for 5 days. Cells showed a linear growth in the first 3 days and a peak on the third day. Then, dead cells increased subsequently (Figure 4B). Therefore, RGD-PEG-PAMAM promoted singly floating hepatic cells to aggregate together in a sphere-like growth.
Integrin plays a role in intracellular signal transduction through FAK.27 Cell proliferation and survival are mediated by the FAK–PI3K–AKT and FAK–Ras–MAPK pathways. The activated AKT or ERK can activate a variety of transcription factors and mediate the activity of FAK-regulated gene expression. We detected the expression of FAK, AKT, and ERK in hepatic cells. As shown in Figure 4C, phosphorylated FAK, AKT, and ERK expression was upregulated after HepG2 and Huh7 cells were treated by RGD-PEG-PAMAM for 16 hours. However, when cells were treated with 50 μM LY294002 for 1 hour prior to the stimulation of RGD-PEG-PAMAM, p-AKT was not expressed at all time points, and p-ERK1/2 was weakly expressed at the 16-hour time point. These results indicated that RGD-PEG-PAMAM promoted hepatic cells proliferation through activating the PI3K–AKT and Ras–MAPK pathways.
Effects of RGD-PEG-PAMAM on the expression of albumin and urea-synthesized key enzymes of cell spheres
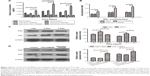
The hepatic cell line Huh7 and nonhepatic cell line 293A were selected in this paper to observe the expression of albumin and urea-synthesized key enzymes of cell spheres in the presence of RGD-PEG-PAMAM in a final concentration of 100 μg/mL. The results showed that for 293A cells, the contents of albumin in all groups were lower at all time points, with insignificant difference; the content of albumin in Huh7 cells was higher than in 293A cells; at 1, 3, and 5 days, the albumin content in Huh7 cells was significantly higher than the group without additives and the PEG-PAMAM group. The albumin content in Huh7 cells was the highest at day 3, ie, up to 9.03 μg/mL, which was 2.22 times that of the group without additives and 2.03 times of the PEG-PAMAM group (Figure 5A). In the presence of ammonium chloride with different concentrations, urea synthesis of Huh7 in the RGD-PEG-PAMAM group was the highest, higher than in the group without additives and the PEG-PAMAM group (P<0.05) (Figure 5B).
Based on this, we further detected the impact of RGD-PEG-PAMAM on the expression of urea-synthesized key enzymes Arg1 and OTC in Huh7 cells sphere. The results (Figure 5C and 5D) showed that the expression of Arg1 and OTC in 3 days was higher than in 1 or 5 days. The difference had statistical significance. In 3 days of culture, the RGD-PEG-PAMAM group was used for comparison with the other two control groups. Results showed that the expression of Arg1 and OTC in the RGD-PEG-PAMAM group was significantly higher than in the group without additives and the PEG-PAMAM group. The difference had statistical significance. RGD-PEG-PAMAM facilitates the aggregation culture of hepatic cells. It can also improve the basic functions of hepatic cells and promote ammonia metabolism of hepatic cells.
Discussion
PAMAM dendrimer contains treelike structures with “branches” radiating from one central core. PAMAM-NH2, one type of PAMAM dendrimer with different functional reactive groups on the external surface, possesses many amine groups on the external surface and contains a large number of amide groups in its branches, which is suitable for PEG modification.28 In this paper, G3-PAMAM was first used to induce PEG transformation, and the RGD-peptide segment was connected to construct new types of dendritic nanomaterials suitable for connecting integrin. The synthesis and structure of the product in this paper have the following characteristics. The material used, G3-PAMAM with the amino terminal, has a small diameter (3.1 nm),28 with weak toxicity and good biological compatibility,29,30 so the toxicity of the product RGD-PEG-PAMAM had minimal impact on the formation of the cultured hepatic cell spheres. Different from the synthesis techniques of other research teams,20 the method we adopted was as follows. Amino acids were linked to PEG2,000 one by one to construct PEG polypeptide, and then an acetyl group was used to protect the polypeptide end. The method for connecting the PEG hydroxyl end with the PAMAM amino terminal can be used to guarantee successful connection. The connection of different amino acids connected behind the RGD-peptide segment has different functions.31 Phenylalanine, which is connected behind RGD, may facilitate the aggregation of hepatic cells. RGD-PEG-PAMAM has a spherical structure and carries more RGD terminals. It can overcome the rigid deficiency of linear connections, and is more conducive to providing a gap for the growth of hepatic cell clusters. These features provide a useful molecular support for spherical dendritic compounds to float on the liquid. Once combined with cells, RGD-PEG-PAMAM can be regarded as a molecular glue, pulling cells together in the cell membrane.
It is important to note that cell toxic damage is also caused by ROS, increased inflammatory marker expression, and DNA damage, dependent upon the chemical nature of the toxicant.25,26 The toxicity produced by PAMAM dendrimers is well correlated with the surface amine groups.32,33 Naha et al25 found that intracellular ROS generation by PAMAM dendrimers is clearly one of the toxic pathways and a clear generation dependence of intracellular increased ROS production (G6 > G5 > G4). This response indicates that the cationic surface amino groups play a direct role in the production of ROS. In addition, Lee et al34 demonstrated that the ROS, cytokine production, and cytotoxicity cascade could be initiated from the internalization of PAMAM dendrimers and their localization in the mitochondria. Therefore, although there may be external stress leading to some degree of indirect toxic response, it is proposed that the principal response is a direct result of internalization of the nanomaterials. In this study, we observed that RGD-PEG-PAMAM-induced cell ROS response was weak. We speculate that there are two relevant factors: 1) the number of amino groups at the end of G3-PAMAM is only half that of G4-PAMAM; 2) the amino groups of G3-PAMAM are replaced by PEG-RGD, which leads the internalization of G3-PAMAM, greatly reduced by the connection of RGD and integrins on the cell surface. Of course, this hypothesis needs to be confirmed by further studies.
Our study shows that RGD-PEG-PAMAM can promote hepatic cell proliferation and affect gene expression. This finding indicates that RGD-PEG-PAMAM can be used as a type of soluble factor to activate an intracellular signaling pathway-like extracellular cell factor to promote the growth of cells. This result is consistent with a previous study that used ligand RGD-peptide segments to construct 3-D cultured hepatic cells of biological material.35,36 Studies have shown that the surface of the hepatic cell has various integrins. The survival of hepatic cells depends on those mediated by β1 integrin when adhering to the liver extracellular matrix.37 Knockdown of β1 integrin will impair the regeneration of the liver,38 and integrin ligand RGD-peptide segments can promote the survival of hepatic cells through activating the β1 integrin–ILK–pAkt pathway.39,40 In addition, RGD-PEG-PAMAM in a final concentration of 100 μg/mL is found to be able to improve the basic functions of hepatic cells and urea-synthesized function. RGD-PEG-PAMAM may be involved in the action of RGD-peptide segments and integrin, and it enhances the action between hepatic cells and matrix, causing hepatic cells to be difficult to remove from the matrix and facilitating the transportation of nutrients.9
The results shown in this paper indicate that the constructed RGD-PEG-PAMAM has biological activities. In addition, in reference to other relevant research (Kim et al pointed out that the synthesized PAMAM polymer can activate the JNK pathway of dental pulp cells and upregulate the expression of related marker genes;41 Jiang et al reported that the synthesized PAMAM polymer can stimulate the proliferation of NIH3T3 cells),20 we can reasonably presume that our product RGD-PEG-PAMAM not only can be used in sphere-aggregate culture of hepatic cells but also has a unique role in the aggregation of other cells. The new hepatic cell-sphere aggregate biological material RGD-PEG-PAMAM is conducive to the simulation of the growth environment of the hepatic cells in vivo and is more suitable for the transportation of nutrients than simple hepatic cell-sphere aggregate cultures. A larger number of cell spheres can be achieved in a short time, rapidly providing adequate functional hepatocytes for the biological artificial liver system.
Conclusion
A new nanostructured PAMAM-dendrimer conjugate (RGD-PEG-PAMAM) decorated by an integrin ligand RGD peptide was successfully constructed. RGD-PEG-PAMAM promotes hepatic cells to aggregate together in a sphere-like growth. Moreover, RGD-PEG-PAMAM improves the basic function and ammonia metabolism of hepatic cells. This study indicates that the hepatocyte sphere treated by RGD-PEG-PAMAM may be a choice for the source of liver cells in biological artificial liver systems, and its potential value needs to be investigated further.
Acknowledgments
This work was supported by the Natural Science Foundation of Fujian Province (2013J01309), the National Natural Science Foundation of China (81470872), and the Construction Project of National Key Clinical Subject of General Surgery. The authors would like to thank Wen Wang from the College of Materials Science and Engineering, Fujian Normal University, for theory and technical support of the synthesis of the RGD-PEG-PAMAM conjugate.
Disclosure
The authors report no conflicts of interest in this work.
References
Nyberg SL. Bridging the gap: advances in artificial liver support. Liver Transpl. 2012;18(Suppl 2):S10–S14. | ||
Zinchenko YS, Schrum LW, Clemens M, Coger RN. Hepatocyte and Kupffer cells co-cultured on micropatterned surfaces to optimize hepatocyte function. Tissue Eng. 2006;12(4):751–761. | ||
Tostões RM, Leite SB, Miranda JP, et al. Perfusion of 3D encapsulated hepatocytes: a synergistic effect enhancing long-term functionality in bioreactors. Biotechnol Bioeng. 2011;108(1):41–49. | ||
Shimada M, Yamashita Y, Tanaka S, et al. Characteristic gene expression induced by polyurethane foam/spheroid culture of hepatoma cell line, Hep G2 as a promising cell source for bioartificial liver. Hepatogastroenterology. 2007;54(75):814–820. | ||
Maringka M, Giri S, Nieber K, Acikgöz A, Bader A. Biotransformation of diazepam in a clinically relevant flat membrane bioreactor model using primary porcine hepatocytes. Fundam Clin Pharmacol. 2011;25(3):343–353. | ||
Yip D, Cho CH. A multicellular 3D heterospheroid model of liver tumor and stromal cells in collagen gel for anti-cancer drug testing. Biochem Biophys Res Commun. 2013;433(3):327–332. | ||
Fujii Y, Nakazawa K, Funatsu K. Intensive promotion of spheroid formation by soluble factors in a hepatocyte-conditioned medium. J Biomater Sci Polym Ed. 2000;11(7):731–745. | ||
Ito M, Taguchi T. Enhanced insulin secretion of physically crosslinked pancreatic beta-cells by using a poly(ethylene glycol) derivative with oleyl groups. Acta Biomater. 2009;5(8):2945–2952. | ||
Du Y, Chia SM, Han R, Chang S, Tang H, Yu H. 3D hepatocyte monolayer on hybrid RGD/galactose substratum. Biomaterials. 2006;27(33):5669–5680. | ||
Du Y, Han R, Wen F, et al. Synthetic sandwich culture of 3D hepatocyte monolayer. Biomaterials. 2008;29(3):290–301. | ||
Esfand R, Tomalia DA. Poly(amidoamine) (PAMAM) dendrimers: from biomimicry to drug delivery and biomedical applications. Drug Discov Today. 2001;6(8):427–436. | ||
Pourjavadi A, Hosseini SH, Alizadeh M, Bennett C. Magnetic pH-responsive nanocarrier with long spacer length and high colloidal stability for controlled delivery of doxorubicin. Colloids Surf B Biointerfaces. 2014;116:49–54. | ||
Yavuz B, Pehlivan SB, Vural I, Unlu N. In vitro/in vivo evaluation of dexamethasone-PAMAM dendrimer complexes for retinal drug delivery. J Pharm Sci. 2015;104(11):3814–3823. | ||
Fu F, Wu Y, Zhu J, Wen S, Shen M, Shi X. Multifunctional lactobionic acid-modified dendrimers for targeted drug delivery to liver cancer cells: investigating the role played by PEG spacer. ACS Appl Mater Interfaces. 2014;6(18):16416–16425. | ||
Shen W, van Dongen MA, Han Y, et al. The role of caveolin-1 and syndecan-4 in the internalization of PEGylated PAMAM dendrimer polyplexes into myoblast and hepatic cells. Eur J Pharm Biopharm. 2014;88(3):658–663. | ||
Li K, Wen S, Larson AC, et al. Multifunctional dendrimer-based nanoparticles for in vivo MR/CT dual-modal molecular imaging of breast cancer. Int J Nanomedicine. 2013;8:2589–2600. | ||
Opina AC, Wong KJ, Griffiths GL, et al. Preparation and long-term biodistribution studies of a PAMAM dendrimer G5-Gd-BnDOTA conjugate for lymphatic imaging. Nanomedicine (Lond). 2015;10(9):1423–1437. | ||
Wu D, Yang J, Li J, et al. Hydroxyapatite-anchored dendrimer for in situ remineralization of human tooth enamel. Biomaterials. 2013;34(21):5036–5047. | ||
Goncalves M, Castro R, Rodrigues J, Tomas H. The effect of PAMAM dendrimers on mesenchymal stem cell viability and differentiation. Curr Med Chem. 2012;19(29):4969–4975. | ||
Jiang LY, Lv B, Luo Y. The effects of an RGD-PAMAM dendrimer conjugate in 3D spheroid culture on cell proliferation, expression and aggregation. Biomaterials. 2013;34(11):2665–2673. | ||
Tang N, Wang Y, Wang X, et al. Stable overexpression of arginase I and ornithine transcarbamylase in HepG2 cells improves its ammonia detoxification. J Cell Biochem. 2012;113(2):518–527. | ||
Park HS, Kim C, Kang YK. Preferred conformations of RGDX tetrapeptides to inhibit the binding of fibrinogen to platelets. Biopolymers. 2002;63(5):298–313. | ||
Nel A, Xia T, Madler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311(5761):622–627. | ||
Lanone S, Boczkowski J. Biomedical applications and potential health risks of nanomaterials: molecular mechanisms. Curr Mol Med. 2006;6(6):651–663. | ||
Naha PC, Davoren M, Lyng FM, Byrne HJ. Reactive oxygen species (ROS) induced cytokine production and cytotoxicity of PAMAM dendrimers in J774A.1 cells. Toxicol Appl Pharmacol. 2010;246(1–2):91–99. | ||
Naha PC, Byrne HJ. Generation of intracellular reactive oxygen species and genotoxicity effect to exposure of nanosized polyamidoamine (PAMAM) dendrimers in PLHC-1 cells in vitro. Aquat Toxicol. 2013;132–133:61–72. | ||
Provenzano PP, Inman DR, Eliceiri KW, Keely PJ. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28(49):4326–4343. | ||
Svenson S, Tomalia DA. Dendrimers in biomedical applications – reflections on the field. Adv Drug Deliv Rev. 2005;57(15):2106–2129. | ||
Boas U, Heegaard PM. Dendrimers in drug research. Chem Soc Rev. 2004;33(1):43–63. | ||
Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. | ||
Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24(24):4385–4415. | ||
Naha PC, Davoren M, Casey A, Byrne HJ. An ecotoxicological study of poly(amidoamine) dendrimers – toward quantitative structure activity relationships. Environ Sci Technol. 2009;43(17):6864–6869. | ||
Maher MA, Naha PC, Mukherjee SP, Byrne HJ. Numerical simulations of in vitro nanoparticle toxicity – the case of poly(amido amine) dendrimers. Toxicol In Vitro. 2014;28(8):1449–1460. | ||
Lee JH, Cha KE, Kim MS, et al. Nanosized polyamidoamine (PAMAM) dendrimer-induced apoptosis mediated by mitochondrial dysfunction. Toxicol Lett. 2009;190(2):202–207. | ||
Xia L, Sakban RB, Qu Y, et al. Tethered spheroids as an in vitro hepatocyte model for drug safety screening. Biomaterials. 2012;33(7):2165–2176. | ||
Shin YM, Jo SY, Park JS, Gwon HJ, Jeong SI, Lim YM. Synergistic effect of dual-functionalized fibrous scaffold with BCP and RGD containing peptide for improved osteogenic differentiation. Macromol Biosci. 2014;14(8):1190–1198. | ||
Pinkse GG, Voorhoeve MP, Noteborn M, Terpstra OT, Bruijn JA, De Heer E. Hepatocyte survival depends on beta1-integrin-mediated attachment of hepatocytes to hepatic extracellular matrix. Liver Int. 2004;24(3):218–226. | ||
Speicher T, Siegenthaler B, Bogorad RL, et al. Knockdown and knockout of β1-integrin in hepatocytes impairs liver regeneration through inhibition of growth factor signalling. Nat Commun. 2014;5:3862. | ||
Pinkse GG, Jiawan-Lalai R, Bruijn JA, de Heer E. RGD peptides confer survival to hepatocytes via the β1-integrin-ILK-pAkt pathway. J Hepatol. 2005;42(1):87–93. | ||
Bray LJ, Suzuki S, Harkin DG, Chirila TV. Incorporation of exogenous RGD peptide and inter-species blending as strategies for enhancing human corneal limbal epithelial cell growth on Bombyx mori silk fibroin membranes. J Funct Biomater. 2013;4(2):74–88. | ||
Kim JK, Shukla R, Casagrande L, et al. Differentiating dental pulp cells via RGD-dendrimer conjugates. J Dent Res. 2010;89(12):1433–1438. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


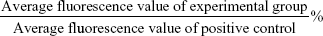


 , RGD-PEG structure linking on the amino terminal of PAMAM. (A) Synthetic scheme for the synthesis of RGD-PEG-PAMAM conjugates. (B) NMR spectra of RGD-PEG-PAMAM, PEG-PAMAM, and PAMAM. The emergence of peptide peaks indicates coupling of peptides to PAMAM.
, RGD-PEG structure linking on the amino terminal of PAMAM. (A) Synthetic scheme for the synthesis of RGD-PEG-PAMAM conjugates. (B) NMR spectra of RGD-PEG-PAMAM, PEG-PAMAM, and PAMAM. The emergence of peptide peaks indicates coupling of peptides to PAMAM.