Back to Journals » International Journal of Nanomedicine » Volume 12
Aptamer-conjugated PEGylated quantum dots targeting epidermal growth factor receptor variant III for fluorescence imaging of glioma
Authors Tang J, Huang N, Zhang X, Zhou T, Tan Y, Pi J, Pi L, Cheng S, Zheng H , Cheng Y
Received 24 January 2017
Accepted for publication 18 April 2017
Published 22 May 2017 Volume 2017:12 Pages 3899—3911
DOI https://doi.org/10.2147/IJN.S133166
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Linlin Sun
Jiaze Tang,1 Ning Huang,1 Xiang Zhang,1,2 Tao Zhou,3 Ying Tan,1,4 Jiangli Pi,5 Li Pi,1 Si Cheng,6 Huzhi Zheng,5 Yuan Cheng1
1Department of Neurosurgery, The Second Affiliated Hospital of Chongqing Medical University, 2Chongqing Key Laboratory of Ultrasound Molecular Imaging, Institute of Ultrasound Imaging, 3Chongqing Key Laboratory of Biochemistry and Molecular Pharmacology, 4Institute of Life Sciences, Chongqing Medical University, 5Key Laboratory on Luminescent and Real-Time Analytical Chemistry, Ministry of Education, College of Chemistry and Chemical Engineering, Southwest University, 6Department of Orthopaedics, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
Abstract: The extent of resection is a significant prognostic factor in glioma patients. However, the maximum safe resection level is difficult to determine due to the inherent infiltrative character of tumors. Recently, fluorescence-guided surgery has emerged as a new technique that allows safe resection of glioma. In this study, we constructed a new kind of quantum dot (QD)-labeled aptamer (QD-Apt) nanoprobe by conjugating aptamer 32 (A32) to the QDs surface, which can specially bind to the tumors. A32 is a single-stranded DNA capable of binding to the epidermal growth factor receptor variant III (EGFRvIII) specially distributed on the surface of glioma cells. To detect the expression of EGFRvIII in human brain tissues, 120 specimens, including 110 glioma tissues and 10 normal brain tissues, were examined by immunohistochemistry, and the results showed that the rate of positive expression of EGFRvIII in the glioma tissues was 41.82%, and 0.00% in normal brain tissues. Besides, the physiochemical properties of QD-Apt nanoparticles (NPs) were thoroughly characterized. Biocompatibility of the NPs was evaluated, and the results suggested that the QD-Apt was nontoxic in vivo and vitro. Furthermore, the use of the QD-Apt in labeling glioma cell lines and human brain glioma tissues, and target gliomas in situ was also investigated. We found that not only could QD-Apt specially bind to the U87-EGFRvIII glioma cells but also bind to human glioma tissues in vitro. Fluorescence imaging in vivo with orthotopic glioma model mice bearing U87-EGFRvIII showed that QD-Apt could penetrate the blood–brain barrier and then selectively accumulate in the tumors through binding to EGFRvIII, and consequently, generate a strong fluorescence, which contributed to the margins of gliomas that were visualized clearly, and thus, help the surgeons realize the maximum safe resection of glioma. In addition, QD-Apt can also be applied in preoperative diagnosis and postoperative examination of glioma. Therefore, these achievements facilitate the use of tumor-targeted fluorescence imaging in the diagnosis, surgical resection, and postoperative examination of glioma.
Keywords: quantum dot, aptamer, EGFRvIII, brain glioma, tumor-targeted imaging, extent of resection
Introduction
Glioma is the most common and lethal malignant brain tumor.1 The GBM patients usually live for only 12–15 months after diagnosis due to lack of effective treatment.2 Surgical resection is the most effective therapy for glioma patients, and maximum safe resection is crucial to improve the prognosis of patients with low- or high-grade gliomas.3–6 However, balancing maximum cytoreduction with preservation of normal brain tissue is complicated due to the infiltrative nature of the tumors. Over 90% of recurrent tumors develop within a 2–3 cm margin of the primary site and are thought to arise from microscopic glioma cells that infiltrate the area surrounding the original tumor.7 The extent of resection has been proven to be one of the most important predictors of OS, PFS, and neurological outcome.8–10 Thus, maximum safe resection becomes tremendously important. Therefore, a technique that can delineate the boundary of tumors to realize maximum safe resection and protect the normal brain tissues is urgently needed. It has been proved that intraoperative fluorescein used to guide the surgical resection can improve the OS and PFS of patients.11 Although literature reports the use of fluorescence, such as 5-aminolevulinate acid, sodium fluorescein, and so on, in guiding the resection of gliomas, defects such as low quantum fluorescence yield and low photostability limit their clinical application.12,13
Nanotechnology plays an increasingly important role in clinical application due to its many unique physicochemical properties and great versatility.14 QDs are fluorescent NPs that have many prominent advantages in the fields of bio-imaging for diagnostic and therapeutic applications. Compared with the conventional fluorescent materials, QDs exhibit many excellent advantages, including water solubility, broad absorption with narrow emission spectra, high quantum fluorescence yield, low photobleaching, and chemical stability.15 Troublesomely, because of the release of Cd2+ from the CdSe core, QDs are toxic in vivo and vitro. However, when ZnS constitutes the shell of QDs and is coated with PEG, the biocompatibility of QDs is greatly improved.16–18 Moreover, coating the NPs with PEG enable them to avoid clearance by reticuloendothelial system and plasma protein adsorption, and thus prolong the blood circulation of NPs in the body, and finally raise their probability of reaching their target tissue.19–21
Gliomas always generate genetic rearrangements and mutations.22 Comprehensive genomic analysis of GBM has identified that the amplification of EGFR gene is the most frequent genetic alteration associated with GBM.23 Moreover, EGFR gene amplifications always couple with gene rearrangements. The most common alteration is seen in EGFRvIII which has a truncated extracellular domain with ligand-independent constitutive activity, resulting in enhancing the proliferation, invasion,24–26 radiation resistance,27 and chemotherapy resistance22,28 of glioma while leading to poor prognosis.29 Unlike EGFR, EGFRvIII has not been detected in normal tissues, but has been identified in tumors.30–32
In recent years, aptamers have been developed as a novel recognition probes constituted with uniquely folded short RNA or ssDNA oligonucleotides which can bind to the targets with high specificity and affinity.33 In addition, compared with antibodies or polypeptides, aptamers have many advantages including lower immunogenicity, thermal stability, ease of modification, higher specificity, smaller size, lower production cost, and no toxicity.34,35 Compared with RNA aptamers, DNA aptamers are more stable and cheaper to synthesize. A32 (DNA), which specially binds to EGFRvIII protein, was first discovered by Tan et al36 and then used to conjugate small interfering RNA and deliver it into U87-EGFRvIII cells.37
To overcome the disadvantages of conventional fluorophores and implement the maximum safe resection, this study engineered a nanoprobe, biotin-aptamer-conjugated streptavidin-PEG-CdSe/ZnS QDs (QD-Apt), for preoperative diagnosis, intraoperative resection, and postoperative examination of glioma. We synthesized and thoroughly characterized the QD-Apt. Furthermore, the nanoprobe was used to label the cell lines and the human brain glioma tissues in vitro. Moreover, orthotopic glioma model in mice was used to verify if the nanoprobe can traverse BBB and specially target the tumor expressing EGFRvIII.
Materials and methods
Reagents
U87 and HUVEC cell lines were purchased from Shanghai Life Academy of Sciences Cell Library (Shanghai, People’s Republic of China). U87-EGFRvIII, a U87 glioma cell line that overexpresses EGFRvIII, was friendly provided by Prof Haizhong Feng at the Shanghai Jiao Tong University (Shanghai, People’s Republic of China). Antibodies against human EGFRvIII were obtained from Biorbyt (Cambridge, UK). Primary antibodies β-actin and GFAP were purchased from Abcam (Cambridge, UK). Horseradish peroxidase-conjugated or fluorescence-conjugated secondary antibodies and DAPI were purchased from Zhongshan Golden Bridge Biotechnology (Beijing, People’s Republic of China). Streptavidin-modified PEG-QDs solution (SA-QDs, λem =605±5 nm) was purchased from Wuhan Jiayuan Quantum Dots Co., Ltd (Wuhan, People’s Republic of China). Biotinylated A32,36 with an ssDNA sequence of 5′-biotin-GCAATGGTACGGTACTTCCTGAATGTTGTTTTTTCTCTTTTCTATAGTACAAAAGTGCACGCTACTTTGCTAA-3′, was synthesized by Sangon Biochemistry Company (Shanghai, People’s Republic of China).
EGFRvIII expression in human gliomas
For this study, 110 de-identified surgical specimens of glioma patients and 10 normal brain tissues were collected from the Second Affiliated Hospital of Chongqing Medical University during January 2008 to April 2015. The collection and the use of the surgical specimens were carried out in accordance with the Institutional Review Board-approved protocols. Moreover, the study was reviewed and approved by the institutional ethics committee at the Second Affiliated Hospital of Chongqing Medical University. All patients signed informed consent to participate in this study and permit the further use of their tumor tissues. The pathological diagnosis and grading were performed according to the World Health Organization classification. IHC was used to detect the expression levels of EGFRvIII in glioma specimens and normal brain tissues.
Characterization of QD-Apt
Biotinylated A32 which specially binds to EGFRvIII was dissolved in tris–ethylenediamine tetraacetic acid buffer and mixed with streptavidin-PEG-CdSe/ZnS QDs (SA-QD) at a concentration ratio of 10:1 and incubated for 24 h at 4°C, and then the aptamer-conjugated PEG-CdSe/ZnS QDs (QD-Apt) was added. The unconjugated aptamer was removed by desalination column (NAP™-5; GE Healthcare, Little Chalfont, UK). The sizes of NPs were analyzed using an NP sizer (Zetasizer Nano ZS; Malvern Instruments Ltd., Malvern, UK). Photoluminescence measurement was performed with a fluorescence analyzer (F-4500; Hitachi, Tokyo, Japan). To validate the connection between aptamer and QDs, QD-Apt nanoprobe solution (10 μL) was injected into the loading hole with SA-QDs (10 μL) in another hole as the control. After electrophoresis for 1 h (10 V/cm), the gel was illuminated with an ultraviolet transilluminator (Bio-Print; Vilber Lourmat) and imaged with Quantity One software.
Cell culture
All cell lines were maintained in a 5% CO2 atmosphere at 37°C in Dulbecco’s Modified Eagle’s Medium High Glucose supplemented with 100 U/mL of penicillin, 100 mg/mL of streptomycin, and 10% fetal bovine serum (Hyclone, Logan, UT, USA).
Western blot
HUVEC, U87, and U87-EGFRvIII cells were lysed with RIPA Lysis Buffer (Beyotime Institute of Biotechnology, Beijing, People’s Republic of China) containing 1 mM phenylmethane sulfonyl fluoride. The concentrations of proteins were measured by the bicinchoninic acid method. Then, equivalent amounts of protein were separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes. Thereafter, the membranes were soaked in PBS containing 5% skimmed milk, and then incubated with primary antibodies overnight at 4°C and horseradish peroxidase-labeled secondary antibodies for 1 h at 37°C. Finally, the proteins were visualized with ECL reagent by Quantity One-4.6 software (Bio-Rad, Hercules, CA, USA).
Reverse transcription-polymerase chain reaction
Total cellular DNA and RNA were extracted using RNAiso Plus (TaKaRa) according to the manufacture’s protocol. The concentrations of nucleic acid samples were measured by a spectrophotometric detector. Thereafter, RNA specimens were reverse-transcribed to synthesize cDNA using the Primescript RT Reagent Kit (TaKaRa). The primer sequence of EGFRvIII was forward 5′-CTTCGGGGAGCAGCGATGCGAC-3′ and reverse 5′-ACCAATACCTATTCCGTTACAC-3′, and the product size was 243 bp; for GAPDH, which was used as a standard, the primer sequence was forward 5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse 5′-GTAGAGGCAGGGATGATGTTCT-3′, and the product size was 132 bp. Amplification conditions were as follows: 95°C for 5 min, followed by 45 cycles at 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s, and lastly, extension at 72°C for 10 min.
Immunohistochemistry
Paraffin-embedded samples were cut and mounted on slides. After dewaxed in xylene and rehydrated in graded alcohols, the sections were antigen-retrieved in 10 mM citrate buffer for 10 min at 100°C in a microwave oven and blocked with 5% goat serum for 30 min. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide, and incubated with EGFRvIII antibody overnight at 4°C. Thereafter, sections were incubated with goat anti-rabbit secondary antibody for 30 min at 37°C, and then immunostained with 3,3′-diaminobenzidine and counterstained with hematoxylin. The primary antibodies were replaced with PBS as the negative controls which were processed along with the samples. Afterward, the slides were reviewed by two independent pathologists using a microscope (DM6000 B; Leica, Wetzlar, Germany). Immunohistochemical staining of EGFRvIII was calculated as both percentage of positive cells and color intensity. According to the percentage of the positive cells, the grades were classified as “0” (negative), “1” (<10%), “2” (10%–50%), and “3” (>50%). The intensity was scored as follows: “0” (absent), “1” (light yellow), “2” (yellowish brown), and “3” (brown). The expression level of EGFRvIII was evaluated by SI, which was calculated using the following formula: SI = proportion score × intensity score. SI of 0, 1–2, 3–4, and ≥6 was classified as negative (-), low expression (+), medium expression (++), and high expression (+++), respectively.
Cell viability assay
U87-EGFRvIII, U87, and HUVEC cells were seeded into 96-well culture plates (5×103 cells/well) and incubated overnight. Meanwhile, the blank group wells were added with 100 μL medium without cells. After 24 h, PBS, QDs, or QD-Apt at different concentrations were added to each group and incubated for 24, 48, and 72 h, respectively. At designated time points, 10 μL of the CCK-8 solution was added to each well and incubated for 2 h. Then, the absorbance values (A450 nm) were measured by a microplate reader (Bio-Rad 680). The cell viability was calculated using the following formula: Viability (%) = (QDs or QD-Apt groups − Blank groups)/(PBS groups − Blank groups ) ×100.
Cell fluorescence imaging in vitro
U87-EGFRvIII, U87, and HUVEC cells were seeded on the coverslips for 24 h in 24-well plates, and then were treated with 10 nM of QDs or QD-Apt for 4 h. For the competition experiment, U87-EGFRvIII cells were simultaneously incubated with QD-Apt and a 10-fold excess of free aptamers. Afterward, these cells were fixed in 4% paraformaldehyde for 20 min, and stained with DAPI for 5 min. A laser scanning confocal microscope was used to observe the fluorescence imaging of those cells. All images were taken under the same parameters, and fluorescence intensities were analyzed by NIS-Elements Viewer software 4.20.
Binding assay by flow cytometry
U87, U87MG-EGFRvIII, and HUVEC cells were trypsinized and washed twice with PBS. Cells (2×105) were incubated with QDs or QD-Apt at a final concentration of 10 nM in 100 μL PBS at 37°C for 4 h. Then, the cells were washed thrice with PBS, suspended in 200 μL PBS, and analyzed by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Fluorescence imaging in human glioma tissues
Paraffin sections of normal brain and glioma tissues with or without EGFRvIII expression were deparaffinized in xylene and dehydrated in a graded ethanol series. Then, antigen retrieval was performed in citric acid buffer, and nonspecific binding was blocked by incubating with 5% goat serum for 30 min. The sections were incubated with GFAP antibody overnight at 4°C, and then with goat anti-rabbit secondary antibody for 1 h at 37°C. Next, the sections were incubated with QD-Apt or QDs at 37°C for 4 h, and then stained with DAPI for 5 min at room temperature. Finally, a laser scanning confocal microscope was used to observe the fluorescence imaging of those slides.
In vivo toxicity study
All animal procedures were approved by the ethics committee of Chongqing Medical University (People’s Republic of China) and conducted according to the European Community Council Directive 010/63/UE. Efforts were made to minimize the number of animals used and their suffering. Besides, animals were maintained in accordance with the guidelines of the National Institute of Nutrition. Male C57BL/6 athymic nude mice (6–8 weeks) were administered with either PEG-QDs or QD-Apt via tail vein injection (5 nM). After 1, 7, 14, 21, and 28 days, the weights of mice were recorded, respectively. Moreover, the brain, heart, liver, spleen, lungs, and kidneys were harvested and stained with hematoxylin and eosin for histopathological study using light microscopy (DM6000 B; Leica).
Orthotopic brain glioma model in mice
Male nude mice aged 6–8 weeks (Laboratory Animal Center, Medical University of Chongqing) were anesthetized, sterilized, and immobilized on a stereotaxic instrument. A sagittal incision was made to expose the bregma. Then, we drilled a hole in the cranium, which was 2.5 mm right lateral and 0.5 mm anterior to the bregma (right hemisphere). U87 and U87-EGFRvIII glioma cells (1×106/5 μL) were injected at a depth of 3.3 mm from the brain surface, respectively. The needle was slowly pushed in for 10 min, and then withdrawn slowly in another 5 min. Tumor growth was monitored by MRI after 2 weeks.
Biodistribution and targeted imaging of glioma in situ
Six nude mice bearing U87-EGFRvIII brain tumors were randomly divided into two groups (1 and 2) with three rats in each group. Similarly, another six nude mice bearing U87 brain tumors were randomly divided into two groups (3 and 4) with three rats in each group. In group 1 and group 3, mice were injected with QD-Apt (5 nmol) via the tail vein. In group 2 and group 4, mice were similarly injected with PEG-QDs (5 nmol). After 6 h of circulation, all mice were anesthetized, and NPs-targeted imaging of glioma was performed in vivo (330 nm excitation filter and 605 nm emission filter) using the Living Image System (Maestro EX IVIS; Cambridge Research Instruments, Woburn, MA, USA). Afterward, every mouse was sacrificed and dissected to isolate the tumor, para-tumor, normal brain, heart, liver, spleen, lungs, and kidneys. These tissues (0.1 g) were cut into small pieces and dissolved in RIPA Lysis Buffer. After centrifugation at 12,000 g for 15 min at 4°C, the supernatant (100 μL) was collected to determine the uptake of nanomaterials using a fluorescence plate reader.38 Furthermore, the tumors and normal brain tissues were frozen and cut into 10-μm sections. After staining with GFAP antibody, fluorescence-conjugated secondary antibodies (green light), and DAPI (blue light), the sections were observed.
Statistical analysis
Statistical analyses were performed by SPSS 19.0. Statistical differences among groups were analyzed by analysis of variance or t-test. P<0.05 was considered statistically significant. All data are shown as mean ± standard deviation.
Results
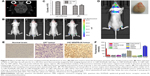
EGFRvIII expression in human glioma tissues and cell lines
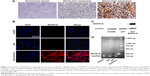
A total of 120 paraffin-embedded clinical specimens, including 10 normal brains and 60 grade I–II and 50 grade III–IV glioma tissues, were examined by immunohistochemical analysis to determine the clinical relevance of EGFRvIII in human glioma. In accord with other literatures, we discovered that 41.82% of the glioma patients expressed EGFRvIII, while the 10 normal tissues had no expression of EGFRvIII (Figure 1A). The high expression (SI ≥6) rate of EGFRvIII in low-grade glioma was 6.67%; however, it was 18.00% in high-grade glioma, and there was a significant difference between the rates (P<0.001).
In addition, to detect the expression of EGFRvIII in HUVEC, U87, and U87-EGFRvIII cell lines, Western blot, RT-PCR, and immunofluorescence analysis were performed. The results demonstrated that there was no expression of EGFRvIII in HUVEC and U87 cells. In contrast, U87-EGFRvIII showed a strong positive expression of EGFRvIII (Figure 1B–D).
Characteristics of QD and QD-Apt
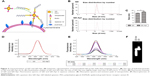
A schematic representation of the preparation of QD-Apt nanoprobe and its recognition of EGFRvIII is shown in Figure 2A. The NP sizer indicated that the sizes of QD-Apt were slightly increased compared with QD, but all of their sizes were about 20 nm (Figure 2B and C). When the aptamer was conjugated to SA-QDs by the biotin-streptavidin cross-linking technique, fluorescence intensity was not significantly changed (Figure 2D). Photoluminescence measurement revealed that the maximum luminescence wavelength of both QD and QD-Apt was 605±5 nm, and their absorption peak was observed at 330 nm (Figure 2E). To identify whether SA-QDs and biotin-aptamer conjugated, electrophoresis was performed. As Figure 2F shows, the velocity of QD-Apt NP was obviously faster than that of QD as QD-Apt had a larger charge-to-mass ratio than QD, which suggested that biotinylated aptamer and streptavidin-QD had been connected.
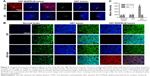
Targeting of bioprobe NPs to glioma cell lines in vitro
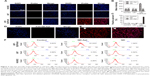
To investigate the characteristics of nanoprobes for tumor targeting in vitro, we used QD and QD-Apt to label the cells and then observed them with a laser scanning confocal microscope. After 4-h incubation with U87-EGFRvIII, the nanoprobes were found to bind to cytomembrane and be uptaken into the cellular cytoplasm (Figure 3A). However, in U87 and HUVEC cells, no expression of EGFRvIII and no significant fluorescence signal were detected, regardless of whether the cells were incubated with QD or QD-Apt. Besides, the fluorescence intensity was blocked effectively when a 10-fold excess of free A32 was added (Figure 3D). Furthermore, flow cytometry was used to validate the labeling effect of the bioprobes, and the results showed that the U87-EGFRvIII incubated with QD-Apt had a high labeling rate (81.53%) which was much higher than the other groups (Figure 3F).
Fluorescence imaging in human glioma tissues
QDs and QD-Apt were used to label the normal brain tissues, EGFRvIII(-) glioma tissues, and EGFRvIII(+) glioma tissues which had been identified by IHC. We discovered that QD-Apt was specially bound to the EGFRvIII(+) glioma tissues. On the contrary, both the normal brain tissues and EGFRvIII(-) glioma tissues were not labeled with QD or QD-Apt (Figure 4A). Based on the above results, we can draw a conclusion that QD-Apt can specially recognize and bind to EGFRvIII(+) glioma tissues.
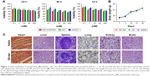
Biodistribution and targeted imaging of glioma in situ
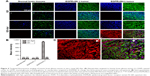
Orthotopic glioma models were used to examine the targeting effect of QD-Apt. After 2 weeks of establishing the model, all the mice were evaluated by MRI (Figure 5A). At 4 weeks of tumor development, a whole-body fluorescence imaging was performed in all mice. After tail vein injection of QDs or QD-Apt for 6 h, the mice were monitored by the IVIS Imaging System (Figure 5B). Intriguingly, the mice harboring U87-EGFRvIII tumor administered with QD-Apt (group 1) had strong fluorescence signals in tumor areas, while the group administered with QDs (group 2) had no obvious fluorescence signals in the tumor region. In addition, the mice bearing U87 tumors (groups 3 and 4) had no significant fluorescence in tumor areas, regardless of whether they were administered with QD-Apt or QDs. Further, a quantitative analysis of the fluorescence intensity in the tumor regions was performed in each group, and the results suggested that the tumors in group 1 mice exhibited a remarkable fluorescence intensity in contrast with the normal brain tissues, while the other groups exhibited no significantly different fluorescence between the tumors and the normal brain tissues (Figure 5C). Moreover, the brains were dissected from mice in group 1 and compared with the corresponding fluorescence imaging picture. As Figure 5D shows, the outline of brain tumor was consistent with that of the fluorescence imaging displayed. In addition, IHC was used to determine the expression of EGFRvIII in the tumors and normal brain tissues of mice. There was a strong expression of EGFRvIII in the mice bearing U87-EGFRvIII tumors, while no expression of EGFRvIII was seen in the mice bearing U87 tumors (Figure 5E). Besides, tumors, para-tumors, normal brain tissues, and other organs were dissected from mice, and their fluorescence intensity was measured. The results suggested that the QD-Apt predominantly accumulated in the tumors (EGFRvIII(+)), liver, and kidneys, and low levels were observed in the para-tumor, normal brain, spleen, heart, and lungs (Figure 5F).
Furthermore, the tumors were frozen and cut into 10-μm sections. Thereafter, the sections were stained with DAPI and then observed by a fluorescence microscope. As shown in the Figure 6A, the group 1 mice had a clear boundary between the tumors and the normal brain tissues. Moreover, the frozen sections of the tumors and the normal brain tissues were labeled with GFAP and DAPI. The normal astrocytes and glioma cells were recognized by GFAP (green), while QD-Apt (red) was only observed in U87-EGFRvIII tumors (Figure 6B).
Safety evaluation of QDs and QD-Apt
The biological safety of QDs in vitro and in vivo is the prerequisite for its application. To detect the toxicity of the nanoprobe, CCK-8 was applied to evaluate the cell viability of HUVEC, U87, and U87-EGFRvIII cells after treatment with various concentrations of QD-Apt for 24, 48, and 72 h. As Figure 7A shows, an unremarkable cytotoxicity was exerted by the nanoprobes at concentrations varying from 0.1 to 100 nM. Further, short- and long-term toxicity was detected in vivo. A total of 12 nude mice were randomly divided into three groups (n=3), and groups 1, 2, and 3 were administered with saline, PEG-QDs, and QD-Apt via tail vein injection, respectively. The body weight was recorded after injection at 1, 7, 14, 21, and 28 days. The results showed that the body weights of the three groups of mice were not significantly different (Figure 7B). Then, the mice were sacrificed, and their major organs (brain, heart, liver, spleen, lungs, and kidneys) were collected to analyze the pathologic changes. The results revealed that no tissue damage, necrosis, or inflammation was observed among the three groups of mice (Figure 7C).
Discussion
Surgical resection is crucial to improve the prognosis of patients with low- or high-grade gliomas,3–6 and hence, maximum safe resection becomes tremendously important. However, it is difficult to resect low- and high-grade gliomas due to their propensity to infiltrate deep into surrounding parenchyma. Therefore, a technique that can allow visualizing the boundary of tumors in order to realize the maximum safe resection and protect the normal brain tissues is urgently needed. Firstly, we need to search for a biomarker which is expressed only in tumors but not in normal tissues. EGFRvIII had been detected in gliomas,24 lung cancer,39 breast cancer,40 squamous cell carcinoma of the head and neck,41 and colorectal cancer,42 but not in normal tissues. In this study, we also validated that EGFRvIII is a tumor-specific antigen; in other words, it is only expressed in tumors but not in normal tissues. Hence, EGFRvIII is an ideal target to treat gliomas. Secondly, similar to many pharmaceuticals, the antibody EGFRvIII cannot cross the BBB due to its large sizes. Therefore, a small molecule which can penetrate the BBB and specially bind to EGFRvIII is needed. Aptamers are oligonucleotides (DNA or RNA) that are developed through an in vitro selection process known as SELEX, and can bind to a wide range of target molecules with a high affinity and specificity. Compared with an antibody, an aptamer has many advantages including lower immunogenicity, thermal stability, ease of modification, higher specificity, smaller size, and lower production cost.34 A32, which was first discovered by Tan et al,36 can specially bind to EGFRvIII protein and is applied as a bioprobe in this study. Thirdly, a material with prominent fluorescent properties and a small size was indispensable in this study. Compared with conventional fluorescent materials, QDs have prominent advantages. However, the toxicity of QDs is an important concern. Nevertheless, QDs with a CdSe core and a ZnS shell, coated with PEG, can not only greatly decrease the toxicity but also show little interaction with tissue constituents and bind to the target more efficiently.17,43 In correspondence with other studies, we confirm that the QDs and QD-Apt was nontoxic and exhibited optimal biocompatibility. Moreover, studies have validated that the safe dose of QDs was 1 μM in vitro and 15 nM in vivo, which was higher than the concentrations used in this study.44,45 Lastly, biotinylated A32 was conjugated with streptavidin-PEG-CdSe/ZnS QDs, and a nanoprobe (QD-Apt) was synthesized. QD-Apt possessed a small size (22 nm), and its fluorescence properties did not change significantly compared with QDs. Applications of many pharmaceuticals and imaging agents are limited by their inability to cross the BBB due to their large sizes. QD-Apt was only about 20 nm in diameter, and small enough to cross the BBB, which was demonstrated in mice using Living Image System in this study. Moreover, accumulated literature reports that the PEGylated NPs which are larger than our nanoprobe can pass the BBB.38
QD-Apt was applied to label the EGFRvIII in vivo and in vitro in this study. The results suggested that QD-Apt could not only specially bind to the glioma cell lines expressing EGFRvIII but also connect to the EGFRvIII(+) glioma tissues with a high specificity in vitro and exhibit strong fluorescence intensity. Therefore, QD-Apt can be applied to detect the EGFRvIII in tumor tissues and as an alternative to IHC. It has been reported that the technique of QD-IHC spectral imaging is more sensitive than conventional IHC.46 In this study, QD-Apt also exhibited excellent performance in gliomas imaging in vivo. QD-Apt was injected via tail vein which then traversed the BBB and remained in circulation for a long period. The U87-EGFRvIII xenograft tumors were labeled with bright fluorescence, and the boundaries of tumors were visualized clearly in situ, which might guide the surgeons to implement the maximum safe resection for gliomas. Moreover, the QD-Apt can also be applied in diagnosis, and pre- and postoperative examination of gliomas. Likewise, the QD-Apt can also be applied to other tumors expressing EGFRvIII, such as lung cancer, breast cancer, and squamous cell carcinoma of the head and neck. Although a number of studies have reported that QDs were used to visualize the gliomas, there still remained some shortcomings. Gd-DOTA-Ag2S QDs were used to image the gliomas by diffusion, but they lacked specific targeting ability and led to low specificity and efficacy.19 A QD-labeled aptamer GBI-10, which can specially bind to tenascin-C, was used as a nanoprobe for imaging glioma cell lines in vitro.47 However, due to poor specificity and lack of verification in vivo, its clinical application was limited. Therefore, the QD-Apt which we engineered can be considered an ideal nanoprobe for imaging gliomas due to its high specificity, nontoxicity, strong fluorescence, and effective traverse of BBB. However, every bean has its black. The QD-Apt can only be applied to the tumors expressing EGFRvIII.
In this study, we combined the advantages of PEG-QDs with the superiorities of aptamers and engineered a nanoprobe (QD-Apt) with fascinating performances. Glioma cells, human brain glioma tissues, and animals were administered with the nanoprobe. Based on the results, QD-Apt exhibited a strong fluorescence signal and specially bound to EGFRvIII protein in vitro and vivo. Moreover, QD-Apt can allow visualizing the boundary of tumors clearly in orthotopic brain tumor mice bearing U87-EGFRvIII gliomas, and so, it can help the surgeons realize the maximum safe resection of gliomas. Therefore, this nanoprobe (QD-Apt) can be viewed as a novel prospect for the molecular diagnosis, image-guided surgery, and postoperative examination of gliomas.
Abbreviations
A32, aptamer 32; BBB, blood–brain barrier; DAPI, 4′,6-diamidino-2-phenylindole; EGFRvIII, epidermal growth factor receptor variant III; GBM, glioblastoma; IHC, immunohistochemistry; MRI, magnetic resonance imaging; NP, nanoparticle; OS, overall survival; PBS, phosphate buffer saline; PEG, polyethylene glycol; PFS, progression-free survival; QD, quantum dot; QD-Apt, QD-labeled aptamer; RT-PCR, reverse transcription-polymerase chain reaction; SI, staining index; ssDNA, single-stranded DNA.
Acknowledgments
The authors thank Prof Haizhong Feng (Shanghai Jiao Tong University) for kindly providing U87-EGFRvIII. They also thank Yanghao Zhou and all others who provided help in this study. This work was supported by grants from the National Natural Science Foundation of China (nos 81201066, 81371674, 81471676, and 81401505) and Chongqing Graduate Student Research Innovation Fund (no CYS15123).
Author contributions
Huzhi Zheng and Yuan Cheng conceived and designed the study. Jiaze Tang, Ning Huang, Xiang Zhang, and Tao Zhou performed the experiments. Ying Tan, Jiangli Pi, Li Pi, and Si Cheng analyzed the data. Jiaze Tang drafted the manuscript. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that no competing financial interests exist in this work.
References
Sturm D, Bender S, Jones DT, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14(2):92–107. | ||
Clarke J, Butowski N, Chang S. Recent advances in therapy for glioblastoma. Arch Neurol. 2010;67(3):279–283. | ||
Kawaguchi T, Sonoda Y, Shibahara I, et al. Impact of gross total resection in patients with WHO grade III glioma harboring the IDH 1/2 mutation without the 1p/19q co-deletion. J Neurooncol. 2016;129(3):505–514. | ||
Altieri R, Zenga F, Fontanella MM, et al. Glioma surgery: technological advances to achieve a maximal safe resection. Surg Technol Int. 2015;27:297–302. | ||
Kashi AS, Rakhsha A, Houshyari M. Overall survival in adult patients with low-grade, supratentorial glioma: ten years’ follow up at a single institution. Electron Physician. 2015;7(3):1114–1120. | ||
Hollon T, Hervey-Jumper SL, Sagher O, Orringer DA. Advances in the surgical management of low-grade glioma. Semin Radiat Oncol. 2015;25(3):181–188. | ||
Wise J. Laser treatment opens temporary window in blood-brain barrier. BMJ. 2016;352:i1154. | ||
Marko NF, Weil RJ, Schroeder JL, Lang FF, Suki D, Sawaya RE. Extent of resection of glioblastoma revisited: personalized survival modeling facilitates more accurate survival prediction and supports a maximum-safe-resection approach to surgery. J Clin Oncol. 2014;32(8):774–782. | ||
Li-Feng C, Yang Y, Xiao-Dong M, et al. Optimizing the extent of resection and minimizing the morbidity in insular high-grade glioma surgery by high-field intraoperative MRI guidance. Turk Neurosurg. Epub 2016 Aug 23. | ||
Sanai N, Polley MY, McDermott MW, Parsa AT, Berger MS. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg. 2011;115(1):3–8. | ||
Neira JA, Ung TH, Sims JS, et al. Aggressive resection at the infiltrative margins of glioblastoma facilitated by intraoperative fluorescein guidance. J Neurosurg. 2016;1–12. [Epub ahead of print]. | ||
Eljamel S. 5-ALA fluorescence image guided resection of glioblastoma multiforme: a meta-analysis of the literature. Int J Mol Sci. 2015;16(5):10443–10456. | ||
Su X, Huang QF, Chen HL, Chen J. Fluorescence-guided resection of high-grade gliomas: a systematic review and meta-analysis. Photodiagnosis Photodyn Ther. 2014;11(4):451–458. | ||
Tonga GY, Saha K, Rotello VM. 25th anniversary article: interfacing nanoparticles and biology: new strategies for biomedicine. Adv Mater. 2014;26(3):359–370. | ||
Wu P, Yan XP. Doped quantum dots for chemo/biosensing and bioimaging. Chem Soc Rev. 2013;42(12):5489–5521. | ||
Bruneau A, Fortier M, Gagne F, et al. In vitro immunotoxicology of quantum dots and comparison with dissolved cadmium and tellurium. Environ Toxicol. 2015;30(1):9–25. | ||
Tang Y, Han S, Liu H, et al. The role of surface chemistry in determining in vivo biodistribution and toxicity of CdSe/ZnS core-shell quantum dots. Biomaterials. 2013;34(34):8741–8755. | ||
Ye L, Yong KT, Liu L, et al. A pilot study in non-human primates shows no adverse response to intravenous injection of quantum dots. Nat Nanotechnol. 2012;7(7):453–458. | ||
Li C, Cao L, Zhang Y, et al. Preoperative detection and intraoperative visualization of brain tumors for more precise surgery: a new dual-modality MRI and NIR nanoprobe. Small. 2015;11(35):4517–4525. | ||
He X, Ma N. An overview of recent advances in quantum dots for biomedical applications. Colloids Surf B Biointerfaces. 2014;124:118–131. | ||
Kolate A, Baradia D, Patil S, Vhora I, Kore G, Misra A. PEG – a versatile conjugating ligand for drugs and drug delivery systems. J Control Release. 2014;192:67–81. | ||
Adamson C, Kanu OO, Mehta AI, et al. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18(8):1061–1083. | ||
Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. | ||
Liu Z, Han L, Dong Y, et al. EGFRvIII/integrin β3 interaction in hypoxic and vitronectin enriching microenvironment promote GBM progression and metastasis. Oncotarget. 2016;7(4):4680–4694. | ||
Sangar V, Funk CC, Kusebauch U, Campbell DS, Moritz RL, Price ND. Quantitative proteomic analysis reveals effects of epidermal growth factor receptor (EGFR) on invasion-promoting proteins secreted by glioblastoma cells. Mol Cell Proteomics. 2014;13(10):2618–2631. | ||
Yin J, Park G, Kim TH, et al. Pigment epithelium-derived factor (PEDF) expression induced by EGFRvIII promotes self-renewal and tumor progression of glioma stem cells. PLoS Biol. 2015;13(5):e1002152. | ||
Lammering G, Hewit TH, Valerie K, et al. EGFRvIII-mediated radioresistance through a strong cytoprotective response. Oncogene. 2003;22(36):5545–5553. | ||
Liu XJ, Wu WT, Wu WH, et al. A minority subpopulation of CD133(+)/EGFRvIII(+)/EGFR(-) cells acquires stemness and contributes to gefitinib resistance. CNS Neurosci Ther. 2013;19(7):494–502. | ||
Jain R, Poisson LM, Gutman D, et al. Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology. 2014;272(2):484–493. | ||
Greenall SA, Donoghue JF, Van Sinderen M, et al. EGFRvIII-mediated transactivation of receptor tyrosine kinases in glioma: mechanism and therapeutic implications. Oncogene. 2015;34(41):5277–5287. | ||
Sampson JH, Archer GE, Mitchell DA, Heimberger AB, Bigner DD. Tumor-specific immunotherapy targeting the EGFRvIII mutation in patients with malignant glioma. Semin Immunol. 2008;20(5):267–275. | ||
Wikstrand CJ, Hale LP, Batra SK, et al. Monoclonal antibodies against EGFRvIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Cancer Res. 1995;55(14):3140–3148. | ||
Sun H, Tan W, Zu Y. Aptamers: versatile molecular recognition probes for cancer detection. Analyst. 2016;141(2):403–415. | ||
Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol Ther Nucleic Acids. 2014;3:e182. | ||
Camorani S, Cerchia L. Oligonucleotide aptamers for glioma targeting: an update. Cent Nerv Syst Agents Med Chem. 2015;15(2):126–137. | ||
Tan Y, Shi YS, Wu XD, et al. DNA aptamers that target human glioblastoma multiforme cells overexpressing epidermal growth factor receptor variant III in vitro. Acta Pharmacol Sin. 2013;34(12):1491–1498. | ||
Zhang X, Liang H, Tan Y, Wu X, Li S, Shi Y. A U87-EGFRvIII cell-specific aptamer mediates small interfering RNA delivery. Biomed Rep. 2014;2(4):495–499. | ||
Huang N, Cheng S, Zhang X, et al. Efficacy of NGR peptide-modified PEGylated quantum dots for crossing the blood-brain barrier and targeted fluorescence imaging of glioma and tumor vasculature. Nanomedicine. 2017;13(1):83–93. | ||
Duan J, Wang Z, Bai H, et al. Epidermal growth factor receptor variant III mutation in Chinese patients with squamous cell cancer of the lung. Thorac Cancer. 2015;6(3):319–326. | ||
Xu W, Bi Y, Zhang J, et al. Synergistic antitumor efficacy against the EGFRvIII+HER2+ breast cancers by combining trastuzumab with anti-EGFRvIII antibody CH12. Oncotarget. 2015;6(36):38840–38853. | ||
Tinhofer I, Klinghammer KF, Weichert W, et al. Interference of amphiregulin and EGFRvIII expression with outcome of patients with squamous cell carcinoma of the head and neck (SCCHN) receiving cetuximab-docetaxel treatment. J Clin Oncol. 2011;29(15 Suppl):5508. | ||
Khelwatty S, Essapen S, Bagwan I, Green M, Seddon A, Modjtahedi H. The impact of co-expression of wild-type EGFR and its ligands determined by immunohistochemistry for response to treatment with cetuximab in patients with metastatic colorectal cancer. Oncotarget. 2017;8(5):7666–7677. | ||
Nekolla K, Kick K, Sellner S, et al. Influence of surface modifications on the spatiotemporal microdistribution of quantum dots in vivo. Small. 2016;12(19):2641–2651. | ||
Kirchner C, Liedl T, Kudera S, et al. Cytotoxicity of colloidal CdSe and CdSe/ZnS nanoparticles. Nano Lett. 2005;5(2):331–338. | ||
Hauck TS, Anderson RE, Fischer HC, Newbigging S, Chan WC. In vivo quantum-dot toxicity assessment. Small. 2010;6(1):138–144. | ||
Xu H, Chen C, He Y, et al. Analysis of cancer marker in tissues with Hadamard transform fluorescence spectral microscopic imaging. J Fluoresc. 2015;25(2):397–402. | ||
Chen XC, Deng YL, Lin Y, et al. Quantum dot-labeled aptamer nanoprobes specifically targeting glioma cells. Nanotechnology. 2008;19(23):235105. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.