Back to Journals » Infection and Drug Resistance » Volume 13
Antimicrobial Susceptibility Pattern and Associated Factors of Pediatric Septicemia in Southern Ethiopia
Authors Ameya G, Weldemedhin T, Tsalla T , Gebremeskel F
Received 22 August 2020
Accepted for publication 7 October 2020
Published 30 October 2020 Volume 2020:13 Pages 3895—3905
DOI https://doi.org/10.2147/IDR.S278293
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Gemechu Ameya,1 Temesgen Weldemedhin,2 Tsegaye Tsalla,3 Feleke Gebremeskel4
1Department of Medical Laboratory Sciences, Kotebe Metropolitan University, Addis Ababa, Ethiopia; 2Department of Public Health, Wolaita Sodo University, Wolaita Sodo, Ethiopia; 3Department of Medical Laboratory Sciences, College of Medicine and Health Sciences, Arba Minch University, Arba Minch, Ethiopia; 4School of Public Health, College of Medicine and Health Sciences, Arba Minch University, Arba Minch, Ethiopia
Correspondence: Gemechu Ameya
Department of Medical Laboratory Sciences, Kotebe Metropolitan University, P.O. Box: 3268, Addis Ababa, Ethiopia
Tel +251-91-783-7681
Email [email protected]
Background: Septicemia is one of the major causes of morbidity and mortality in pediatric patients throughout the world. Drug-resistant pathogens are one of the major challenges to control. The study aimed to identify the major etiological agents, antimicrobial susceptibility pattern and associated factors of septicemia among pediatric patients in southern Ethiopia.
Methods: A cross-sectional study was conducted on pediatric patients. Blood samples were cultured and antimicrobial susceptibility testing was conducted by Kirby-Bauer disc diffusion techniques. Data were collected by pre-tested questionnaire to identify potential associated factors of septicemia. A bivariate logistic regression analysis was used and adjusted odds ratio with 95% CI at ≤ 0.05 level of significance was computed to determine the presence and strength of the association.
Results: Of 238 participants, 27 (11.3%) of them had a positive blood culture. Staphylococcus aureus (32.2%), coagulase negative Staphylococci (25%), and Klebsiella pneumoniae (14.3%) were the predominant isolates. The isolated bacteria showed high rates of resistance to amoxicillin, ceftriaxone, streptomycin and ampicillin. Multi-drug resistance (MDR) was observed in 82.1% of the isolates. Being infant [AOR=4.18, 95% CI, (1.3– 13.0)], admission > 10 days [AOR=5.54, 95% CI, (1.51– 20.41)], burn [AOR=3.55, 95% CI, (1.02– 12.38)] and wound cases [AOR=5.52, 95% CI, (1.50– 20.34)] were associated with pediatric septicemia.
Conclusion: Gram positive bacteria were the predominant isolates and majority of isolates were MDR pathogens. Very young age, prolonged hospital stays, burn and wound cases were associated with pediatric septicemia. Establishing antibiotic stewardship is mandatory to minimize the high prevalence of drug resistance.
Keywords: antimicrobial susceptibility, multi-drug resistance, pediatric, septicemia
Introduction
Septicemia is caused by the presence of microorganisms within the bloodstream and their dissemination throughout the body with evidence of systemic responses towards those microbes with variable severity.1 The introduction of bacteria to the bloodstream can be from the lungs, genitourinary tract, gastrointestinal tract, skin or soft tissue.2 The pathogenesis of the case may vary depending on the virulence of the pathogen, the portal of entry, the susceptibility and response of the host, and the temporal evolution of the condition.2 The predominant etiologic agents and their antimicrobial resistance rate may vary with geographic area and time period.3 This systemic dissemination of microbes can cause a life-threatening illness that gets worse quickly from their spreading and releasing of toxins in the blood. As a result, septicemia requires rapid and aggressive diagnosis and antimicrobial treatment.4
Globally, 31.5 million cases occur each year and septicemia is the leading cause of death worldwide in the pediatric population resulting in an estimated 7.5 million deaths annually.5,6 In sub-Saharan African countries, septicemia is an important cause of illness and death in children, accounting for 30–70% of illness, making it a significant health problem.5,7 Septicemia is very common in children and can lead to complications such as shock, multi-organ failure and death.4
Multiple factors may be associated with pediatric sepsis. It may vary based on geographic area and health-care setup. Some of the identified associated factors of pediatric sepsis are presence of indwelling intravenous devices, use of steroids and immunomodulators, prolonged hospitalization, chronic antibiotic therapy, surgery, burns or bedsores and serious injuries.8–10 Underlying diseases such as chronic kidney disease,11 hematologic malignancies (HMs),12 immune suppressed individuals and HIV/AIDS2 can be associated with septicemia.
Multi-drug resistance in various bacterial pathogens has reached a pandemic level during the last two decades.13 This resistance pattern varies in accordance with geographic and regional location, health-care setup, and existing practice.14,15 The timely and appropriate use of antimicrobial drugs is very important and the only way to treat septicemia. However, antibiotics resistance is a fast-growing problem in developing countries.7 Inappropriate treatment of sepsis aggravates the disease which leads to death of patients and emergence of new drug- resistant strains. This has become a serious health problem with many economic and social impacts all over the world.15 The increasing frequency of antimicrobial resistance found in bloodstream infections is of great concern for African countries where access to care and broad-spectrum antimicrobials is often limited.16 The infections caused by multi-drug resistant strains result in prolonged hospital stays, increased risk of death, and require treatment with more expensive antibiotics.16 Research conducted in Ethiopia revealed that drug resistance is increasing to commonly used antibiotics in the country.7
Only a few studies have been conducted to investigate the epidemiology and to provide viable alternative approaches in the management of septicemia in Ethiopia.1,5,7,15,17 No research has been conducted on pediatric septicemia in the study area. The aim of this study is to identify the etiological agents and antimicrobial susceptibility patterns in septicemia and its associated factors among pediatric patients attending Arba Minch general hospital, southern Ethiopia.
Materials and Methods
Study Design and Setting
An institutional based cross-sectional study design was conducted on pediatric patients attending the pediatric emergency, inpatient and outpatient departments of Arba Minch general hospital of southern Ethiopia. Arba Minch general hospital is in Arba Minch town, which is in Gamo zone, a regional state in in the south of the country. The hospital serves more than 1.5 million people.
Study Population
The study participants were sampled from pediatric patients in ranging in age from 1 month to 15 years old who were visiting the emergency, inpatient and outpatient departments of Arba Minch General Hospital during the data collection period (September 2018 to January 2019). Pediatric patient with clinical symptoms of septicemia were included in the study. Study participants who took antibiotic treatment within the last two weeks during the data collection time were excluded. Pediatric patients without care givers during the data collection period were also excluded from the study.
Sample Size Determination and Sampling Procedure
Sample size was determined using a single population proportion formula. The prevalence of septicemia and associated factors and multi-drug resistance proportion were considered to determine the maximum sample size. The maximum sample size was obtained from a study conducted in Addis Ababa, Ethiopia,1 in which the hospital admission was 16.9% (p= 0.17), with margin of error (d= 0.05) and 95% confidence interval; the total sample size became 217. Then 10% non-respondent rate was added and the final sample size became 238.
A systematic random sampling technique was used to select the study participants. The skip interval was computed by taking the previous number of patients who visited the pediatric wards in a similar time frame. The k value was calculated by N/n formula; while N was the number of patients who visited the hospital in the similar period and n was the sample size. Then an integer between 1 to k was randomly selected to determine a random start point. Finally the sampling was done by every Kth interval.
Data Collection Procedures and Instrument
Semi-structured pretested questionnaire was used to collect data related to socio-demographic, socio-economic, clinical characteristics and potential associated factors after reviewing previous similar studies.1,5,7,15,17 Other relevant information such as previous antibacterial therapy, application of indwelling medical device, prolonged hospital stays, medical conditions and other factors for each patient were collected from patient charts. The data were collected by face to face interview.
Major Laboratory Examinations
Blood Specimen Collection
The vein puncture site was disinfected with 70% alcohol and 2% tincture of iodine before collecting. A venous blood sample of 5 mL was collected for children older than 2 years and 2–3 mL for 2-year-old and younger patients. For each patent two samples were collected from different sites aseptically. The blood sample of each pediatric patient was collected before the beginning of antibiotic treatment.
Blood Culture
The collected blood samples were inoculated into blood culture bottles of tryptic soya broth (Oxoid, Hampshire, UK) by maintaining a minimum of 1:10 blood to broth ratio. The bottles were labeled with each patient’s identification number, date and time of collection. The bottles containing specimens were transported to laboratory within 30 min at room temperature. Then blood culture bottles were incubated at 37°C and inspected daily for the presence of visible sign of microbial growth such as floccular deposits on the top of the layer, sub-surface turbidity, hemolysis, surface pellicle, gas production and/or coagulation of broth for 7 consecutive days. If any sign of microbial growth was observed in a blood culture bottle, sub-culture was done according to the method stated below.
Sub-Culture of Primary Blood Culture
For blood cultures that showed signs of microbial growth, sub-cultures were made onto blood culture plate (BAP), chocolate culture plate (CAP), and MacConkey & Mannitol salt agar (Oxoid, Hampshire, UK). The MacConkey & Mannitol salt agar plates were incubated in aerobic atmosphere whereas the blood and chocolate agar plates in candle jar at 37°C for 24–48 hours. Bacterial growths on the sub-cultured plate were isolated by their characteristic appearance such as colony morphology, effect on culture media, Gram reaction and other specific identification panels for biochemical reactions.
Bacteria Identification
The pure cultures of isolated organisms from the sub-culture plates were subjected to biochemical tests for further identification of the pathogen. The biochemical tests were done according to their Gram reaction. Gram negative organisms were identified using biochemical tests such as indole test, carbohydrate fermentation reaction in triple sugar iron agar, citrate utilization, urease test, motility test, lysine decarboxylase, lysine deaminase, and oxidase test. For Gram positive bacteria biochemical tests such as catalase, coagulase, and mannitol fermentation were used for identification according to methods described in CLSI.18
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing for all bacterial isolates was done by disc diffusion assay on Mueller-Hinton agar plates according to Kirby-Bauer technique.18 Standard inoculums were adjusted to 0.5 McFarland standard and four drug disks were used in a single 110 mm plate.
The following antimicrobial disks were used in the given concentrations: ampicillin (AMP) (10 μg), amoxicillin-clavulanic acid (AUG) (30 μg), ceftriaxone (CTR) (30 μg), cephalothin (CLT) (30 μg), ciprofloxacin (CPR) (5 μg), chloramphenicol (CHL) (30 μg), doxycycline (DOX) (30 μg), erythromycin (ERY) (15 μg), gentamicin (GEN) (10 μg), kanamycin (K) (30 μg), cefoxitin (CXT) (30 μg), penicillin (PEN) (10 IU), streptomycin (STR) (10 μg), tetracycline (TET) (30 μg), trimethoprim-sulphamethoxazole (COT) (1.25/23.75μg) and amoxicillin (AMC) (30 μg). The result was interpreted by measuring zone of inhibition as sensitive, intermediate, and resistant according to the standardized CLSI.
Quality Control
Quality control of the questionnaires was done before starting the actual data collection. Pre-test questionnaire was done on 5% of the sample size one week before data collection. The completeness of data was checked before data entry. Standard operating procedures were strictly followed throughout the laboratory analysis. Quality control of the culture media was done whenever a new batch of media was prepared. Visual inspections for cracks in media, unequal fill, hemolysis, evidence of freezing, presence of air bubbles and any sign of contamination were conducted before inoculating the culture media. Performance of the culture media was checked by standard known bacteria specifically Escherichia coli ATCC 25,922, S. aureus ATCC 25,923, Enterococcus faecalis (ATCC-29,212) and Pseudomonas aeruginosa (ATCC-27,853).
Data Processing and Analysis
Data were analyzed using SPSS version 20.0 software package. Data completeness and consistency were checked by running cross tabulation of each variable. A bivariate logistic regression was performed to show any association between independent variables and the outcome variable (septicemia). A variable with p-value ≤ 0.25 in the bivariate logistic regression were included in multivariable logistic regression analysis to identify variables that independently associated with the outcome variable. The presence and strength of association between independent variables and outcome variable were computed by adjusted odds ratio with 95% CI and Hosmer–Lemeshow test was used to test the fitness of model.
Ethical Approval and Consent to Participate
The study was conducted in accordance with the Declaration of Helsinki. An ethical clearance was obtained from Arba Minch University, College of Medicine and Health Sciences institutional ethical review board. Before data collection, a permission letter was obtained from the relevant departments of the Arba Minch general hospital. Prior to sample collection participants were informed clearly about the objective and procedure of the study. Written informed consent and assent was obtained from all participants and their care givers. Participation in the study was fully voluntary. All information obtained in this study was kept confidential at all levels and utilized only for the study. For positive findings, we communicated with the attending physician for appropriate treatment of the pediatric patients.
Results
Socio-Demographic Characteristics of Study Participants
A total of 238 pediatric patients participated in the study. Out of these participants, 120 (50.4%) were females. The mean age and standard deviation of the participants were 2.25-year-old ± 1.024. The infants were slightly above one-quarter while children with age range from 1 to 4 years old were more than one-third of the participants. Underweight children (BMI < 18.5kg/m2) accounted for 18% of the participants. More than half of the study participants were urban dwellers (Table 1).
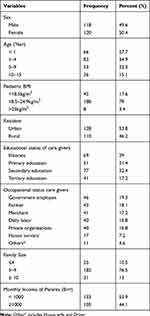 |
Table 1 Socio-Demographic & Socio-Economic Characteristics of Study Participants and Their Care Givers |
Out of all pediatric care givers about 29% of them were illiterate (unable to read or write). The majority of the care givers were governmental employees, followed by farmers. The monthly income of 56% of care givers was less than 1000 Ethiopian Birr (< USD 40). Nearly three out of four care givers had a family size of 5– 9 while about 13% of them had 10 and more (Table 1).
Clinical Characteristics of Study Participants
Among study participants, slightly less than two-thirds of the patients attended the outpatients department while the others were admitted in pediatrics emergency and pediatrics inpatient ward. From all admitted pediatric patients, only 21 (24.7%) were admitted for more than 10 days at the time of data/sample collection. The length of hospital stay from the date of admission to blood sample collection was 1–21 days with a mean of 3 days admission. Usage of intravenous device during treatment was observed in 61 (25.6%) of the study participants and about 4.2% of the participants had used antibiotics for a prolonged time. Among the study participants, 2.5%were HIV/AIDS positive (Table 2).
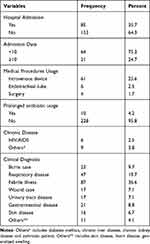 |
Table 2 Clinical Characteristics of Study Participants |
Magnitude and Bacterial Isolates
Out of 238 study participants, 27 (11.3%) blood culture results were positive for different bacterial species. From the positive cultures 28 bacteria were identified and one of the blood cultures showed mixed growth. Gram positive bacterial species were the predominant (57.1%) isolate. The Gram positive bacteria isolated were S. aureus (32.2%) and coagulase negative Staphylococci (CONS) (25.0%). Among Gram negative bacteria K. pneumoniae (14.3%) and E. coli (10.7%) were predominantly isolatedfollowed by Salmonella species (7.1%), Proteus mirabilis (7.1%) and P. aeruginosa (3.6%) (Figure 1).
 |
Figure 1 Distribution of bacterial species isolated in study participants. |
Factors Associated with Pediatric Septicemia
All socio-demographic and clinical factors of study participants were analyzed for their association to septicemia by using bivariate analysis with backward logistic regression. Age group between 1 to 11 months had a statistically significant association to septicemia [AOR= 4.18, 95 CI, (1.34, 13.00)]. Participants who were admitted to hospital admission for more than 10 days were about 6 times more likely to develop septicemia [AOR= 5.54, 95 CI, (1.51–20.41)]. Patients with burn injuries were about 3.5 times more likely to develop septicemia compared with the other participants [AOR=3.55, 95 CI, (1.02–12.38)]. Participants with wounds were about 5.5 times more likely to develop septicemia compared with their counterparts [AOR=5.52, 95 CI, (1.50–20.34)] (Table 3).
 |
Table 3 Factors Associated with Pediatric Septicemia in Study Participants |
Antimicrobial Susceptibility Patterns of Bacterial Isolates
Fifteen antimicrobial drug disks were used to check susceptibility pattern of Gram positive bacterial isolates. All Gram positive bacterial isolates showed resistance to ceftriaxone, high resistance to amoxicillin (93.8), ciprofloxacin, tetracycline and erythromycin. On the other hand, the lowest resistance rate was observed against cefoxitin, chloramphenicol and amoxicillin-clavulanic acid while resistance was not observed to cephalothin. More than 77.8% of S. aureus isolates were resistant to amoxicillin, ceftriaxone, doxycycline, tetracycline, erythromycin, ciprofloxacin, penicillin, ampicillin and trimethoprim-sulfamethoxazole. In contrast the lowest resistance rate was observed for chloramphenicol (11.1%), and amoxicillin-clavulanic acid. From the total S. aureus isolates about 22.2% were methicillin resistant S. aureus (MRSA). Coagulase negative staphylococci isolates were highly resistant to ceftriaxone and amoxicillin, however the lowest resistance was observed to chloramphenicol, ampicillin, cefoxitin and doxycycline. All CONS isolates showed no resistance to cephalothin and amoxicillin-clavulanic acid (Table 4).
 |
Table 4 Antimicrobial Susceptibility Pattern of Gram-Positive Bacteria Isolated from Study Participants |
A total of 12 different antibacterial drug disks were used to examine susceptibility patterns of Gram negative bacterial isolates. All Gram negative isolates showed resistance to streptomycin and amoxicillin. High resistance was observed against cephalothin, ampicillin, trimethoprim-sulfamethoxazole, ciprofloxacin, doxycycline, tetracycline, amoxicillin-clavulanic acid and gentamicin. K. pnemoniae were resistant to amoxicillin, ampicillin, cephalothin, amoxicillin-clavulanic acid and streptomycin. P. aeroginosa isolates were only sensitive to gentamicin and intermediate to ciprofloxacin and doxycycline while the isolate was resistant to the rest of all the tested antibiotics. No resistance was observed to ciprofloxacin, doxycycline and trimethoprim- sulfamethoxazole by E. coli and Salmonella species while highresistance to amoxicillin, streptomycin and gentamicin was observed. P. mirabilus was highly resistant to amoxicillin, tetracycline, doxycycline and streptomycin but sensitive to ciprofloxacin, chloramphenicol, trimethoprim-sulfamethoxazole and ceftriaxone (Table 5).
 |
Table 5 Antimicrobial Susceptibility Pattern of Gram Negative Bacteria Isolated from Study Participants |
From the total of 28 isolates tested for antimicrobial susceptibility tests, multiple drug resistance (MDR) was observed in 23 (82.1%). Among the MDR isolates, 12 (three-quarters) of the isolates were from Gram positive and 11 (91.7%) isolates were from Gram negative bacteria. All isolated K. pnemoniae, E. coli, P. aeruginosa and P. mirabilis were MDR from Gram negative bacteria. All S. aureus isolates and nearly half of CONS were MDR from Gram positive bacterial (Figure 2). In general, trimethoprim-sulfamethoxazole, ciprofloxacin and chloramphenicol were effective drugs against Gram negative bacteria. However, Gram negative isolates were highly resistant to streptomycin, amoxicillin-clavulanic acid, ampicillin, amoxicillin and cephalothin. Cefoxitin, amoxicillin-clavulanic acid, cephalothin and chloramphenicol were effective drugs for Gram positive bacteria but ceftriaxone and amoxicillin were ineffective.
 |
Figure 2 Multi-drug resistance isolates among identified bacteria from blood sample. |
Discussion
The results of this study showed that the overall culture positivity rate of septicemia identified from pediatric patients was 11.3% [95% CI (7.9%, 16%)] which was similar with studies conducted in Trinidad (10.1%)3, Addis Ababa, Ethiopia (8.9% and 12.4%)1, 4, Zanzibar (9.6%)19 and Uganda (14%).20 The present study finding was also in line with global epidemiology (8.2%).21 Bacterial sepsis remains a major cause of pediatric morbidity and mortality despite advances in laboratory diagnosis and treatment.22
A relatively higher rate of septicemia was reported compared with studies conducted in Mekelle, Ethiopia (7.4%),15 Nepal (7.28%,23 7.48%,24 7.7%25), Europe and Australia/New Zealand.21 The reason of this variation may be due to study differences in the socio-demographic factors and health-care system. North America, Europe and Australia/New Zealand showed lower pediatric sepsis prevalence which may be related to good implementation of infection prevention, early identification of cases and effective treatment of identified cases. On the other hand, the findings of present study were lower than the study conducted in India (27%)26 and Nigeria (16.4%).27 The possible reason for the lower prevalence in our study may be differences in the socio-demographic and socio-economic status of the participants and difference in the number of the study participants.
In the current study Gram positive organisms were more frequently isolated than Gram negative organisms. A similar finding was observed in the study conducted in Gondar, Ethiopia7 and in Cape Town, South Africa.16 Staphylococcus aureus was the most prevalent etiological agent for bloodstream bacterial infections and this finding is comparable with other studies conducted in different areas.15,17,28 Coagulase negative staphylococci (CONS) was the other common cause of bacterial infection in the bloodstream which was consistent with other studies conducted in India, Egypt and Ethiopia.5,29,30 The high prevalence of CONS might be in association with an increasing burden of nosocomial infection in hospitalized patients. The pathogen was previously considered as a culture contaminant but is now recognized to be an important cause of bloodstream infections.31,32 In the present study K. pneumoniae was the predominant pathogen among Gram negative organisms followed by E. coli. Similar findings was observed in studies conducted in Ethiopia and Nigeria.1,7,27,33
In the current study different factors were found to be associated with septicemia. Participants in the infant age group were about 4 times more likely to developsepticemia compared with children above 5 years old. This finding is in concordance with other research conducted in Nepal and the USA.22,34 The possible reason for this is the infants have less immunity compared with the older children to protect themselves from the invasion of pathogens. In addition to this the bodies of infants are less colonized by normal microbiota. The presence of well-established resident normal microbiota has a significant role in protection from pathogenic bacteria.35
Prolonged hospitalization was the other factor associated with septicemia in the present study. The finding is in line with another study conducted in Ethiopia.1 This may be associated with nosocomial infection. The associated factors to the burden of septicemia are increasing worldwide due to the high number of patients admitted to hospital for prolonged periods. Septicemia accounts for 10–20% of all nosocomial infections and is one of the common causes of mortality in the hospitalized patients.7 The high number of hospitalizations is due to increasing levels of chronic illness and to medical procedures such as surgery and intravenous devices which could be risk factors for infection.
Burn and wound cases had a statistically significant association with bloodstream infections in this study. Our finding of association of septicemia with burn cases was in agreement with studies conducted in the USA.10,34 Patients with wounds had a statistically significant association with bloodstream infection and this may be due to transepidermal water loss and blood flow from their initial wounding and in the later stages and post-operatively.36 The main reason for this high burden of pediatric sepsis may be a lack of continuous evaluation or surveillance in the management system and also the likelihood of implementing internationally accepted management of cases in low income countries.5,23,37 Unavailability of laboratory setup for routine examination of blood culture and antimicrobial susceptibility tests is also another challenge to overcome to prevent the development of sepsis in wound and burn cases.4,26
The antimicrobial resistance pattern was varied among isolated pathogens. In the present study all Gram positive organisms showed resistance for ceftriaxone. Similar findings were observed in studies conducted in Egypt and Nigeria.27,29 In the current study the majority of isolated organisms of Gram positive and Gram negative bacteria were resistant to amoxicillin and ampicillin, similar to studies conducted in Egypt and Ghana.28,29 Most of the Gram positive organisms were resistant to tetracycline and erythromycin in the present study. A similar finding was observed in a study conducted in Egypt.29 Most of the Gram negative organisms were resistant to cephalothin, gentamicin, streptomycin and amoxicillin-clavulanic acid which showed similarity to studies conducted in Nigeria and India.30,33 The possible explanation for such high resistance might be uncontrolled use of antimicrobial drugs in the study area.
K. pneumoniae, E. coli, Salmonella species, P. mirabilis and P. aeruginosa showed 100% resistance for amoxicillin and streptomycin. This finding is in line with other studies conducted in Ethiopia.1,2 P. aeruginosa was only sensitive to gentamicin and the isolate was resistant to the other tested drugs in the present study. This finding was similar to Ethiopian and Indian studies.5,7,26 However, this study finding was different from a study conducted in Gondar, Ethiopia that showed P. aeruginosa was resistant to gentamycin.2 The observed difference may be due to difference in the use of antibiotic drugs and the presence of microbiology laboratory.
In the current study multi-drug resistance was observed in 82.1% of isolates which was similar to other studies conducted in Ethiopia which showed resistance of 89.1%, 80% and 65%.1,5,17 The present study had a higher multi-drug resistance rate compared with several other studies.23,25,38 Multi-drug resistance was observed to be higher in Gram negative bacteria than Gram positive bacteria. This finding was in line with other studies conducted in Ethiopia and north India.1,38 All S. aureus. K. pneumoniae, E. coli, P. aeruginosa and P. mirabilis strains showed multi-drug resistance similar to studies conducted in Ethiopia and Nepal.1,5,25 The observed multi-drug resistance may be due to uncontrolled use of antimicrobial drugs in the study area. Treatment of bacterial infection without drug sensitivity test is the other possible reason for the observed high prevalence.
Conclusion
The study showed that Gram positive bacteria were the predominant etiologic agents in pediatric septicemia. Staphylococcus aureus, CONS, K. pneumoniae and E. coli were the commonest causative agents. Infants, prolonged hospitalization, burns, and wound cases were independently associated with pediatric septicemia. Gram positive bacteria were more resistant to ceftriaxone and amoxicillin while Gram negative bacteria were resistant to amoxicillin, ampicillin and streptomycin. High multi-drug resistance rates were observed in most of the isolates. Establishing hospital antibiotic stewardship is essential to minimize the high prevalence of drug resistance. Working on the identified associated factors is vital to minimize the observed high prevalence of pediatric septicemia.
Abbreviations
AIDS, acquired immunodeficiency syndrome; ATCC, American Type Culture Collection; BMI, body mass index; CLSI, Clinical Laboratory Standards Institute; CONS, coagulase negative Staphylococci; COR, crude odds ratio; MDR, multi-drug resistance; SPSS, Statistical Package for Social Sciences.
Author Details
GA ([email protected]), TM ([email protected]), TT ([email protected]), FG ([email protected]).
Acknowledgment
We would like to thank Arba Minch University Colleges of Medicine and Health Sciences department of medical laboratory science for giving us material support during laboratory analysis. We are very grateful to Arba Minch general hospital staff for cooperating during data collection.
Disclosure
The authors declare that they have no competing interests.
References
1. Hailu M, Mulugeta G, Asraty D. Prevalence and antimicrobial resistance pattern of bacterial isolates mong children suspected for septicemia and urinary tract infections at Tikur Anbessa Specialized Hospital. Int J Sci Eng Res. 2016;7(10):1431–1444.
2. Alebachew G, Teka B, Endris M, Shiferaw Y, Tessema B. Etiologic agents of bacterial sepsis and their antibiotic susceptibility patterns among patients living with human immunodeficiency virus at Gondar University Teaching Hospital, Northwest Ethiopia. Biomed Res Int. 2016;2016:5371875. doi:10.1155/2016/5371875.
3. Orrett FA, Changoor E. Bacteremia in children at a regional hospital in Trinidad. Int J Infect Dis. 2007;11(2):145–151.
4. Negussie A, Mulugeta G, Bedru A, et al. Bacteriological profile and antimicrobial susceptibility pattern of blood culture isolates among septicemia suspected children in selected Hospitals Addis Ababa, Ethiopia. Int J Biol Med Res. 2015;6(1):4709–4717.
5. Kumalo A, Kassa T, S Mariam Z, Deresse Daka D, Tadesse AH. Bacterial profile of adult sepsis and their antimicrobial susceptibility pattern at Jimma University Specialized Hospital, South West Ethiopia. Health Sci J. 2016;10(2):3.
6. Ruth A, McCracken CE, Fortenberry JD, Hall M, Simon HK, Hebbar KB. Pediatric severe sepsis: current trends and outcomes from the pediatric health information systems database. Pediatr Crit Care Med. 2014;15(9):828–838. doi:10.1097/PCC.0000000000000254
7. Dagnew M, Yismaw G, Gizachew M, et al. Bacterial profile and antimicrobial susceptibility pattern in septicemia suspected patients attending Gondar University Hospital, Northwest Ethiopia. BMC Res Notes. 2013;6:283. doi:10.1186/1756-0500-6-283
8. Singh AK, Venkatesh V, Pal Singh R, Singh M. Bacterial and antimicrobial resistance profile of bloodstream infections: a hospital-based study. CHRISMED J Health Res. 2014;1:140–144.
9. Jagdish L, Naik TB, Gupta RK, Jais M. Etiology of blood culture from septicemia cases and their antibiotic susceptibility pattern at a tertiary care hospital. Microbiol Res (Pavia). 2016;3(4):436–440.
10. Wardhana A, Djan R, Halim Z. Bacterial and antimicrobial susceptibility profile and the prevalence of sepsis among burn patients at the burn unit of Cipto Mangunkusumo Hospital. Ann Burns Fire Disasters. 2017;30(2):107–115.
11. Jamil B, Bokhari MT, Saeed A, et al. Bacteremia: prevalence and antimicrobial resistance profiling in chronic kidney diseases and renal transplant patients. J Pak Med Assoc. 2016;66(6):705–709.
12. Trecarichi EM, Pagano L, Candoni A, et al. Current epidemiology and antimicrobial resistance data for bacterial bloodstream infections in patients with hematologic malignancies: an Italian multicentre prospective survey. Clin Microbiol Infect. 2015;21(4):337–343. doi:10.1016/j.cmi.2014.11.022
13. Kalpesh Gohel AJ. Bacteriological profile and drug resistance patterns of blood culture isolates in a Tertiary Care Nephrourology Teaching Institute. Biomed Res Int. 2014;1:153747.
14. Onyedibe KI, Bode-Thomas F, Nwadike V, et al. High rates of bacteria isolates of neonatal sepsis with multidrug resistance patterns in Jos, Nigeria. Ann Pediatr Child Health. 2015;3(2):1052.
15. Wasihun AG, Wlekidan LN, Gebremariam SA, et al. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle Hospital, Northern Ethiopia. Springerplus. 2015;4(1):314. doi:10.1186/s40064-015-1056-x
16. Crichton H, O’Connell N, Rabie H, Whitelaw AC, Dramowski A. Neonatal and paediatric bloodstream infections: pathogens, antimicrobial resistance patterns and prescribing practice at Khayelitsha District Hospital, Cape Town, South Africa. S Afr Med J. 2018;108(2):99–104. doi:10.7196/SAMJ.2018.v108i2.12601
17. GEyesus T, Moges F, Eshetie S, Yeshitela B, Abate E. Bacterial etiologic agents causing neonatal sepsis and associated risk factors in Gondar, Northwest Ethiopia. BMC Pediatr. 2017;17(1):137. doi:10.1186/s12887-017-0892-y
18. Wayne PA. Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing: 28th Ed. Informational Supplement, M100. Clinical and Laboratory Standards Institute (CLSI); 2018. Available from: https://clsi.org/media/1930/m100ed28_sample.pdf.
19. Onken A, Said AK, Jørstad M, Jenum PA, Blomberg B, Galdiero M. Prevalence and antimicrobial resistance of microbes causing bloodstream infections in Unguja, Zanzibar. PLoS One. 2015;10(12):e0145632. doi:10.1371/journal.pone.0145632
20. Kajumbula H, Fujita A, Mbabazi O, et al. Antimicrobial Drug resistance in blood culture isolates at a Tertiary Hospital, Uganda. Emerg Infect Dis. 2018;24(1):174–175. doi:10.3201/eid2401.171112
21. Weiss SL, Fitzgerald JC, Pappachan J, et al. Global epidemiology of pediatric severe sepsis: the sepsis prevalence, outcomes, and therapies study. Am J Respir Crit Care Med. 2015;191(10):1147–1157. doi:10.1164/rccm.201412-2323OC
22. Bhayani K, Pathak AN, Bhargava V, Kothari N, Saxena J, Jyoti A. Study of infectious pathogens and their antibiotics susceptibility in sepsis patients in Santokba Durlabhji Memorial Hospital and Research Center, Jaipur. Int J Biosolution. 2013;3(1):116–120.
23. Parajuli NP, Parajuli H, Pandit R, Shakya J, Khanal RP. Evaluating the trends of bloodstream infections among pediatric and adult patients at a Teaching Hospital of Kathmandu, Nepal: role of drug resistant pathogens. Can J Infect Dis Med Microbiol. 2017;8763135.
24. Shrestha S. Neonatal sepsis: bacteriological profile and antibiotic sensitivity pattern in Nepal Medical College. Nepal Med Coll J. 2015;17:43–46.
25. Prakash Simkhada SR. Bacteriological profile and antibiotic susceptibility pattern of blood culture isolates from Patients Visiting Tertiary Care Hospital. Glob J Med Res. 2016;16:1.
26. Negussie A, Mulugeta G, Bedru A, et al. Bacteriological profile and antimicrobial susceptibility pattern of blood culture isolates among septicemia suspected children in a Rural Tertiary Care Hospital. Int J Biol Med Res. 2015;6(1):4709–4717.
27. Nwadioha SI, Nwokedi EOP, Kashibu E, Odimayo MS, Okwori EE. A review of bacterial isolates in blood cultures of children with suspected septicemia in a Nigerian tertiary Hospital. Afr J Microbiol Res. 2010;4(4):222–225.
28. Aku FY, Akweongo P, Nyarko K, et al. Bacteriological profile and antibiotic susceptibility pattern of common isolates of neonatal sepsis, Ho Municipality, Ghana-2016. Matern Health Neonatol Perinatol. 2018;4:2. doi:10.1186/s40748-017-0071-z
29. Mohsen L, Ramy N, Saied D, et al. Emerging antimicrobial resistance in early and late emerging antimicrobial resistance in early and late onset. Antimicrob Resist Infect Control. 2017;6:63. doi:10.1186/s13756-017-0225-9
30. Nazir A, Sana I, Peerzada BY, Farooq T. Study of prevalence and antimicrobial susceptibility pattern in blood isolates from a tertiary care hospital in North Kerala, India. Int J Curr Microbiol App Sci. 2014;3:655–662.
31. Karunakaran R, Raja NS, Ng KP, Navaratnam P. Etiology of blood culture isolates among patients in a multidisciplinary teaching hospital in Kuala Lumpur. J Microbiol Immunol Infect. 2007;40(5):432–437.
32. Piette A, Verschraegen G. Role of coagulase-negative staphylococci in human disease. Vet Microbiol. 2009;134(1–2):45–54. doi:10.1016/j.vetmic.2008.09.009
33. Shittu Mujeeb O, Orisadare Olayiwola P, Jikeme Osemeke E, Shittu Bashirat T, Bello Lateef A, Oluremi Adeolu S. Antibiotic susceptibility pattern of bacteria isolates in neonates at a Children Hospital, Nigeria. J Med Sci Clin Res. 2014;2(10):2576–2583.
34. Aliyu S, Cohen B, Liu J, Larson E. Prevalence and risk factors for bloodstream infection present on hospital admission. J Infect Prev. 2018;19(1):37–42.
35. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141.
36. Koskela M Wound healing and skin in severe sepsis. Medicine. 2016. Available from: http://jultika.oulu.fi/files/isbn9789526214269.pdf.
37. Rabirad N, Mohammadpoor M, Lari AR, Shojaie A, Bayat R, Alebouyeh M. Antimicrobial susceptibility patterns of the gram-negative bacteria isolated from septicemia in Children’s Medical Center. J Prev Med Hyg. 2014;55:23–26.
38. Gupta S, Kashyap B. Bacteriological profile and antibiogram of blood culture isolates from a tertiary care hospital of North India. Trop J Med Res. 2016;19:94–99. doi:10.4103/1119-0388.185426
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
