Back to Journals » Clinical Ophthalmology » Volume 16
Analysis of Glaucoma Associated Genes in Response to Inflammation, an Examination of a Public Data Set Derived from Peripheral Blood from Patients with Hepatitis C
Authors Player JK , Riordan SM , Duncan RS, Koulen P
Received 3 March 2022
Accepted for publication 2 June 2022
Published 23 June 2022 Volume 2022:16 Pages 2093—2103
DOI https://doi.org/10.2147/OPTH.S364739
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Jacob K Player,1,* Sean M Riordan,1,* R Scott Duncan,1 Peter Koulen1,2
1Vision Research Center, Department of Ophthalmology, School of Medicine, University of Missouri – Kansas City, Kansas City, MO, 64108, USA; 2Department of Biomedical Sciences, School of Medicine, University of Missouri – Kansas City, Kansas City, MO, 64108, USA
*These authors contributed equally to this work
Correspondence: Peter Koulen, Vision Research Center, Department of Ophthalmology, School of Medicine, University of Missouri – Kansas City, 2411 Holmes Street, Kansas City, MO, 64108, USA, Tel +1 816-235-6773, Email [email protected]
Introduction: Glaucoma is the second leading cause of blindness worldwide and despite its prevalence, there are still many unanswered questions related to its pathogenesis. There is evidence that oxidative stress and inflammation play a major role in disease progression. Glaucoma patients from several studies showed altered gene expression in leukocytes, revealing the possibility of using peripheral biomarkers to diagnose or stage glaucoma. The fact that glaucoma is associated with gene expression changes in tissues distant from the retina underscores the possible involvement of systemic oxidative stress and inflammation as potential contributing or compounding factors in glaucoma.
Methods: We assembled a list of oxidative stress and inflammatory markers related to glaucoma based on a review of the literature. In addition, we utilized publicly available data sets of gene expression values collected from peripheral blood mononuclear cells and macrophages from two patient groups: those chronically infected by the hepatitis C virus and those who have cleared it. Activation of the innate immune response can render cells or tissues more responsive to a second delayed proinflammatory stimulus. Additional gene expression data from these cells after subsequent polyinosinic:polycytidylic acid treatment, used to elicit an acute inflammatory response, allowed for the investigation of the acute inflammatory response in these groups. We used fold-change comparison values between the two patient groups to identify genes of interest.
Results: A comparison analysis identified 17 glaucoma biomarkers that were differentially expressed in response to HCV-mediated inflammation. Of these 17, six had significant p-values in the baseline vs treated values. Expression data of these genes were compared between patients who had cleared the Hepatitis C virus versus those who had not and identified three genes of interest for further study.
Discussion: These results support our hypothesis that inflammation secondary to Hepatitis C virus infection affects the expression of glaucoma biomarker genes related to the antioxidant response and inflammation. In addition, they provide several potential targets for further research into understanding the relationship between innate responses to viral infection and inflammatory aspects of glaucoma and for potential use as a predictive biomarker or pharmacological intervention in glaucoma.
Keywords: in silico analysis, inflammation, expression analysis, peripheral blood mononuclear cells
Introduction
After cataracts, glaucoma is the second leading cause of blindness worldwide, estimated to have affected 76 million individuals in 2020 and to affect approximately 112 million by 2040.3 Glaucoma comprises a spectrum of progressive optic neuropathies characterized by retinal ganglion cell (RGC) death and optic nerve degeneration with many of its forms associated with elevated intraocular pressure (IOP).4 The role of inflammation as a critical factor in the pathogenesis and disease progression of glaucoma is widely known,5–9 and some of the treatments for glaucoma currently used are anti-inflammatory in nature.10 Indeed, a recent study investigated the prevalence of autoimmune disease in patients with primary open-angle glaucoma and identified a higher prevalence of autoimmune disease in the POAG population compared to controls (10.1% vs 17.4%).11 Work on animal models of early-stage ocular hypertension also showed evidence of genes related to neuroinflammation responding in the retina.12 These data support the idea of an inflammatory-centered genetic screen for early detection of glaucoma.12
Numerous viral infections, including herpes simplex virus and varicella zoster virus infections affecting ocular tissues and resulting in uveitis, generate predisposing conditions for glaucoma and result in secondary glaucoma for a large fraction of these patient populations.13,14 In addition to herpes simplex virus infection, Epstein-Barr virus infections,15,16 and potentially, based on preclinical data, Zika virus infections,17 may play a role in the development of glaucoma resulting from virally-induced anterior uveitis. Also, an association between autoimmune hepatitis and uveitis was uncovered.18
Despite these connections, there remain questions as to the mechanism of secondary glaucomas resulting from infection. Specifically, whether the development of secondary glaucoma could be due to the activation of key genetic pathways for disease development. Therefore, we attempted to establish whether genes that have been previously associated with glaucoma pathology are responsive to an inflammatory state caused by an active viral infection. As a first step in this line of questioning, we investigated a publicly available database of gene expression in peripheral blood samples collected from patients infected with Hepatitis C Virus (HCV). HCV causes a blood-borne chronic inflammatory disease of the liver that is estimated to affect >184 million individuals worldwide.19 Although long-term chronic infection with HCV can be fatal, the disease is highly treatable if detected. However, the infection often remains undetected or undiagnosed, and therefore untreated. Spontaneous clearance of the virus occurs in approximately 25% of cases.20
The study by Qian et al contains baseline (mock) and simulated viral infection (treatment) gene expression datasets from heparinized blood taken from individuals exposed to hepatitis C.21 The patient population was divided into two groups: those with chronic hepatitis C infection (VL+) and those who have cleared the virus (VL-). The study participants consisted of 32 VL+ and 37 VL- individuals that were recruited through Yale University School of Medicine and the South Central Rehabilitation Center in New Haven, CT, USA. Ages in the study group ranged from 23 to 63, the racial background of the study group was mixed, consisting of white, black, and Hispanic groups, and the study was moderately balanced between males and females (38.4% female). Only 10 samples from each group were chosen at random for use in microarray analysis due to limitations in sample size.21
To assess the innate immune portion of the inflammatory response of the individuals, samples were treated with either synthetic double-stranded RNA or polyinosinic-polycytidylic acid (poly(I:C)) to mimic viral infection, or a control.21 Poly(I:C) treatment is a commonly used method to simulate viral infections22 in blood samples. RNA samples were collected from both mock and treated samples for transcriptional analysis using an Illumina HumanHT-12 v4 Beadchip whole human genome expression array. Fold-changes between mock and treated samples were then calculated to identify inflammatory response genes.21
The results of our meta-analysis indicated that there is indeed a shortlist of glaucoma-associated genes that respond to viral infection and inflammation. In addition, the study population we used has the potential to provide unique insights into not only how the inflammatory response may affect gene expression in glaucoma-associated genes but also how that expression might be influenced by the presence of chronic inflammation. The presence of a VL+ and a VL- group provides a useful comparison for the differential effects of chronic inflammation, as well as a control for the effects specific to the previous infection with HCV.
This study provides a first step towards determining whether there is a genetic component in secondary glaucoma due to viral infection, opening the door for future studies in both human and animal models as to whether a genetic component exists in the development of secondary glaucoma and whether there exist actionable targets for medical intervention. In addition, a clinical study could be conceived to test whether genetic changes in leukocytes present in the blood could be used to confirm or possibly predict the development of a diagnosis of glaucoma, allowing for earlier intervention and better medical outcomes.
Materials and Methods
Biomarker Catalog Construction
A literature review was conducted to populate a catalog of potential biomarkers for glaucoma in humans. Glaucoma of any type was considered for the purpose of this search. The search for scientific literature was conducted using both the Google Scholar™ academic search engine and the PubMed® database. Search parameters were narrowed to exclude any results not published within the past 17 years (prior to 2005), with the earliest study used dating from 2007. Search terms used included “glaucoma” and “biomarker” or “biomarkers”, with corresponding Boolean operators used as stated. Fifty unique potential genetic biomarkers were identified from seven original studies and two review articles. To avoid discrepancies resulting from varying nomenclature between studies, a list of all abbreviated gene symbol aliases reported for each gene was compiled using the GeneCards® database, resulting in a total of 372 unique gene symbol identifiers for use in cross-referencing. A list of 50 genes was then compiled along with relevant information regarding each gene from each study, including whether differential expression was observed, whether a genetic variant was identified, and whether a gene was an autoantibody target, as this would relate to the immune response. The biological source of each sample was also listed.
Fold-Change Analysis
Tables S1 and S2 in the supplemental file 1 of the work by Qian et al21 contain a list of log2 fold-changes for differentially expressed poly(I:C) responsive genes in macrophages and peripheral blood mononuclear cells (PBMCs), for both the VL- and VL+ groups. These values were filtered using the biomarker alias list described above to identify 17 unique glaucoma biomarker genes that were differentially expressed in response to exposure to an inflammatory stimulus. Fold-changes for the VL- group were then isolated and assembled into a new table, and fold-changes ≥1.5 or ≤ −1.5 were used to classify glaucoma-related peripheral inflammatory response genes. Differences between the fold-changes for the VL- and VL+ groups for these 17 genes were then assessed and filtered for statistical significance (P ≤ 0.05), leaving 6 genetic biomarkers corresponding to 5 unique genes. These data were then isolated and assembled into a new table.
Results
Glaucoma Biomarker Catalog
To date, a disease-causing gene or genes have not been definitively identified for glaucoma, but oxidative stress and inflammation are known contributors. Numerous candidate genes related to oxidative stress have been identified in recent years that are thought to contribute to the development and progression of this debilitating condition. Studies have determined that primary open-angle glaucoma (POAG) patients had reduced blood total antioxidant status and higher aqueous humor levels of superoxide dismutase, glutathione peroxidase 1 (GPX1), and catalase genes.23 Studies have identified SNPs in GPX1 and other antioxidant enzymes associated with POAG.24 Elevated reactive oxygen species (ROS) generated by nicotinamide adenine dinucleotide phosphate (NADPH) oxidase and inflammation are associated with glaucoma and retinopathies and depletion of superoxide dismutase and GPX1 in the eyes of hypertensive animals.25
Furthermore, chronic inflammation is thought to play a key role in the progression of glaucomatous damage and vision loss. Single nucleotide polymorphisms (SNPs) in genes encoding innate immune response proteins have been associated with some forms of glaucoma in certain populations.26 For example, SNPs in the IL-10 gene are associated with an increase in the risk of POAG in the Chinese Han population.27 Similarly, SNPs in the toll-like receptor 4 (TLR4) gene confer a slightly elevated risk for POAG in northern Chinese.28 Serum alterations of TH1 and TH2 cytokines are associated with glaucoma, suggesting that inflammation may contribute to POAG.29 A T-helper cytokine imbalance may modulate the immune microenvironment in glaucomatous eyes and thus influence optic neuropathy.30,31
We conducted a literature review to identify 50 glaucoma biomarker genes identified in clinical studies of patients with glaucoma that could be contributing to oxidative stress and inflammation. A list of these genes, the reference where they were found, and the type of cells or tissue the genetic material was collected from are presented in Table 1. As blood is the most common source of biological material for these types of studies, the majority of cell and tissue type listed is derived from blood products. In addition, we have also indicated the type of finding related to the gene identified in each study. Information in this column includes changes in expression versus control patients, sequence variants, and whether the gene is an autoantibody target as that would be especially relevant for our study.
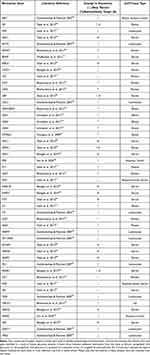 |
Table 1 Glaucoma Biomarker Gene Catalogue |
Glaucoma-Associated Biomarker Genes in Poly(I:C) Stimulated Macrophage and PBMCs
Studies have determined that activation of the innate immune response can render cells or tissues more responsive to a second delayed proinflammatory stimulus.1,2 To determine what effect innate immune receptor stimulation of macrophages from HCV cleared patients had on the expression of glaucoma-associated biomarker genes, we filtered the list of poly(I:C) responsive genes in the VL- group using the glaucoma biomarker gene list from Table 1. Of the 17 differentially expressed genes in the VL- group we identified, only two were from macrophage samples, with the rest coming from PBMC samples. Among these 17, only three had log2 fold-changes ≥1.5 or ≤ −1.5. All three of the genes with greater or less than a 1.5 log2 fold-change were from the PBMC group (Table 2).
 |
Table 2 (VL-) Log2 Fold-Changes in Poly I:C-Responsive Glaucoma Biomarker Genes |
Stimulation of macrophage or PBMCs from VL- patients with the TLR3 agonist, poly(I:C), led to a decrease in GPX1, GAS7, MMP9, ABCA1, GPX4, GPX3, and CAT. Since many of these genes encode proteins involved in the antioxidant response, it is possible that this may represent an increase in oxidative stress in these cells. Stimulation of macrophages or PBMCs from VL- patients with poly(I:C) led to an increase in NCAM, ITK, NFκB1, HSPA1B, HSPD1, and EDN1, suggesting a potential increase in the immune response in these cells (Table 2).
Comparison of Poly(I:C) Response Genes Between Active and Cleared HCV Infections
Comparison of the fold-changes before and after poly(I:C) treatment from the VL- and VL+ groups yielded 6 genes with statistically significant differences in expression (Table 3). Of these 6, all were from PBMC samples, with only GPX1 from both the VL- and VL+ groups and EDN1 from the VL+ group reporting a significant fold change (≥1.5 or ≤ −1.5). These results suggest that HCV clearance led to a partial reversal in gene expression changes for EDN1, CAT, and NCAM, but not a complete reversal. HCV clearance increased the gene expression level for ABCA1 and GPX1 (Table 3).
 |
Table 3 Comparison of the Fold-Changes from the (VL-) and (VL+) Groups |
With genes identified as glaucoma biomarkers in the literature and evidence of glaucoma-associated immune response genes in peripheral blood samples, the next step is to investigate whether these genes respond in the eyes of a glaucoma disease model. There are several key areas of the eye in glaucoma that are likely to be directly affected by the differential expression of each of the genes identified in Table 2, and these areas and their corresponding genes have been identified below (see Figure 1). Although the expression levels of these genes and their corresponding proteins in these locations may differ significantly from those in PBMC samples, this provides a useful point of departure for further research seeking to better understand the relationship between glaucoma and inflammation.
 |
Figure 1 Regions of the eye expressing glaucoma biomarker genes found to be responsive to Poly(I:C) challenge in peripheral blood samples. Regions of the eye are listed along with the encoded proteins likely to affect them from the differentially expressed genes identified in Table 2. Differentially expressed genes were detected using microarray analysis of PBMCs taken from human blood samples of patients infected with HCV and then challenged with poly(I:C). Red indicates decreased PBMC gene expression compared to baseline in response to poly(I:C) challenge, while green indicates increased expression compared to baseline. Please note that the expression levels were not found in the regions indicated in the figure and do not represent them. That is beyond the scope of the current study. The regional classifications demonstrate areas where glaucoma-related immune response genes/proteins may be investigated in future studies. |
Discussion
The results support the hypothesis that inflammation affects the expression of glaucoma biomarker genes in PBMCs, and to a much lesser extent, in macrophages. This adds to a growing body of work suggesting that, in some cases, inflammation may play an important role in glaucoma pathogenesis and/or pathology. Some local and systemic infections, and the drugs that treat them, have caused reversible forms of glaucoma. It is currently unclear how glaucoma can affect gene expression in distant tissues such as leukocytes, but studies have shown that glaucoma is associated with increases in the level of blood metabolites (citrate), oxidized serum proteins, circulating micro RNAs, and circulating proteins such as autoantibodies.26,32–40 It is conceivable that some of these factors may be responsible for the gene expression changes observed in leukocytes from glaucoma patients, but research in this area is limited.
Additionally, our results indicate that although glaucoma biomarker genes are similarly expressed in response to an inflammatory stimulus for both VL- and VL+ groups, the levels of expression may be modulated by the presence of chronic inflammation, which may provide interesting avenues for future research.
Oxidative stress is closely linked to inflammation and has been reported to play a role in glaucoma-associated progressive optic nerve damage23,41 and RGC death.42 GPX1, GPX3, GPX4, and CAT all encode proteins that can reduce oxidative stress, and all were downregulated in response to poly(I:C) stimulation, although the degree of this downregulation varied in significance. In the VL+ group, CAT downregulation was nearly double that of the VL- group in PBMCs, which may indicate that it could pose a particular risk to those with chronic inflammation. However, the relationships between these genes and glaucoma, while interesting, require further investigation to fully understand.
GPX1, for example, which had a significant log2 fold-change, has been shown previously to be down-regulated in glaucomatous PBMC samples, but was also shown to be elevated in the aqueous humor in the same study.23,41 GPX1 was also upregulated in RGCs in a rat model of glaucoma in another study.23 The fact that it was differentially expressed in response to biological stress in all samples and tissues, but in different ways, is further evidence of its utility as a glaucoma biomarker and the need to better understand its role.
Irregular immune activity has also been increasingly recognized as being associated with disease progression in glaucoma patients.43 Several different studies have cited notable changes in the immune system that have occurred in the eyes of glaucoma patients, many of which are consistent with or similar to those found in autoimmune disorders.44 ITK, NCAM1, and NFkappaB1 expression are all tied directly to the immune response. NCAM1 is well known for its role in the expansion of T and B lymphocytes and natural killer cells, and ITK is known to play an important role in T-cell proliferation and differentiation. NFkappaB1 encodes a transcription regulator that is responsive to a variety of different immunologically important insults, and irregular NFkappaB1 activity has also been associated with inflammation, autoimmunity, and a variety of immunodeficiencies.45 Expression of each of these three genes was upregulated, and although no individual fold-changes were ≥1.5 or ≤ −1.5, taken together, their altered expression suggests that dysregulation of the immune system could play an interesting pathological role in glaucoma that merits further investigation.
Glaucoma is most commonly associated with elevated IOP, which leads to optic nerve cupping and RGC death, ultimately resulting in visual impairment. This build-up of pressure typically results when aqueous humor generation or inflow exceeds aqueous outflow via the trabecular meshwork (TM) and uveoscleral outflow pathway. The TM consists of a dynamic series of tissues with different pore sizes, with the smallest measuring at 1–2 µm,46 and is responsible for ~75% of the resistance to aqueous humor outflow.47
ABCA1 works as a pump in the cellular lipid removal pathway, and recent studies have made a strong case for its interaction with annexin A1 as its primary pathogenic role in glaucoma.48,49 Interestingly, ABCA1 was differentially expressed less than half-as-much in the VL+ group than in the VL- group. MMP9 has been shown to be active in the TM50 and functions as a secreted proteinase that specializes in the breakdown of the extracellular matrix. The observed downregulation of MMP9 could potentially contribute in part to decreased TM maintenance and a gradual rise in IOP.
EDN1 activity has perhaps some of the most substantive connections to glaucoma. It has been shown in several studies to not only be associated with glaucoma but also directly cause RGC death after injection in animal models, and to have protective effects when blocked as a therapeutic target.51,52 EDN1 was also more heavily expressed in the VL+ group, again indicating a possible unique risk for those suffering from chronic inflammation.
Considered together, the possible risks associated with all these glaucoma biomarker genes strongly support the stance that glaucoma is a highly multifactorial disease and that studies of its pathology and of possible therapeutic interventions should not limit themselves to the TM alone. Differential expression of these genes in response to an inflammatory stimulus also provides additional evidence that inflammation should be considered a possible risk factor for glaucoma through a variety of pathways.53–69 Future work should focus on using these identified genes to investigate the role of inflammation as a possible multi-hit contributor35 to glaucoma pathology/pathogenesis, as well as on the comorbidity of glaucoma in those experiencing chronic inflammation.
Abbreviations
RGC, retinal ganglion cell; IOP, intraocular pressure; HCV, Hepatitis C Virus; VL+, patients with chronic HCV infection; VL-, patients who have cleared HCV infection; Poly(I:C), polyinosinic-polycytidylic acid; PBMC, Peripheral blood mononuclear cells; POAG, Primary open-angle glaucoma; GPX1, glutathione peroxidase 1; ROS, reactive oxygen species; NADPH, nicotinamide adenine dinucleotide phosphate; SNP, single nucleotide polymorphism; TLR4, Toll-like receptor 4; TM, Trabecular meshwork; TLR3, Toll-like receptor 3; GAS7, Growth Arrest-Specific Protein 7; MMP9, Matrix Metallopeptidase 9; ABCA1, ATP Binding Cassette Subfamily A member 1; GPX4, glutathione peroxidase 1; GPX3, glutathione peroxidase 3; CAT, Catalase; NCAM, Neural Cell adhesion molecule 1; ITK, IL2 Inducible T Cell Kinase; NFκB1, Nuclear Factor Kappa B Subunit 1; HSPA1B, Heat Shock Protein Family A (Hsp70) Member 1B; HSPD1, Heat Shock Protein Family D (Hsp60) Member 1; EDN1, Endothelin 1.
Disclosure
The authors report no conflicts of interest in relation to this work and declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The research reported in this publication was supported in part by grants from the National Eye Institute (EY022774, EY031248) of the National Institutes of Health (PK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support by the Felix and Carmen Sabates Missouri Endowed Chair in Vision Research, a Challenge Grant from Research to Prevent Blindness and the Vision Research Foundation of Kansas City, is gratefully acknowledged.
References
1. Levy JH. The human inflammatory response. J Cardiovasc Pharmacol. 1996;27(Suppl 1):S31–7. doi:10.1097/00005344-199600001-00008
2. Conti P, Caraffa A, Tetè G, et al. Mast cells activated by SARS-CoV-2 release histamine which increases IL-1 levels causing cytokine storm and inflammatory reaction in COVID-19. J Biol Regul Homeost Agents. 2020;34(5):1629–1632. doi:10.23812/20-2edit
3. Allison K, Patel D, Alabi O. Epidemiology of glaucoma: the past, present, and predictions for the future. Cureus. 2020;12(11):e11686. doi:10.7759/cureus.11686
4. Weinreb RN, Aung T, Medeiros FA. The pathophysiology and treatment of glaucoma: a review. JAMA. 2014;311(18):1901–1911. doi:10.1001/jama.2014.3192
5. Cohen LP, Pasquale LR. Clinical characteristics and current treatment of glaucoma. Cold Spring Harb Perspect Med. 2014;4(6):a017236–a017236. doi:10.1101/cshperspect.a017236
6. Sng CC, Ang M, Barton K. Uveitis and glaucoma: new insights in the pathogenesis and treatment. Prog Brain Res. 2015;221:243–269. doi:10.1016/bs.pbr.2015.06.008
7. Harun-Or-Rashid M, Inman DM. Reduced AMPK activation and increased HCAR activation drive anti-inflammatory response and neuroprotection in glaucoma. J Neuroinflammation. 2018;15(1):313. doi:10.1186/s12974-018-1346-7
8. Shestopalov VI, Spurlock M, Gramlich OW, Kuehn MH. Immune responses in the glaucomatous retina: regulation and dynamics. Cells. 2021;10(8):Aug. doi:10.3390/cells10081973
9. Williams PA, Marsh-Armstrong N, Howell GR, Lasker II. Neuroinflammation in glaucoma: a new opportunity. Exp Eye Res. 2017;157:20–27. doi:10.1016/j.exer.2017.02.014
10. Doucette LP, Walter MA. Prostaglandins in the eye: function, expression, and roles in glaucoma. Ophthalmic Genet. 2017;38(2):108–116. doi:10.3109/13816810.2016.1164193
11. Lorenzo MM, Devlin J, Saini C, et al. The prevalence of autoimmune diseases in patients with primary open-angle glaucoma undergoing ophthalmic surgeries. Ophthalmol Glaucoma. 2022;5(2):128–136. doi:10.1016/j.ogla.2021.08.003
12. Gramlich OW, Godwin CR, Wadkins D, Elwood BW, Kuehn MH. Early functional impairment in experimental glaucoma is accompanied by disruption of the GABAergic system and inceptive neuroinflammation. Int J Mol Sci. 2021;22(14):7581. doi:10.3390/ijms22147581
13. Miserocchi E, Waheed NK, Dios E, et al. Visual outcome in herpes simplex virus and varicella zoster virus uveitis: a clinical evaluation and comparison. Ophthalmology. 2002;109(8):1532–1537. doi:10.1016/s0161-6420(02)01113-2
14. Thean JH, Hall AJ, Stawell RJ. Uveitis in Herpes zoster ophthalmicus. Clin Exp Ophthalmol. 2001;29(6):406–410. doi:10.1046/j.1442-9071.2001.d01-29.x
15. Morris RW, Dunbar MT. Atypical presentation and review of the ICE syndrome. Optometry. 2004;75(1):13–25. doi:10.1016/s1529-1839(04)70007-x
16. Keorochana N, Treesit I, Funarunart P. Characteristics and clinical outcomes of hypertensive anterior uveitis. Ocul Immunol Inflamm. 2020;28(4):538–548. doi:10.1080/09273948.2019.1587471
17. Singh PK, Kasetti RB, Zode GS, Goyal A, Juzych MS, Kumar A. Zika virus infects trabecular meshwork and causes trabeculitis and glaucomatous pathology in mouse eyes. mSphere. 2019;4(3). doi:10.1128/mSphere.00173-19
18. Lim LL, Scarborough JD, Thorne JE, et al. Uveitis in patients with autoimmune hepatitis. Am J Ophthalmol. 2009;147(2):332–338 e1. doi:10.1016/j.ajo.2008.08.019
19. Thrift AP, El-Serag HB, Kanwal F. Global epidemiology and burden of HCV infection and HCV-related disease. Nat Rev Gastroenterol Hepatol. 2017;14(2):122–132. doi:10.1038/nrgastro.2016.176
20. Grebely J, Prins M, Hellard M, et al. Hepatitis C virus clearance, reinfection, and persistence, with insights from studies of injecting drug users: towards a vaccine. Lancet Infect Dis. 2012;12(5):408–414. doi:10.1016/S1473-3099(12)70010-5
21. Qian F, Bolen CR, Jing C, et al. Impaired toll-like receptor 3-mediated immune responses from macrophages of patients chronically infected with hepatitis C virus. Clin Vaccine Immunol. 2013;20(2):146–155. doi:10.1128/CVI.00530-12
22. Fortier ME, Kent S, Ashdown H, Poole S, Boksa P, Luheshi GN. The viral mimic, polyinosinic: polycytidylic acid, induces fever in rats via an interleukin-1-dependent mechanism. Am J Physiol Regul Integr Comp Physiol. 2004;287(4):R759–66. doi:10.1152/ajpregu.00293.2004
23. Tang B, Li S, Cao W, Sun X. The association of oxidative stress status with open-angle glaucoma and exfoliation glaucoma: a systematic review and meta-analysis. J Ophthalmol. 2019;2019:1803619. doi:10.1155/2019/1803619
24. Sobot V, Stamenkovic M, Simic T, et al. Association of GSTO1, GSTO2, GSTP1, GPX1 and SOD2 polymorphism with primary open angle glaucoma. Exp Eye Res. 2022;214:108863. doi:10.1016/j.exer.2021.108863
25. Santana-Garrido A, Reyes-Goya C, Fernandez-Bobadilla C, et al. NADPH oxidase-induced oxidative stress in the eyes of hypertensive rats. Mol Vis. 2021;27:161–178.
26. Kim CD, Gudiseva HV, McGeehan B, et al. Association of the SNP rs112369934 near TRIM66 gene with POAG endophenotypes in African Americans. Genes. 2021;12(9):1420. doi:10.3390/genes12091420
27. Zhang Y, Yang A, Huang J. Identification of gene changes induced by dexamethasone in the anterior segment of the human eye using bioinformatics analysis. Med Sci Monit. 2019;25:5501–5509. doi:10.12659/MSM.915591
28. Liu H, Qi S, He W, Chang C, Chen Y, Yu J. Association of single-nucleotide polymorphisms in TLR4 gene and gene-environment interaction with primary open angle glaucoma in a Chinese northern population. J Gene Med. 2020;22(1):e3139. doi:10.1002/jgm.3139
29. Huang P, Qi Y, Xu YS, et al. Serum cytokine alteration is associated with optic neuropathy in human primary open angle glaucoma. J Glaucoma. 2010;19(5):324–330. doi:10.1097/IJG.0b013e3181b4cac7
30. Wong M, Huang P, Li W, Li Y, Zhang SS, Zhang C. T-helper1/T-helper2 cytokine imbalance in the iris of patients with glaucoma. PLoS One. 2015;10(3):e0122184. doi:10.1371/journal.pone.0122184
31. Jiang B, Harper MM, Kecova H, et al. Neuroinflammation in advanced canine glaucoma. Mol Vis. 2010;16:2092–2108.
32. Barbosa-Breda J, Himmelreich U, Ghesquiere B, Rocha-Sousa A, Stalmans I. Clinical metabolomics and glaucoma. Ophthalmic Res. 2018;59(1):1–6. doi:10.1159/000479158
33. Beutgen VM, Perumal N, Pfeiffer N, Grus FH. Autoantibody biomarker discovery in primary open angle glaucoma using Serological Proteome Analysis (SERPA). Front Immunol. 2019;10:381. doi:10.3389/fimmu.2019.00381
34. Chang D, Sha Q, Zhang X, et al. The evaluation of the oxidative stress parameters in patients with primary angle-closure glaucoma. PLoS One. 2011;6(11):e27218. doi:10.1371/journal.pone.0027218
35. Duncan RS, Rohowetz L, Vogt A, Koulen P. Repeat exposure to polyinosinic: polycytidylic acid induces TLR3 expression via JAK-STAT signaling and synergistically potentiates NFkappaB-RelA signaling in ARPE-19 cells. Cell Signal. 2020;66:109494. doi:10.1016/j.cellsig.2019.109494
36. Engin KN, Yemisci B, Yigit U, Agachan A, Coskun C. Variability of serum oxidative stress biomarkers relative to biochemical data and clinical parameters of glaucoma patients. Mol Vis. 2010;16:1260–1271.
37. Fraenkl SA, Muser J, Groell R, et al. Plasma citrate levels as a potential biomarker for glaucoma. J Ocul Pharmacol Ther. 2011;27(6):577–580. doi:10.1089/jop.2011.0062
38. Hindle AG, Thoonen R, Jasien JV, et al. Identification of candidate miRNA biomarkers for glaucoma. Invest Ophthalmol Vis Sci. 2019;60(1):134–146. doi:10.1167/iovs.18-24878
39. Romano GL, Platania CB, Forte S, Salomone S, Drago F, Bucolo C. MicroRNA target prediction in glaucoma. Prog Brain Res. 2015;220:217–240. doi:10.1016/bs.pbr.2015.04.013
40. Tezel G, Thornton IL, Tong MG, et al. Immunoproteomic analysis of potential serum biomarker candidates in human glaucoma. Invest Ophthalmol Vis Sci. 2012;53(13):8222–8231. doi:10.1167/iovs.12-10076
41. Bodh SA, Kumar V, Raina UK, Ghosh B, Thakar M. Inflammatory glaucoma. Oman J Ophthalmol. 2011;4(1):3–9. doi:10.4103/0974-620X.77655
42. Vohra R, Tsai JC, Kolko M. The role of inflammation in the pathogenesis of glaucoma. Surv Ophthalmol. 2013;58(4):311–320. doi:10.1016/j.survophthal.2012.08.010
43. Tezel G, Wax MB. Glaucoma. Chem Immunol Allergy. 2007;92:221–227. doi:10.1159/000099273
44. Tezel G. The immune response in glaucoma: a perspective on the roles of oxidative stress. Exp Eye Res. 2011;93(2):178–186. doi:10.1016/j.exer.2010.07.009
45. Lorenzini T, Fliegauf M, Klammer N, et al. Characterization of the clinical and immunologic phenotype and management of 157 individuals with 56 distinct heterozygous NFKB1 mutations. J Allergy Clin Immunol. 2020;146(4):901–911. doi:10.1016/j.jaci.2019.11.051
46. Wah LP, Singh D, Abdul Ghani S, Shoeb Ahmad S. The dynamics of aqueous humor outflow—A major review. US Ophthalmic Review. 2014;07(02). doi:10.17925/usor.2014.07.02.137
47. Goel M, Picciani RG, Lee RK, Bhattacharya SK. Aqueous humor dynamics: a review. Open Ophthalmol J. 2010;4:52–59. doi:10.2174/1874364101004010052
48. Luo J, Wang S, Zhou Z, Zhao ZY. Ad- and AAV8-mediated ABCA1 gene therapy in a murine model with retinal ischemia/reperfusion injuries. Mol Ther Methods Clin Dev. 2021;20:551–558. doi:10.1016/j.omtm.2021.01.012
49. Li L, Xu L, Chen W, et al. Reduced Annexin A1 secretion by ABCA1 causes retinal inflammation and ganglion cell apoptosis in a murine glaucoma model. Front Cell Neurosci. 2018;12:347. doi:10.3389/fncel.2018.00347
50. Mohd Nasir NA, Agarwal R, Krasilnikova A, Sheikh Abdul Kadir SH, Iezhitsa I. Effect of dexamethasone on the expression of MMPs, adenosine A1 receptors and NFKB by human trabecular meshwork cells. J Basic Clin Physiol Pharmacol. 2020;31(6). doi:10.1515/jbcpp-2019-0373
51. Howell GR, Macalinao DG, Sousa GL, et al. Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J Clin Invest. 2011;121(4):1429–1444. doi:10.1172/JCI44646
52. Marola OJ, Syc-Mazurek SB, Howell GR, Libby RT. Endothelin signaling in glaucomatous neurodegeneration. Invest Ophthalmol Vis Sci. 2019;60(9):661.
53. van Koolwijk LM, Ramdas WD, Ikram MK, et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012;8(5):e1002611. doi:10.1371/journal.pgen.1002611
54. Coca-Prados M, Escribano J, Ortego J. Differential gene expression in the human ciliary epithelium. Prog Retin Eye Res. 1999;18(3):403–429. doi:10.1016/S1350-9462(98)00026-3
55. Prasanna G, Dibas A, Hulet C, Yorio T. Inhibition of Na(+)/K(+)-ATPase by endothelin-1 in human nonpigmented ciliary epithelial cells. J Pharmacol Exp Ther. 2001;296(3):966–971.
56. Golubnitschaja O, Wunderlich K, Decker C, Monkemann H, Schild HH, Flammer J. Molecular imaging of perfusion disturbances in glaucoma. Amino Acids. 2002;23(1–3):293–299. doi:10.1007/s00726-001-0141-3
57. Agapova OA, Ricard CS, Salvador-Silva M, Hernandez MR. Expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in human optic nerve head astrocytes. Glia. 2001;33(3):205–216. doi:10.1002/1098-1136(200103)33:3<205::aid-glia1019>3.0.co;2-d
58. Hu C, Niu L, Li L, et al. ABCA1 regulates IOP by modulating Cav1/eNOS/NO signaling pathway. Invest Ophthalmol Vis Sci. 2020;61(5):33. doi:10.1167/iovs.61.5.33
59. Chen Y, Lin Y, Vithana EN, et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat Genet. 2014;46(10):1115–1119. doi:10.1038/ng.3078
60. van Oterendorp C. Endocrine dysfunction in open angle glaucoma. [Endokrine Auffalligkeiten beim Offenwinkelglaukom]. Klin Monbl Augenheilkd. 2021;238(2):128–131. doi:10.1055/a-1306-1033
61. Janssen SF, Gorgels TG, Ramdas WD, et al. The vast complexity of primary open angle glaucoma: disease genes, risks, molecular mechanisms and pathobiology. Prog Retin Eye Res. 2013;37:31–67. doi:10.1016/j.preteyeres.2013.09.001
62. Joe MK, Tomarev SI. Expression of myocilin mutants sensitizes cells to oxidative stress-induced apoptosis: implication for glaucoma pathogenesis. Am J Pathol. 2010;176(6):2880–2890. doi:10.2353/ajpath.2010.090853
63. Mayer U. Comparative investigations of catalase activity in different ocular tissues of cattle and man. Albrecht Von Graefes Arch Klin Exp Ophthalmol. 1980;213(4):261–265. doi:10.1007/BF00417548
64. Micera A, Quaranta L, Esposito G, et al. Differential protein expression profiles in glaucomatous trabecular meshwork: an evaluation study on a small primary open angle glaucoma population. Adv Ther. 2016;33(2):252–267. doi:10.1007/s12325-016-0285-x
65. Moazzeni H, Mirrahimi M, Moghadam A, Banaei-Esfahani A, Yazdani S, Elahi E. Identification of genes involved in glaucoma pathogenesis using combined network analysis and empirical studies. Hum Mol Genet. 2019;28(21):3637–3663. doi:10.1093/hmg/ddz222
66. Beutgen VM, Schmelter C, Pfeiffer N, Grus FH. Autoantigens in the trabecular meshwork and glaucoma-specific alterations in the natural autoantibody repertoire. Clin Transl Immunol. 2020;9(3):e01101. doi:10.1002/cti2.1101
67. Wang DY, Ray A, Rodgers K, et al. Global gene expression changes in rat retinal ganglion cells in experimental glaucoma. Invest Ophthalmol Vis Sci. 2010;51(8):4084–4095. doi:10.1167/iovs.09-4864
68. Zanon-Moreno V, Asensio-Marquez EM, Ciancotti-Oliver L, et al. Effects of polymorphisms in vitamin E-, vitamin C-, and glutathione peroxidase-related genes on serum biomarkers and associations with glaucoma. Mol Vis. 2013;19:231–242.
69. Fairless E, Kooragayala K, Karakulah G, et al. Distinct expression of heat shock proteins in mouse models of retinal degeneration. Invest Ophthalmol Vis Sci. 2016;57(12):2258.
70. Golubnitschaja O, Flammer J. What are the biomarkers for glaucoma? Surv Ophthalmol. 2007;52 Suppl 2(6):S155–61. doi:10.1016/j.survophthal.2007.08.011
71. Nath AP, Ritchie SC, Byars SG, et al. An interaction map of circulating metabolites, immune gene networks, and their genetic regulation. Genome Biol. 2017;18(1):146. doi:10.1186/s13059-017-1279-y
72. Bhattacharya SK, Lee RK, Grus FH. Molecular biomarkers in glaucoma. Invest Ophthalmol Vis Sci. 2013;54(1):121–131. doi:10.1167/iovs.12-11067
73. Ghaffariyeh A, Honarpisheh N, Heidari MH, Puyan S, Abasov F. Brain-derived neurotrophic factor as a biomarker in primary open-angle glaucoma. Optom Vis Sci. 2011;88(1):80–85. doi:10.1097/OPX.0b013e3181fc329f
74. Schmelter C, Perumal N, Funke S, Bell K, Pfeiffer N, Grus FH. Peptides of the variable IgG domain as potential biomarker candidates in primary open-angle glaucoma (POAG). Hum Mol Genet. 2017;26(22):4451–4464. doi:10.1093/hmg/ddx332
75. Fourgeux C, Martine L, Bjorkhem I, et al. Primary open-angle glaucoma: association with cholesterol 24S-hydroxylase (CYP46A1) gene polymorphism and plasma 24-hydroxycholesterol levels. Invest Ophthalmol Vis Sci. 2009;50(12):5712–5717. doi:10.1167/iovs.09-3655
76. Sun C, Zhang H, Jiang J, et al. Angiogenic and inflammatory biomarker levels in aqueous humor and vitreous of neovascular glaucoma and proliferative diabetic retinopathy. Int Ophthalmol. 2020;40(2):467–475. doi:10.1007/s10792-019-01207-4
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
