Back to Journals » Cancer Management and Research » Volume 11
An efficient method that combines the ThinPrep cytologic test with E6/E7 mRNA testing for cervical cancer screening
Authors Pan D , Zhang CQ, Liang QL , Hong XC
Received 11 December 2018
Accepted for publication 23 April 2019
Published 24 May 2019 Volume 2019:11 Pages 4773—4780
DOI https://doi.org/10.2147/CMAR.S197749
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Rituraj Purohit
Dan Pan,1,* Chan-Qiong Zhang,1,* Qi-Lian Liang,2 Xiao-Cui Hong2
1Department of Pathology, the Third Clinical Institute Affiliated to Wenzhou Medical University, Wenzhou People’s Hospital, Wenzhou, Zhejiang, People’s Republic of China; 2Oncology Center, Affiliated Hospital of Guangdong Medical University, Zhanjiang, People’s Republic of China
*These authors contributed equally to this work
Background: Cervical cancer is strongly associated with persistent human papillomavirus (HPV) infections. The ThinPrep cytologic test (TCT), HPV DNA detection, and E6/E7 mRNA testing are widely used to screen for cervical abnormalities.
Purpose: This study aimed to find a suitable method for cervical cancer diagnosis (but not for cervical cancer distant metastasis), especially among women whose TCT results are atypical squamous cells of undetermined significance (ASCUS) or worse (including ASCUS).
Patients and methods: A total of 301 samples from Wenzhou People’s Hospital from June 2014 to September 2017 were collected, we conducted comparative analysis of the diagnostic performance of several conventional screening methods both individually and in combination.
Results: We compared the sensitivity, specificity, positive predictive value, negative predictive value, and Youden index retrospectively estimated not only by single TCT, HPV DNA detection, or E6/E7 mRNA testing but also by combination methods, such as TCT+HPV DNA, TCT+E6/E7 mRNA, or TCT+HPV DNA+E6/E7 mRNA. Screening under TCT+E6/E7mRNA was confirmed with relatively higher sensitivity of 76.1% (95% CI: 0.659–0.841), specificity of 74.6% (95% CI: 0.681–0.803), and the highest Youden index of 0.507.
Conclusion: The joint screening methods showed relatively reliable specificity and sensitivity for cervical disease screening, and detection by TCT+E6/E7 mRNA has the potential to be a widely used clinical method for cervical cancer screening.
Keywords: ThinPrep cytologic test, human papillomavirus DNA, E6/E7 mRNA, cervical cancer
Introduction
Cervical cancer is the third most common type of cancer and the fourth for mortality rate in the world among women.1 Cervical intraepithelial neoplasia (CIN) is a premalignant cervical disease caused by persistent high-risk human papillomavirus (HR-HPV) infection. Among HR-HPVs, the HPV 16 and 18 subtypes account for 65–75% of cervical cancers, while the other high-risk genotypes account for the rest.2–4 HPV screening (secondary prevention) remains a key strategy for identifying cervical precancerous lesions and invasive cervical cancer precursors.5 Women over 30 years of age or women with ambiguous cytology results are recommended to undergo HPV DNA testing.6,7 In addition, HR-HPV genotype expression encodes two main transformation genes of E6 and E7 proteins, which are necessary for the immortality of human primary keratinocytes. These two carcinogenic genes are evenly preserved and highly expressed in cervical cancer cells, and their continuous expression is responsible for HPV initial cervical oncogenesis.
The E6 and E7 proteins were initially identified through their capability to target the p53 and retinoblastoma protein (pRb) tumor suppressor pathways in host cells, thereby interfering with cell cycle regulation. E7 stimulates the cell cycle via its capability to bind and inactivate cellular pRb, whereas E6 binds to p53 that leads to degradation of p53 via the proteasomal pathway.8,9 A considerable number of modalities have focused on screening, such as the ThinPrep cytologic test (TCT), HPV DNA detection, and HPV mRNA testing. Although the TCT is widely used in clinical practice, it is easy to cause false negative or false positive results, especially among atypical squamous cells of undetermined significance or worse (ASCUS+) (including ASCUS) cases, due to subjective factors, such as operation and reading. The TCT has a high specificity for cervical disease diagnosis, but its sensitivity is poor, and it is susceptible to the quality and subjective factors of the specimen. HR-HPV screening can compensate for the lack of liquid-based cytology, and can be combined with liquid-based cytology screening to improve sensitivity. The negative predictive value (NPV) of HR-HPV E6/E7 mRNA detection is high, and the two methods can make up for each other’s deficiencies, thus significantly reducing the rate of missed diagnosis of CIN I, CIN II, CIN III, and cervical cancer. However, the specificity of HPV DNA detection is low, and the virus activity cannot be affirmed by HPV DNA detection. A large number of unnecessary colposcopy and biopsy procedures are also required. HPV mRNA testing has also been applied for screening, but its specificity and sensitivity are affected by age.10,11 As a result, the TCT, HPV DNA detection, and HPV mRNA testing are not only unable to predict the progression of cervical lesions and risk assessment but also can increase the patients’ mental burden. Thus, a combination of specific and sensitive methods should be adopted in cervical cancer triage in the future.
In this retrospective study, we have taken the pathology diagnosis as the golden standard to assess the performances of the TCT, HPV DNA detection, the E6/E7mRNA test, and their joint detection.
Materials and methods
Case selection
We conducted a retrospective analysis of women who attended the outpatient department of Wenzhou People’s Hospital from June 2014 to September 2017. The data from this study were derived from the clinical information obtained from patients after normal clinical examination. We performed a retrospective analysis of anonymized information which did not include further intervention for the patients, so patient consent was waived. This study was reviewed and approved by the Ethics Committee of Wenzhou People’s Hospital, and complied with the Declaration of Helsinki. The selected cases were cervical abnormalities diagnosed by histological analysis. All cases were screened by the TCT, HPV DNA, and E6/E7 mRNA. The cases whose cytologic results were ASCUS+ (including ASCUS) were collected. The women who participated in the study ranged in age from 20 to 82 years, and the mean age was 45.46 years. They mainly complained about contact bleeding, vaginal discharge abnormalities, genital itching, and other symptoms. Criteria for selecting the subjects were as follows: 1) not pregnant currently; 2) no evidence of any immunodeficiency; and 3) no history of therapy neoplasms.
TCT
An endocervical brush slowly inserted into the cervical canal to a depth of approximately 1 cm should be at the junction of the squamous epithelium and columnar epithelium at the outer cervix, and with a certain pressure rotated 5–10 times in a clockwise or counterclockwise direction. Cervical samples must be immediately transferred to the TCT medium and prepared for ThinPrep specimens following the manufacturer’s instructions. Cytology slides were produced automatically by ThinPrep 2000 (Hologic) and diagnosed by two cytology experts. The TCT results were classified according to the 2001 Bethesda System (TBS), namely, negative for intraepithelial lesions or malignancy; ASCUS; atypical squamous cells, cannot exclude high-grade squamous intraepithelial lesion; low-grade squamous intraepithelial lesion (LSIL); high-grade squamous intraepithelial lesion; squamous carcinoma; and adenocarcinoma. Results higher than ASCUS (ASCUS+) were regarded as positive.
HPV E6/E7 mRNA detection
We used the QuantiVirus HPV E6/E7 mRNA assay (Kodia, Xinxiang, China), a nucleic acid hybridization procedure based on branched-chain DNA hybridization technology, to identify HPV-positive patients. Hybridization was performed in a microplate with the samples and corresponding probes (14 high-risk types, namely, HPVs 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68). Both the viral mRNA and the preamplified probe hybridize with the solid-phase coated probe, and the amplified probe binds to the preamplified probe to form a branched DNA complex. The luminescent probe labeled with alkaline phosphatase was hybridized to the solid-phase complex and the luminescence value of the sample was detected by the QuantiVirus luminescence instrument. Finally, the results were calculated by Diacarta software and recorded as relative light units. If the signal was greater than or equal to the luminescence threshold, then the result for the corresponding patient was positive. Otherwise, the result was negative.
HPV DNA test
HPV DNA testing using the HPV Nucleic Acid Test Kit (Tellgen, Shanghai, China) via PCR technology was conducted to identify 27 genotypes in the Luminex 200 system.12 The HPV DNA chip was not only capable of detecting various HPV subtypes simultaneously but also showed higher sensitivity and specificity than those of traditional cytology. The results for HPV DNA detection were divided into HPV–, HPV 16/18+ (positive results for either type 16 or type 18 with or without the 25 other types), and HR-HPV+ (positive for 17 HR-HPVs with or without the 10 other low-risk HPV types).
Histological analysis
Women with positive cervical cytology detected either by HPV DNA, the TCT, or E6/E7 mRNA tests were referred to further examination and treatment. Cervical samples were collected from cervical biopsies, conical resections, or total hysterectomy. The tissue specimens, which were fixed by formalin and embedded by paraffin, were stained with H&E for histopathological examination. The results were diagnosed by two senior pathologists according to the World Health Organization (Fourth Edition 2014) pathological diagnostic criteria. The histological results were defined as CIN I, CIN II, CIN III, and cancer. Cases lower than CIN I (including CIN I, CIN−) were considered negative, whereas those higher than CIN II (including CIN II, CIN II+) were positive.
Statistical analysis
The performance of any single detection or joint detection method was assessed using specificity, sensitivity, positive predictive value (PPV), NPV, and Youden index.
Statistical analysis was performed by two statistical packages, namely, IBM SPSS 19.0.1 for Windows (IBM Corporation, Armonk, NY, USA). Statistical analyses were performed using a chi-square test. All P-values reported were two-sided, and P<0.05 was considered statistically significant.
Results
Among 301 cases, the numbers of ASCUS, LSIL, atypical squamous cells, cannot exclude high-grade squamous intraepithelial lesion, and high-grade squamous intraepithelial lesion were 153, 65, 23, and 60, respectively (summarized in Table 1). The positive rate of CIN II+ increased with an increase in the TCT grade, which showed consistency of the TCT results and histologic diagnosis results to some extent. However, numerous cases could not be screened correctly by the TCT. For example, if a case is LSIL, they will be referred for colposcopy for further diagnosis. However, from Table 1, the positive rate of LSIL was only 24.6%, which means 75.4% of cases have no need to refer to further diagnosis, which induces a waste of time and resources. This condition also reflects the low specificity of the TCT.
 | Table 1 The cases of CIN I– and CIN II+ among different TCT grades |
Screening results of TCT, HR-HPV, HPV 16/18, and E6/E7 mRNA
We analyzed the performances of the TCT, HR-HPV, HPV 16/18, and E6/E7 mRNA (summarized in Tables 2–5). A comparison of the TCT and HR-HPV methods indicated that HR-HPV shows the highest sensitivity of 95.65% (95% CI: 0.886–0.986) and the lowest specificity of 15.31% (95% CI: 0.109–0.211). Screening by HPV 16/18 is more specific than by HR-HPV, which suggests that HPV types 16 and 18 are more correlated with high-grade CIN.
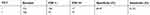 | Table 2 The performance of the TCT |
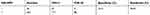 | Table 3 The performance of the HR-HPV |
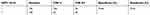 | Table 4 The performance of HPV 16/18 |
 | Table 5 The performance of E6/E7 mRNA |
Screening results of TCT+HR-HPV, TCT+HPV 16/18, TCT+E6/E7 mRNA, and TCT+HR-HPV+E6/E7 mRNA
We analyzed the performances of the TCT, HPV DNA, and E6/E7 mRNA and did not acquire a perfect method with both high specificity and high sensitivity. We therefore then analyzed the joint detection performances of TCT+HR-HPV, TCT+HPV 16/18, TCT+E6/E7 mRNA, and TCT+E6/E7 mRNA+HR-HPV (summarized in Tables 6–9). The positive rate of these methods increased along with an increase in pathological grade. The TCT+HPV 16/18 method had high specificity of 90.90% (95% CI: 0.860–0.943) but remarkable low sensitivity of 45.65% (95% CI: 0.353–0.563), which cannot be applied in clinical screening for cervical cancer. In addition, the specificity and sensitivity of TCT+HR-HPV were 71.29% (95% CI: 0.646–0.772) and 79.34% (95% CI: 0.694–0.868), respectively. The corresponding rates for TCT+E6/E7 mRNA were 74.64% (95% CI: 0.681–0.803) and 76.10% (95% CI: 0.659–0.841). These latter two joint methods display high rates of specificity and sensitivity.
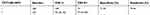 | Table 6 The performance of TCT+HR-HPV |
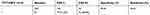 | Table 7 The performance of TCT+HPV 16/18 |
 | Table 8 The performance of TCT+E6/E7 mRNA |
 | Table 9 The performance of TCT+E6/E7 mRNA+HR-HPV |
We summarized all of the data (Table 10) and found that, compared with other methods, the TCT+E6/E7 mRNA method presented high rates of specificity, sensitivity, PPV, and NPV, namely, 74.64%, 76.10%, 56.91%, and 87.64%, respectively, and the highest Youden index of 0.507. TCT+HR-HPV is similar to TCT+E6/E7 mRNA, with specificity, sensitivity, PPV, NPV, and Youden index of 71.29%, 79.34%, 54.88%, 88.69%, and 0.506, respectively. Compared with E6/E7 mRNA, the P-value is <0.05, showing a significant difference. Although the method of TCT+E6/E7 mRNA+HR-HPV had a high Youden index, it is inconvenient for clinical screening with many operations.
 | Table 10 Comparison of different screening methods |
Discussion
HPV infection is a major risk factor for cervical cancer.13,14 Almost 100% of high-grade CIN and cervical cancer are associated with HPV infection, which is consistent with our results presented in Tables 3 and 4. The majority of HPV infections are transient infections and will be cleared within several months by the immune system.15 If the viral infection continues to persist latently in a group of patients, it can result in an increased risk of acquiring epithelial cell aberration and subsequently progressing to cancers.16 Such progression takes approximately 10–15 years.17
The TCT grade of ASCUS is difficult to decide on, ie, whether to refer to further diagnosis, and it has become a significant problem for doctors. The TCT, HPV DNA detection, and E6/E7 mRNA are widely applied for primary screening. However, subjective factors, such as operation and reading of TCT results, lead to false negative or false positive results. Furthermore, the specificity of HR-HPV DNA detection and E6/E7 testing is low, which causes a large number of overdiagnoses and overtreatment that can increase the mental burden of patients. A better method must thus be adopted for primary screening, especially in ASCUS+ (including ASCUS). The viral oncogenes E6 and E7 are responsible for the occurrence of cervical cancer caused by HPV. E6 and E7 oncogene transcription can be monitored directly through the detection of E6 and E7 mRNA transcripts or proteins,18 or indirectly via detecting the p16 expression of the host cell,19 which is upregulated by the HR-HPV E7 protein. Wallace et al compared the specificity between the E6/E7 mRNA assay and the HPV DNA assay and determined that the former has better specificity than that of the latter.20 Cuschieri et al compared the clinical performance of an HPV DNA and an RNA-based test in women with cytological abnormalities, and found that the RNA-based test was more specific than HPV DNA in the total cohort (p<0.001).21 HR-HPV DNA test results have lower predictive value for cervical cancer because they cannot distinguish between an overinfection or a persistent infection, and cervical cancer is based on persistent infection. Although HPV DNA testing has high clinical sensitivity, its specificity is low, and the vast majority of positive results are nonhistologically high lesions caused by short-term infections. Studies have shown that HPV E6/E7 mRNA detection has greater advantages in specificity than HPV DNA genotyping.22
The study group consisted of 301 women, collected from Wenzhou People’s Hospital from June 2014 to September 2017. The sensitivity values of the TCT, HR-HPV, HPV 16/18, and E6/E7 mRNA were 81.52% (95% CI: 0.718–0.886), 95.65% (95% CI: 0.886–0.986), 57.61% (95% CI: 0.469–0.677), and 92.39% (95% CI: 0.844–0.966), respectively. HR-HPV DNA, which owned the highest sensitivity, was especially suitable for the screening of CIN in the female population, which is consistent with the reports. Considering the low specificity of HR-HPV DNA, which was only 15.31% (95% CI: 0.109–0.211), we further defined HPV 16/18 as positive, and its specificity was dramatically increased from 15.31% to 74.64% with a decrease in sensitivity simultaneously. HPV 16/18 with a Youden index of 0.322 is generally better than HR-HPV with a Youden index of 0.11. However, this method is still not perfect, and a new detection method should be applied to reduce the high misdiagnosis rate. The specificities of the TCT and E6/E7 mRNA were 65.07% (95% CI: 0.581–0.714) and 33.01% (95% CI: 0.268–0.399), respectively, which were still far from a reliable CIN screening method. We tested joint methods such as TCT+HR-HPV, TCT+HPV 16/18, TCT+E6/E7 mRNA, and TCT+HR-DNA+E6/E7 mRNA, and then measured the sensitivity and specificity of each combined method to compensate for specificity, sensitivity, and diagnostic efficacy. Compared with the single methods, a significant difference in diagnosis, especially in the joint method of TCT+E6/E7 mRNA, which had higher specificity of 74.64% (95% CI: 0.681–0.659), higher sensitivity of 76.10% (95% CI: 0.659–0.841), and the highest Youden index of 0.507, was observed. They have the potential to provide a more scientific and efficient screening program.
The results inferred that cervical precancerous lesions are associated with the level of E6/E7 mRNA transcription; in other words, the transcription of HPV E6/E7 mRNA is closely related to the degree of lesion progression. E6 and E7 are the most important oncogenes in all HPVs, and the proteins of E6 and E7 can extend the life of infected cells and induce cancer by degrading p53 and pRb, which are well-known tumor suppressors. HPV E2 can regulate oncogene expression, and deficiency of E2 increases oncogene expression. When the HPV genome integrates into the host cell’s DNA, the E2 gene product will be disrupted, which results in the early abnormality and following cervical lesions.23,24 The E6 and E7 mRNAs, as transcription products of the oncogenes and the translation templates of the E6 and E7 proteins, reflect the gene expression activity and are related to the severity of the cervical diseases.25 Although the prevalence of E6 and E7 mRNA does not present any differences between the CIN I− and CIN II+ groups, E6/E7 mRNA might play a role in reflecting the activity of HPVs. A causal relationship exists between HR-HPV and almost all cases of cervical cancer and their precursors. The risk of progression to cancer also varies between different HR-HPV genotypes, and types 16 and 18 remain the most common in cervical lesions and are widely detected in screening cervical cancer. The high-risk types of the top three infection rates in this study were HPVs 16, 52, and 58. HPVs 16 and 18 played an important role in diagnosing high-grade CIN; the specificity of HR-HPV was only 15.31% (95% CI: 0.109–0.211), whereas that of HPV 16/18 was 74.60% (95% CI: 0.681–0.803), which suggested that HPV 16 and 18 rather than other HR-HPVs are the main HPV subtypes associated with cervical disease. This finding is consistent with previous studies, which have found that almost all cervical cancers are HPV related, and 70% of which are caused by HPV 16 and HPV 18 infections. HPV infection is therefore closely related to precancerous lesions of the cervix, and a correlation may exist between the viral infection rate and the degree of disease.
Conclusion
In summary, we found that combined diagnosis by the TCT and E6/E7 mRNA, compared with single tests, had a better diagnostic performance, especially when the specificity of the TCT and E6/E7 mRNA diagnosis were not ideal. The combination of the TCT and E6/E7 may exert an important influence on the reflection of malignant progression of cervical cancer and precancerous lesions, and may be a good indicator of the degree of cervical deterioration.
Ethics approval and consent to participate
This study was approved by the Hospital Ethics Committee of the Wenzhou People’s Hospital. As a retrospective study, written consents were not required.
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107
2. Muñoz N, Bosch FX, Castellsagué X, et al. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer. 2004;111(2):278–285. doi:10.1002/ijc.20244
3. Martins AE, Lucena-Silva N, Garcia RG, et al. Prevalence of human papillomavirus infection, distribution of viral types and risk factors in cervical samples from human immunodeficiency virus-positive women attending three human immunodeficiency virus-acquired immune deficiency syndrome reference centres in northeastern Brazil. Mem Inst Oswaldo Cruz. 2014;109(6):738–747.
4. Wright TC, Stoler MH, Behrens CM, et al. Primary cervical cancer screening with human papillomavirus: end of study results from the ATHENA study using HPV as the first-line screening test. Gynecol Oncol. 2015;136(2):189–197. doi:10.1016/j.ygyno.2014.11.076
5. Zhang S, McNamara M, Batur P. Cervical cancer screening: what‘s new? Updates for the busy clinician. Am J Med. 2018;131:702 e1–e5. doi:10.1016/j.amjmed.2017.12.017
6. Guo M, Lin CY, Gong Y, et al. Human papillomavirus genotyping for the eight oncogenic types can improve specificity of HPV testing in women with mildly abnormal Pap results. Mod Pathol. 2008;21(8):1037–1043. doi:10.1038/modpathol.2008.93
7. Tota JE, Bentley J, Blake J, et al. Introduction of molecular HPV testing as the primary technology in cervical cancer screening: acting on evidence to change the current paradigm. Prev Med. 2017;98:5–14. doi:10.1016/j.ypmed.2016.11.029
8. Graham SV. The human papillomavirus replication cycle, and its links to cancer progression: a comprehensive review. Clin Sci (Lond). 2017;131(17):2201–2221. doi:10.1042/CS20160786
9. Hoppe-Seyler K, Bossler F, Braun JA, Herrmann AL, Hoppe-Seyler F. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. 2018;26(2):158–168. doi:10.1016/j.tim.2017.07.007
10. Varnai AD, Bollmann M, Bankfalvi A, et al. Predictive testing of early cervical pre-cancer by detecting human papillomavirus E6/E7 mRNA in cervical cytologies up to high-grade squamous intraepithelial lesions: diagnostic and prognostic implications. Oncol Rep. 2008;19(2):457–465.
11. Coquillard G, Palao B, Patterson BK. Quantification of intracellular HPV E6/E7 mRNA expression increases the specificity and positive predictive value of cervical cancer screening compared to HPV DNA. Gynecol Oncol. 2011;120(1):89–93. doi:10.1016/j.ygyno.2010.09.013
12. Zhang CQ, Huang CG, Zheng X, Pan D. Prevalence of human papillomavirus among Wenzhou women diagnosed with cervical intraepithelial neoplasia and cervical cancer. Infect Agent Cancer. 2018;13:37. doi:10.1186/s13027-018-0211-8
13. Berman TA, Schiller JT. Human papillomavirus in cervical cancer and oropharyngeal cancer: one cause, two diseases. Cancer. 2017;123(12):2219–2229. doi:10.1002/cncr.30588
14. Coutlée F, Ratnam S, Ramanakumar AV, et al. Distribution of human papillomavirus genotypes in cervical intraepithelial neoplasia and invasive cervical cancer in Canada. J Med Virol. 2011;83(6):1034–1041. doi:10.1002/jmv.22081
15. Stanley M. Pathology and epidemiology of HPV infection in females. Gynecol Oncol. 2010;117:S5–10. doi:10.1016/j.ygyno.2010.01.024
16. Egawa N, Egawa K, Griffin H, Doorbar J. Human papillomaviruses; epithelial tropisms, and the development of neoplasia. Viruses. 2015;7(7):3863–3890. doi:10.3390/v7072802
17. Chen HC, Schiffman M, Lin CY, et al. Persistence of type-specific human papillomavirus infection and increased long-term risk of cervical cancer. J Natl Cancer Inst. 2011;103(18):1387–1396. doi:10.1093/jnci/djr283
18. Benevolo M, Vocaturo A, Caraceni D, et al. Sensitivity, specificity, and clinical value of human papillomavirus (HPV) E6/E7 mRNA assay as a triage test for cervical cytology and HPV DNA test. J Clin Microbiol. 2011;49(7):2643–2650. doi:10.1128/JCM.02570-10
19. Duvlis S, Popovska-Jankovic K, Arsova ZS, et al. HPV E6/E7 mRNA versus HPV DNA biomarker in cervical cancer screening of a group of Macedonian women. J Med Virol. 2015;87(9):1578–1586. doi:10.1002/jmv.24199
20. Cuschieri K, Cubie H, Graham C, et al. Clinical performance of RNA and DNA based HPV testing in a colposcopy setting: influence of assay target, cut off and age. J Clin Virol. 2014;59(2):104–108. doi:10.1016/j.jcv.2013.12.001
21. Munkhdelger J, Choi Y, Lee D, et al. Comparison of the performance of the NucliSENS EasyQ HPV E6/E7 mRNA assay and HPV DNA chip for testing squamous cell lesions of the uterine cervix. Diagn Microbiol Infect Dis. 2014;79(4):422–427. doi:10.1016/j.diagmicrobio.2014.04.004
22. Wallace NA, Galloway DA. Novel functions of the human papillomavirus E6 oncoproteins. Annu Rev Virol. 2015;2(1):403–423. doi:10.1146/annurev-virology-100114-055021
23. Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445(1–2):138–168. doi:10.1016/j.virol.2013.04.013
24. Fontecha N, Basaras M, Hernáez S, et al. Assessment of human papillomavirus E6/E7 oncogene expression as cervical disease biomarker. BMC Cancer. 2016;16(1):852.
25. Bzhalava D1, Guan P, Franceschi S, Dillner J, Clifford G. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology. 2013;445(1–2):224–231. doi:10.1016/j.virol.2013.07.015
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
