Back to Journals » Infection and Drug Resistance » Volume 13
Acinetobacter baumannii Efflux Pumps and Antibiotic Resistance
Authors Abdi SN , Ghotaslou R , Ganbarov K , Mobed A , Tanomand A , Yousefi M, Asgharzadeh M, Kafil HS
Received 21 August 2019
Accepted for publication 13 December 2019
Published 12 February 2020 Volume 2020:13 Pages 423—434
DOI https://doi.org/10.2147/IDR.S228089
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Joachim Wink
Seyyed Naser Abdi, 1, 2 Reza Ghotaslou, 3 Khudaverdi Ganbarov, 4 Ahmad Mobed, 1 Asghar Tanomand, 5 Mehdi Yousefi, 3 Mohammad Asgharzadeh, 6 Hossein Samadi Kafil 1
1Drug Applied Research Center, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran; 2Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran; 3Stem Cell Research Center, Tabriz University of Medical Sciences, Tabriz, Iran; 4Department of Microbiology, Baku State University, Baku, Republic of Azerbaijan; 5Department of Basic Sciences, Maragheh University of Medical Sciences, Maragheh, Iran; 6Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
Correspondence: Hossein Samadi Kafil Email [email protected]
Abstract: Acinetobacter baumannii is a nosocomial pathogen and gram-negative coccobacillus that is responsible for opportunistic infections, pneumonia, and infections of the urinary tract, bloodstream, skin, and soft tissue. This bacterium poses a major public health problem due to inducing resistance to several drugs, isolates, multidrug treatment, and occasionally pan drugs. Drug resistance is not only a major concern caused by A. baumannii but also is considered as the main challenge in many other pathogens. Several factors such as the efflux pump are associated with antibiotic resistance, biofilm production, and genetic mutations. In this review, A. baumannii is introduced in then some of the practical works conducted on the existing efflux pump are reviewed. The importance of the efflux pump is considered in this paper in relation to the antibiotic resistance and mechanisms developed for the inhibition of these pumps as well.
Keywords: Acinetobacter baumannii, efflux pumps, antibiotic resistance, biofilm, transporters
Introduction
Acinetobacter baumannii is a genus of gram-negative bacteria from the extensive class of Gammaproteobacteria. This class of bacteria has appeared as a major nosocomial pathogen with the ability to cause some diseases such as pneumonia and bacteremia. The mortality rate of this bacterium reaches 60% in vulnerable patients.1,2 Among the multi-drug resistant strains, A. baumannii is a paramount pathogen in clinical terms, particularly in hospital-acquired infections.3,4 Overall, this bacterium is the most associated with nosocomial infections, particularly in intensive care units (ICUs). Horizontal acquisition of resistance genes is the main factors involved in the emergence of multidrug resistance (MDR). Nevertheless, it has been recently identified that improved expression of chromosomal genes for efflux systems plays a significant role in MDR.5 The main mechanisms to confer resistance to a different class of antibiotics in A. baumannii include aminoglycoside, β-lactamases, modifying enzymes, permeability defects, alteration of target sites, and multidrug efflux pumps. Antibiotics released from the cell lead to a decrease in drug accumulation and a rise in the minimum inhibitory concentrations (MICs). In this regard, efflux pumps lead to the release of antibiotics from the cell efflux, which reduces drug accumulation and increases the MIC.
Each efflux pump contains three components: the outer membrane channel, the periplasmic lipoprotein, and the inner membrane transporter. Four classes of efflux pumps including major facilitator superfamily (MFS), the resistance-nodulation cell division (RND) family, small multidrug resistance (SMR) family, and multidrug and toxic compound extrusion (MATE) family are associated with A. baumannii antimicrobial resistance. Of these several pumps, the MFS and RND families of transporters have been studied in detail. AdeABC and RND-type efflux pump are not only related to aminoglycoside resistance but also involved in the resistance to many other antibiotics such as tigecycline lactams, chloramphenicol, erythromycin, and tetracycline as well. From the five superfamilies of pumps, resistance-nodulation-division (RND) systems are the most important ones in multiple resistant A. baumannii. This system generally demonstrates an extensive substrate variety as well as antibiotics, dyes, biocides, detergents, and antiseptics.
Epidemiology and Treatment of A. baumannii
The most important aspect of the infection with A. baumannii strains is their resistance to entirely known antibiotics, suggesting the need for urgent action by the global health care community. Due to the high antibiotic resistance rate, this pathogen can survive for a long time in the hospital environment and spread nosocomial.6 Several risk factors are important in the relation of Acinetobacter spp infections; eg, length of hospital stay, contact to an ICU, mechanical ventilation, acquaintance to antimicrobial agents, and current surgery. General environmental contamination is frequently established. In this regard, infection incidences have been found in respiratory care equipment, wound care procedures, and some patient care items.7 In humans, A. baumannii has been isolated from all cultivable places. Surprisingly, A. baumannii can form a part of the bacterial microbiota of the skin, mainly in moist regions such as the groin and toe webs. More than 40% of healthy adults can have colonization of the skin and mucous membranes. This rate is even higher among hospital staff members.8,9 Treating multidrug-resistant bacteria has remained as the top priority in the clinician’s practice in caring for hospitalized patients. A. baumannii has established to be a progressively significant and serious species in healthcare-associated infections.10 Carbapenems have been considered for the treatment of choice for severe infections.11 In addition, a growing number of carbapenem-resistant Acinetobacter isolates have been reported globally. Since the past three decades, the use of colistin has been restricted due to its toxicity and the expansion of novel antibiotics with improved care outlines. Conversely, the growing occurrence of multidrug-resistant A. baumannii in addition to a lack of new antimicrobial agents has revived attention in the use of colistin regarding its worthy activity against this organism.12
Efflux Pumps and Mechanisms of Function
A. baumannii is an important and opportunistic pathogen that plays a major role in the pathogenicity of humans. This bacterium can attach to the surfaces in the hospital environment and survive easily for a long time in adverse conditions. A. baumannii is accountable for a severe nosocomial infection, particularly in the intensive care units.13 The option of surviving in natural niches, and in the hospital environment could also be associated with the efflux pump mechanisms. Mechanisms of efflux commonly appear in all cells and play the physiological role.14 For example, in a prokaryote cell, the main functions include evasion of such naturally produced molecules and removal of metabolic products and toxins. Efflux pumps could also be complicated in an early stage of infection, such as adhesion to host cells and colonization. Outstandingly, they remove generally used antibiotics from the cell in the therapy of infections caused by these bacteria.15 Efflux-pump genes and proteins are existent in all organisms. Several studies have shown that efflux pumps in bacteria can confer reduced susceptibility to antibiotics; however, such decreases do not permanently affect clinical levels of antibiotic resistance. Efflux pumps can be specific for one substrate or can transport a range of structurally dissimilar compounds as well as antibiotics of different chemical classes. Such pumps that transport numerous compounds can be related to multidrug resistance (MDR). Several efflux pumps have clinical significance since they can reduce the fatality of bacterial infection by the agent(s) of choice.16 These pumps exist in nearly all bacterial species. Encoding genes of this class of proteins can be located on chromosomes or plasmids.17
Classification of Efflux Pumps
There are several efflux transporters, which are grouped using phylogenetic classification, based on protein sequences. One classification method for all transporter proteins has been proved by the Milton group.18 Five major families of an efflux transporter in the prokaryotic kingdom have been recognized. The families characterized for A. baumannii (Figure 1) are major facilitator superfamily (MF), Tet(A), Tet(B), Comal, multidrug and toxic efflux (MATE) AbeM, resistance nodulation-division (RND), AdeABC, small multidrug resistance (SMR), ATP binding cassette (ABC), and MacB from A. baumannii. All these systems use the proton motive power as an energy source. These ABC families operate ATP hydrolysis to determine the export of substrates. Transporters that efflux multiple substrates, such as antibiotics, have not progressed in response to the stresses of the antibiotic era. All bacterial genomes studied cover numerous different efflux pumps; the findings reveal their ancestral origins. It has been reported that almost 10% of all bacterial genes are elaborate in transport and a large amount of these encode efflux pumps.19 Additionally, transporters differ in their subcellular organization, eg, the RND pumps, which all are exporters of drugs and toxic cations located on the inner membrane (IM). Since these transporters interact with the periplasmic adaptor protein and the outer membrane (OM) channel, they produce a tripartite complex spanning the IM, the periplasm, and the OM. In E. coli, AcrABTolC, and P. aeruginosa MexAB-Opr members of the superfamily, ABC, MATE, and MFS are structured as a tripartite complex. In this mechanism, drug exertion occurs directly into the external medium. Therefore, the return of drugs needs the slow traversal of the OM and an effective permeability barrier projected for tripartite transporters.20 Accordingly, these pumps are far extra efficient in creating obvious resistance to antibiotics, particularly in AcrB of E. coli.21 In comparison, non-structured pumps occurring as single-component or “singlet” pumps in the IMs such as most of MFS and SMR pumps are less operative in creating a noticeable reduction in susceptibility. The main reason for this shortcoming is that the drug molecules are released only into the periplasm and can freely diffuse back into the cytosol because most antibiotics are comparatively lipophilic molecules that can cross the phospholipid bilayer region of the IM. Conversely, RND pumps are supposed to capture antibiotics from the periplasm.22,23 Efflux systems contributing to antibiotic resistance have been defined from some of the clinically imperative bacteria, as well as Campylobacter jejuni (Cme ABC),24 P. aeruginosa comprise MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM,24 Streptococcus pneumonia PmrA,25 Salmonella typhimurium Acr B,26 and Staphylococcus aureus NorA.27 It is noteworthy that all these efflux systems export multiple antibiotics.28
 |
Figure 1 Five main MDEPs with their well-known examples and antibiotic substrates. Data from Yılmaz and Özcengiz.91 Abbreviations: OMP, outer membrane protein; SMR, the small multidrug resistance family; MFS, the major facilitator superfamily; ABC, ATP-binding cassette superfamily; MATE, the multidrug and toxic compound extrusion family; RND, the resistance-nodulation division family. |
Efflux Pumps in Bacterial Structure
he efflux transporter (AdeB) arrests its substrates both from inside the phospholipid bilayer of the inner membrane or the cytoplasm22 and then carry them into the extracellular medium via OMP (AdeC).29 The periplasmic protein AdeA mediates the collaboration among AdeB and AdeC components. Drug transport, by different pump families, is determined by the transmembrane electrochemical gradient of protons. Some of the RND families are proton antiporters that use proton gradient to force efflux and trade one H+ ion for just a single drug molecule.30 Figure 2 depicts the scheme of the structure of the AdeABC.31 This system was revealed to be associated with declined susceptibility to a broad spectrum of antimicrobials as well as aminoglycosides, tetracycline, erythromycin, chloramphenicol, trimethoprim, fluoroquinolones, some β-lactams, and ethidium bromide.32–34
 |
Figure 2 Schematic organization of the ade gene cluster. Data from Wieczorek et al.13 |
In most of the transport modes, all MFS transporters share the same structural fold. The structural core contains several transmembrane helices (TM1-TM12) that are planned into two structurally similar domains; ie, the N-domain (TM1-TM6) and the C-domain TM7-TM12.35 This was first revealed in P. aeruginosa36 and was rediscovered in E. coli in the past decade.37 The latter study37 made a significant point that the involvement of some singlet pumps may have escaped detection because of covering specificities. Thus, in case of the double deletion of an MFS pump, MdfA, SMR pump, and EmrE, the E. coli is susceptible. It is even further susceptible when deleting the AcrB mutant from cationic agents with intracellular targets, alike acriflavine or ethidium. Recent findings revealed that RND pumps are relatively ineffective in arresting drugs from the cytosol.38 Consequently, the singlet pumps frequently show a central role in resistance to agents with intracellular targets. For example, the plasmid-encoded TetA pump creates significant tetracycline resistance.39 Additionally, the synergistic mechanism can sometimes be quite effective. Although the MFS-type MDR pumps typically do not perform the main role in resistance, only the drug pumps of the Tet group, which are commonly plasmid-encoded, are clinically imperative in creating tetracycline-specific resistance in numerous bacterial species. In current terminology, TetA denotes to the MFS exporter and the phylogenetic group, to which it is appropriately specified by the group in parentheses, as in TetA(B).40 TetA pumps existing in gram-negative bacteria contain several phylogenetic groups, while the 14-TMS Tet pumps present in gram-positive bacteria have at least 3 groups.41 The plasmid-encoded TetA pumps were the earliest bacterial drug efflux pumps recognized.42 Biochemical research via the Yamaguchi group indicated that their substrate was the magnesium salt of tetracycline.43 Moreover, cysteine-scanning mutagenesis monitored by tagging studies recognized the residues essential for substrate binding and proton translocation.44
Acinetobacter baumannii Efflux Pumps
In A. baumannii, AdeB is the multidrug transporter protein and AdeA and AdeC are in the MFP and OMP, respectively.45 Accordingly, in A. Baumannii, AdeB is the multidrug transporter protein, AdeA is the MFP, and AdeC is the OMP.32 Briefly, the efflux transporter (AdeB) captures its substrates either from the phospholipid bilayer of the inner membrane or the cytoplasm22 and subsequently transports them into the extracellular medium via OMP(AdeC).29 Efflux pump plays an important role in the antimicrobial resistance of A. baumannii. Nevertheless, the function of the Emr pump system and the relationship between Emr and drug resistance have not been characterized in A. baumannii. Among them, A1S_1772 (emrB) and A1S_1773 (emrA) were demonstrated to be co-transcribed as a single operon. Surprisingly, the Emr pump systems in A. baumannii contribute to adaptation to osmotic stress and resistance to colistin.46 Approximately, all efflux pumps of gram-negative bacteria are described in A. baumannii. Here, we describe these state-of-the-art efflux pumps:
RND Transporters
The AdeABC pump of A. baumannii BM4454, which causes resistance to several antibiotic classes such as aminoglycosides, consists of proteins including AdeA, AdeB, and AdeC. AdeB is a member of the RND superfamily.47 RND transporters employed in the inner membrane and form transporter cover complexes in gram-negative bacteria together with periplasmic membrane fusion proteins (MFPs) and outer membrane factors (OMFs). Since these macromolecular complexes are tripartite units, they perform as authoritative machines that eject multiple antibiotics through outer membranes. Contemporary, several studies have been conducted on the structures of the RND, MFP, and OMF components. AdeABC pump in wild A. baumannii has mysterious control by the AdeRS two-component system.47 In a relevant study, susceptibility testing and phenotypic evaluation confirmed the role of active efflux in mediating decreased susceptibility to biocides. Inactivation of either the adeB or adeJ transporter gene led to increased susceptibility to biocides. Moreover, RTPCR analysis exhibits increased adeB and adeJ expression in clinical isolates. This study was the first one establishing the role of efflux pumps in facilitating reduced susceptibility to disinfectants and other chemical substrates in A. baumannii.48 Multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) were applied to detect the existence and overexpression of the three Ade efflux systems. They also analyzed the sequences of the regulators AdeRS, a two-component system for AdeABC and AdeL, and a LysR-type regulator for AdeFGH. The results of this study showed that the incidence of AdeABC efflux pump overexpression in MDR A. baumannii is caused by a variety of single mutations in the corresponding two-component regulatory systems.49 In A. baumannii, AdeB is the multidrug transporter protein, AdeA is the MFP, and AdeC is the OMP.32 The efflux transporter (AdeB) arrests its substrates both from inside the phospholipid bilayer of the inner membrane or the cytoplasm.22 Next, it transfers them into the extracellular medium via OMP (AdeC).29 The periplasmic protein AdeA mediates the collaboration among AdeB and AdeC components. Drug transport by different pump families is determined by the transmembrane electrochemical gradient of protons. Some of the RND families are proton antiporters that use the proton gradient to force efflux and trade one H+ ion for just a single drug molecule.30 Figure 2 depicts the scheme of the structure of the AdeABC.31 This system was revealed to be associated with declined susceptibility to a broad spectrum of antimicrobials as well as aminoglycosides, tetracycline, erythromycin, chloramphenicol, trimethoprim, fluoroquinolones, some β-lactams, and ethidium bromide.32–34 The knockdown of AdeB can reduce the resistance to antibiotics in isolates with confirmed other genetic-based mechanisms.50
MFS Transporters: Selective
The major facilitator superfamily (MFS) is the unique large group of secondary active transporters that is carried from bacteria to humans.51 Additionally, MFS proteins transport a wide range of substrates through biological membranes and show a crucial role in multiple physiological processes.52 MFS transporters can be categorized into more than 70 families on the foundation of sequence homology.53 Depending on the transport mode and basic principle MFS transporters, they can be classified into three main groups including uniporters transport with a single substrate; symporters transport with a substrate in association by a coupling ion (typically protons); and antiporters transport as substrate with a co-substrate in opposed directions, where the binding of one is dependent on the prior release of the other.54 Uniporters need no external energy participation; nevertheless, in general, they can merely transport their substrates down their concentration gradient, while symporters and antiporters are able to employ the energy stored in the concentration gradient of their link ion or co-substrate to transport substrates contrary to their concentration gradient. In A. baumannii, the main efflux pumps are Tet(A) and Tet(B). The efflux determinative Tet(A) confers resistance to tetracycline and Tet(B) to Minocycline and tetracycline.55 MFS transporter-like open reading frame (ORF) of 453 bp was recognized in a pathogenic strain A. baumannii AIIMS 7 and its relationship with adherence and biofilm formation was explored. Recent findings revealed that the recognized MFS transporter-like ORF (PMT) is linked not only with adherence but also with biofilm formation and probable eDNA release in A. baumannii AIIMS 7.56 The results of another study indicated that efflux is an imperative mechanism of fosfomycin resistance and AbaF is elaborate in fosfomycin resistance in A. baumannii. AbaF also appears to play a role in biofilm formation and virulence of A. baumannii. The MFS transporter AbaF dynamically effluxes out Fosfomycin and adapts the cells resistant. Moreover, its expression is upregulated on exposure to fosfomycin. AbaF is expressed in fosfomycin-resistant clinical strains of A. baumannii. These strains have reduced susceptibility to fosfomycin in the presence of efflux inhibitors.57
SMR Transporters
The proton-motive force-driven SMR transporters are members of the drug/metabolite transporter (DMT) superfamily. They are very small and each has just four TMSs. Accordingly, dissimilar MFS transporters apparently act as monomers, SMR transporters. Moreover, they interchange the H+ received by the pumping out of either mono-cationic and ethidium or di-cationic such as parquets complexes.58 A study was conducted on chromosomally encoded putative drug efflux pump of the SMR family, named AbeS, from a multidrug-resistant strain of A. baumannii to reveal its role in antimicrobial resistance. Expression of the cloned abeS gene in hypersensitive E. coli host KAM32 resulted in declined susceptibility to various classes of dyes, antimicrobial agents, and detergents. The deletion of the abeS gene in A. baumannii confirmed its role in deliberating resistance to these complexes.59
ABC Transporters
The ABC transporters are functionally various and mediate ATP-dependent export or import of solutes.60 The structures of ABC transporters reveal transmembrane domains (TMDs) that contain substrate-binding domains (NBDs)-binding pockets and nucleotide that bind and hydrolyze ATP to drive the transportation cycle.61,62 The ABC exporters can be divided into heterodimeric and homodimeric groups, with the latter being particularly related to inherent and acquired antibiotic resistance in gram-positive bacteria (eg, EfrCD, LmrCD, and BmrCD). Although most homodimeric ABC transporters are thought to have two same nucleotide-binding sites, heterodimeric ABC transporters contain a degenerate binding site that does not use ATP hydrolysis.63 Most of the transporters involved in drug efflux belong to the ABC family and are seen in fungi and animal cells.64 In gram-negative bacteria, there are only rare cases of ABC family drug efflux pumps. In this regard, MsbA, the exporter of biosynthetic intermediates of lipopolysaccharide (LPS), was revealed to pump out drugs and erythromycin when extra expressed in Lactococcus lactis.65 The ideal bacterial ABC drug exporter is MacB of E. coli, which acts together with the periplasmic adaptor MacA and the OM channel TolC (Table 1).66 MacAB-TolC leads to an increase in the number of macrolide MICs in the presence of these pumps.66 Recently, crystal structures of ABC transporters have been reported for various organisms for both substrate exporters and importers. In type I bacterial ABC exporters for antibiotics and cytotoxic agents, the minimum functional unit has two transmembrane domains (TMDs) and two cytosolic nucleotide-binding domains (NBDs).67,68
 |
Table 1 Efflux Pump Families in Acinetobacter baumannii with Strains Which Pumps Described for the First Time |
MATE Transporters
Nowadays, the MATE transporters have developed a portion of a new superfamily,18 ie, the multidrug/oligosaccharide-lipid/polysaccharide (MOP) flippase superfamily, owing to their linking to transporters like the LPS flippase RfbX. These transporters are prevalent in bacteria and are similarly found in advanced animals and plants. For the first time, these transporters were recognized as a Na+/cationic antiporter agent named NorM from Vibrio parahaemolyticus.69 The main role of these pumps is to excrete components such as cationic dyes, fluoroquinolones, and aminoglycosides in the periplasmic space. Several of these transporters applied a gradient of Na+ and H+ as the energy source. The crystal structure of NorM from Vibrio cholerae points toward an outward-facing conformation with two gateways open to the outer leaflet of the IM.70 The AbeM protein showed sequence homology more than 90% similarity and an extra 70% identity with the sequence of ACIAD0429, which was assessed to be a NorM homolog of A. baumannii ATCC 19606 (Table 1). AbeM may be one of the MATE families of multidrug efflux pumps. The presence of AbeM efflux pump confers more than a 4-fold increment in the MICs of ciprofloxacin, norfloxacin, ofloxacin, gentamicin, triclosan, daunorubicin, doxorubicin, ethidium bromide, and rhodamine 6G. Furthermore, it generates a reproducible 2-fold increase in the MICs of kanamycin, chloramphenicol, erythromycin, trimethoprim, and tetraphenylphosphonium (TPPCl).71 Sequence analysis has shown that AbeM is a member of the MATE family of pumps. AbeM was found to be as an H+-coupled multidrug efflux pump and a unique member of the MATE family. Different types of efflux pumps in the most important strain of A. baumannii are introduced in Table 1. As can be seen, efflux pumps have key roles in adherence, biofilm formation, and multidrug resistance (MDR). Another important point from Table 1 is that E. coli strains extensively used as a host bacterium for gene cloning purposes.
Genetic Organization
The encoded genes for the AdeABC efflux pump are located on the bacterial chromosome.47 Generally, the genes are organized as a definite construction, or are attached and directly organized such as adeA, adeB, and adeC genes (Figure 2). Also, the encoding gene of the periplasmic auxiliary protein is positioned adjacent to the encoding gene of transporter protein, which is placed adjacent to the OMP. There are two regulatory genes, known as adeS and adeR, which are thoroughly associated with proteins of two-component regulatory systems. These genes are transcribed in the opposite direction to each other and are contained upstream from adeA. The double section system consists of signal transduction ways in bacteria that reply to environmental conditions.32,47 The protein AdeR (regulator) contains several amino acids. As a usual transcriptional manager, protein AdeS (sensor kinase) is smaller and reveals the activity of bacterial histidine kinase. Altogether, they adjust target gene expression in response to incentives. The sensor protein displays the environmental conditions and triggers or deactivates the response regulator protein, which controls the expression of the efflux pump.32,47 Double component regulation systems normally facilitate the adaptive responses of the bacterial cells to an extensive range of environmental stimuli.72 This system is related to diverse pumps of RND family performing in gram-negative bacteria among the AdeABC of A. baumannii.73,74 The state of the pump expression is controlled by such a system and needs the existence of the stimulus and a substrate in the environment of the cell. This substrate triggers the pump in order to eliminate the substrate around the cell. Antibiotics can act as inducers and regulate the expression of several efflux pumps at the stage of gene transcription or mRNA translation, by cooperating with regulatory systems.75 Few of pumps are autonomous of environmental stimuli and persistently active the distinct level. Such activity is defined as constitutive. Several studies have confirmed that the assembly of the AcrA, AcrB, and TolC proteins into a functional AcrAB-TolC pump is constitutively occurring in the existence or nonexistence of substrate molecules.76 The AdeABC pump of A. baumannii can modify the expression from inductive to the high level of the constitutive type, comprising the activity of the pump on the very high level named as overexpression. As a result of various mutations or inactivation by insertion sequences in the local regulatory genes, it is possible to activate and deactivate inactivator adeR in A. baumannii.47 Recently categorized proteobacterial antimicrobial compound efflux (PACE) transporter family is more important than A.baumanni and should be considered in future studies77 because they contain highly conserved in other pathogenic bacteria. Although their physiological role remains to be determined, their participation in drug resistance seems to be accidental.78
Efflux Pump Inhibitors
Efflux pump inhibitors (EPIs) are compounds that strengthen the activity of antibiotics against MDR bacteria.79 Many natural compounds and plant extracts have been evaluated for their resistance changing activities especially by reversing the natural resistance of bacteria to certain, antibiotics.79,80 Figure 3 illustrates the structures of the main known Efflux pump inhibitors. Lethal doses of antibiotics can activate numerous processes that allow bacteria to survive antibiotic exposure. Epigenetic studies also may play a part in upregulating efflux transporters. The perceived occurrence of persistence by a subpopulation of bacteria, which may be metabolically inactive survives initial exposure to antibiotics, may genetically match the drug-susceptible group or complicate strategies for treatment.81 Surprisingly, E. coli-persevered cells are reported to have improved efflux activity and lower intracellular antibiotic concentrations.82 Efflux pumps have a role in the formation of hetero-resistance. This function has been considered by subpopulations of bacteria that have a resistant phenotype, regarding clinical levels of resistance in each isolate.83 In various cases, MDR clinical isolates of bacteria display multiple mechanisms of resistance.40 For instance, fluoroquinolone-resistant MDR E. coli was found to improve efflux, reduce permeability, and decrease gyrase sensitivity to the drug (GyrA S83L).84 Extra-expression of efflux pumps is one mechanism that can donate to multidrug resistance. Forwarding protein synthesis to make efflux pumps is metabolically serious; therefore, it is perhaps not surprising that a metabolic cost can be related to drug resistance.85 Even though extra-expression of pumps might refer to drug tolerance, too much extra-expression is able in some cases to be unfavorable to the fitness and virulence of the bacterium, as perceived for case in point in an S. Typhimurium mutant overexpressing AcrAB.86 Additionally, mutations in pumps can be benefited to alter activity. Besides, modified binding pockets in AcrB have been detected to confer clinically appropriate MDR developed in a patient during the treatment of an infection by S. Typhimurium. A G288D substitution altered the substrate specificity of the pump, conferring ciprofloxacin resistance. Briefly, in drug-resistant clinical isolates of C. jejuni, mutations in the drug-binding pocket of the RND transporter CmeB have been recognized.87 The most important drugs, including omeprazole, reserpine, and verapamil, are well recognized in pharmaceutical drugs. Omeprazole is prescribed to treat gastric ulcers because of its capability to block H+ pumps. Therefore, it is recommended as a potential inhibitor of EP families using the H+ gradient to discharge antimicrobial drugs for the cytosol as well as MFS, SMR, and RND. Reserpine and verapamil are the blockers of the P-glycoprotein, an ABC EP found in humans. As ABC EPs are not defined as main contributors to antibiotic resistance, a low impression of these two molecules on clinical A. baumannii strains can be expected.88,89 In summary, efflux pumps obviously decrease the activity of many antibiotic families. Inhibition of the involved pumps can restore the drug susceptibility of resistant strains and might confirm the clinical activity of some traditional antibiotics.90 Using these inhibitors, one should consider the toxicity risk of these inhibitors for eukaryotic cells. This problem is attributed to their similar structure with that of human efflux pumps and bacterial ones.
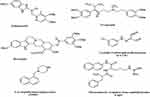 |
Figure 3 Structures of the main developed Efflux pump inhibitors. |
Conclusion
Acinetobacter spp. are among the most important hospital opportunistic pathogens throughout the world. Antibiotic resistance of this bacterium to a wide range of drugs has led to increased mortality rates. The data collected in this review show that efflux pumps play a distinctive role in relation to drug resistance. Among several inhibitors associated with these pumps, some are reviewed in this article. Studies demonstrate that any changes in the behavior of the pumps can affect antibiotic resistance. To ascertain whether synthetic compounds of natural materials can affect these pumps, further studies are needed. The efflux pumps are influenced by numerous genes. Any change in the expression of these genes may affect their performance. In summary, the most important challenge in Acinetobacter spp. infection is their drug resistance. Efflux pumps are the most important factor in the development of this procedure. Using various compounds, we can inhibit these pumps that can help us for control of infection. Efflux pumps are very important structures in bacteria. Since any changes in their performance can affect antibiotic resistance, it is worth paying more attention to the performance of these pumps.
Acknowledgment
This study was supported by Faculty of Medicine, Tabriz University of Medical Sciences with grant number 64650. We thank all staff of DARC and Imam Reza Hospital for their collaboration.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Blanchard C, Barnett P, Perlmutter J, Dunman PM. Identification of Acinetobacter baumannii serum-associated antibiotic efflux pump inhibitors. Antimicrob Agents Chemother. 2014;03535–14.
2. Zahedi Bialvaei A, Samadi Kafil H, Ebrahimzadeh Leylabadlo H, Asgharzadeh M, Aghazadeh M. Dissemination of carbapenemases producing Gram negative bacteria in the Middle East. Iran J Microbiol. 2015;7(5):226–246.
3. Mullié C, Bouharkat B, Guiheneuf R, Serra C, Touil-Meddah AT, Sonnet P. Efflux pumps in Acinetobacter baumannii: role in antibiotic resistance and interest of efflux pump inhibitors as additional therapeutic weapons. In: Méndez-Vilas A, editor. Antimicrobial Research: Novel Bioknowledge and Educational Programs. Formatex Research Center; 2017.
4. Bialvaei AZ, Samadi Kafil H. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin. 2015;31(4):707–721. doi:10.1185/03007995.2015.1018989
5. Coyne S, Courvalin P, Périchon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011;55(3):947–953. doi:10.1128/AAC.01388-10
6. Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–582. doi:10.1128/CMR.00058-07
7. Eliopoulos GM, Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46(8):1254–1263. doi:10.1086/529198
8. Manchanda V, Sanchaita S, Singh N. Multidrug resistant acinetobacter. J Glob Infect Dis. 2010;2(3):291. doi:10.4103/0974-777X.68538
9. Shiralizadeh S, Taghizadeh S, Asgharzadeh M, et al. Urinary tract infections: raising problem in developing countries. Rev Med Microbiol. 2018;29(4):159–165.
10. Fishbain J, Peleg AY. Treatment of Acinetobacter infections. Clin Infect Dis. 2010;51(1):79–84. doi:10.1086/652975
11. Bialvaei AZ, Kafil HS, Asgharzadeh M, Yousef Memar M, Yousefi M. Current methods for the identification of carbapenemases. J Chemother. 2016;28(1):1–19. doi:10.1179/1973947815Y.0000000063
12. Katragkou A, Roilides E. Successful treatment of multidrug-resistant Acinetobacter baumannii central nervous system infections with colistin. J Clin Microbiol. 2005;43(9):4916–4917. doi:10.1128/JCM.43.9.4916-4917.2005
13. Wieczorek P, Sacha P, Hauschild T, Zórawski M, Krawczyk M, Tryniszewska E. Multidrug resistant Acinetobacter baumannii–the role of AdeABC (RND family) efflux pump in resistance to antibiotics. Multidrug resistant Acinetobacter baumannii–the role of AdeABC (RND family) efflux pump in resistance to antibiotics. Folia Histochem Cytobiol. 2008;46(3):. doi:10.2478/v10042-008-0056-x
14. Vila J, Martí S, Sanchez-Céspedes J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J Antimicrob Chemother. 2007;59(6):1210–1215. doi:10.1093/jac/dkl509
15. Ribera A, Ruiz J, Jiminez de Anta MT, Vila J. Effect of an efflux pump inhibitor on the MIC of nalidixic acid for Acinetobacter baumannii and Stenotrophomonas maltophilia clinical isolates. J Antimicrob Chemother. 2002;49(4):697–698. doi:10.1093/jac/49.4.697
16. Piddock LJ. Multidrug-resistance efflux pumps? Not just for resistance. Nat. Rev. Microbiol. 2006;4(8):629.
17. Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun. 2014;453(2):254–267. doi:10.1016/j.bbrc.2014.05.090
18. Saier MH
19. Saier MH
20. Nikaido HJ. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob. Agents Chemother. 1989;33(11):1831.
21. Ma D, Cook D, Alberti M, Pon N, Nikaido H, Hearst JE. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175(19):6299–6313. doi:10.1128/jb.175.19.6299-6313.1993
22. Aires JR, Nikaido H. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J Bacteriol. 2005;187(6):1923–1929. doi:10.1128/JB.187.6.1923-1929.2005
23. Nikaido HJ. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 1996;178(20):5853.
24. Pumbwe L, Piddock LJV. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol Lett. 2002;206(2):185–189. doi:10.1111/j.1574-6968.2002.tb11007.x
25. Gill MJ, Brenwald NP, Wise R. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance inStreptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43(1):187–189.
26. Nikaido H, editor. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin Cell Dev Biol. 2001; Elsevier.
27. Kaatz GW, SM S. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39(12):2650–2655.
28. Webber M, Piddock LJV. The importance of efflux pumps in bacterial antibiotic resistance. J Antimicrob Chemother. 2003;51(1):9–11. doi:10.1093/jac/dkg050
29. Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Three’s company: component structures bring a closer view of tripartite drug efflux pumps. Curr Opin Struct Biol. 2004;14(6):741–747. doi:10.1016/j.sbi.2004.10.003
30. Paulsen I. Multidrug efflux pumps and resistance: regulation and evolution. Curr Opin Microbiol. 2003;6(5):446–451. doi:10.1016/j.mib.2003.08.005
31. Wieczorek P, Sacha P, Hauschild T, Zórawski M, Krawczyk M, Tryniszewska E. Multidrug resistant Acinetobacter baumannii–the role of AdeABC (RND family) efflux pump in resistance to antibiotics. Folia Histochem Cytobiol. 2008;46(3):257–267. doi:10.2478/v10042-008-0056-x
32. Magnet S, Courvalin P, Lambert TJ. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45(12):3375–3380. doi:10.1128/AAC.45.12.3375-3380.2001
33. Vila J, Martí S, Sanchez-Céspedes JJ. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J Antimicrob Chemother. 2007;59(6):1210–1215. doi:10.1093/jac/dkl509
34. Fournier P-E, Vallenet D, Barbe V, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;2(1):e7.
35. Quistgaard EM, Löw C, Guettou F, Nordlund PJ. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat Rev Mol Cell Biol. 2016;17(2):123.
36. Krishnamoorthy G, Leus IV, Weeks JW, Wolloscheck D, Rybenkov VV, Zgurskaya HI. Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria. mBio. 2017;8(5):e01172–17. doi:10.1128/mBio.01172-17
37. Zwama M, Yamasaki S, Nakashima R, Sakurai K, Nishino K, Yamaguchi AJ. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat Commun. 2018;9(1):124.
38. Venter H, Mowla R, Ohene-Agyei T, Ma SJ. RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front Microbiol. 2015;6(377).
39. Li X-Z, Nikaido H. Antimicrobial drug efflux pumps in Escherichia coli. In: Efflux-Mediated Antimicrobial Resistance in Bacteria. Drugs. 2009;69(12):1555–1623.
40. Li X-Z, Plésiat P, Nikaido HJ. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev. 2015;28(2):337–418. doi:10.1128/CMR.00117-14
41. Sapunaric F, Aldema-Ramos M, McMurry L. Tetracycline Resistance: Efflux, Mutation, and Other Mechanisms. In: White D, Alekshun M, McDermott P, editors. Frontiers in Antimicrobial Resistance. Washington, DC: ASM Press;2005:3–18. doi:10.1128/9781555817572.ch1.
42. McMurry L, Petrucci RE, Levy SB. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A. 1980;77(7):3974–3977. doi:10.1073/pnas.77.7.3974
43. Yamaguchi A, Udagawa T, Sawai TJ. Transport of divalent cations with tetracycline as mediated by the transposon Tn10-encoded tetracycline resistance protein. J Biol Chem. 1990;265(9):4809–4813.
44. Tamura N, Konishi S, Yamaguchi AJ. Mechanisms of drug/H+ antiport: complete cysteine-scanning mutagenesis and the protein engineering approach. Curr Opin Chem Biol. 2003;7(5):570–579. doi:10.1016/j.cbpa.2003.08.014
45. Marger MD, Saier MH
46. Lin M-F, Lin -Y-Y, Lan C-Y. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J Microbiol. 2017;55(2):130–136. doi:10.1007/s12275-017-6408-5
47. Marchand I, Damier-Piolle L, Courvalin P, Lambert T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii Is regulated by the AdeRS two-component system. Antimicrob Agents Chemother. 2004;48(9):3298–3304. doi:10.1128/AAC.48.9.3298-3304.2004
48. Rajamohan G, Srinivasan VB, Gebreyes WA. Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J Antimicrob Chemother. 2009;65(2):228–232. doi:10.1093/jac/dkp427
49. Yoon E-J, Courvalin P, Grillot-Courvalin C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother. 2013;57(7):2989–2995. doi:10.1128/AAC.02556-12
50. Abdi SN, Ghotaslou R, Asgharzadeh M, et al. AdeB efflux pump gene knockdown by mRNA mediated peptide nucleic acid in multidrug resistance Acinetobacter baumannii. Microb Pathog. 2020;138:1.
51. Radestock S, Forrest LR. The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J Mol Biol. 2011;407(5):698–715. doi:10.1016/j.jmb.2011.02.008
52. Yan N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem Sci. 2013;38(3):151–159. doi:10.1016/j.tibs.2013.01.003
53. Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH. The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279(11):2022–2035. doi:10.1111/j.1742-4658.2012.08588.x
54. Ehrnstorfer IA, Manatschal C, Arnold FM, Laederach J, Dutzler RJ. Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family. Nat Commun. 2017;8:14033.
55. Marti S, Fernandez-Cuenca F, Pascual A, et al. Prevalence of the tetA and tetB genes as mechanisms of resistance to tetracycline and minocycline in Acinetobacter baumannii clinical isolates. Enferm Infecc Microbiol Clin. 2006;24(2):77.
56. Sahu PK, Iyer PS, Gaikwad MB, Talreja SC, Pardesi KR, Chopade BA. An MFS transporter-like ORF from MDR Acinetobacter baumannii AIIMS 7 is associated with adherence and biofilm formation on biotic/abiotic surface. Int J Microbiol. 2012;2012.
57. Sharma A, Sharma R, Bhattacharyya T, Bhando T, Pathania R. Fosfomycin resistance in Acinetobacter baumannii is mediated by efflux through a major facilitator superfamily (MFS) transporter—AbaF. J Antimicrob Chemother. 2016;72(1):68–74. doi:10.1093/jac/dkw382
58. Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem. 1995;270(12):6856–6863. doi:10.1074/jbc.270.12.6856
59. Srinivasan VB, Rajamohan G, Gebreyes WA. Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53(12):5312–5316. doi:10.1128/AAC.00748-09
60. Narenji H, Gholizadeh P, Aghazadeh M, Rezaee MA, Asgharzadeh M, Kafil HS. Peptide nucleic acids (PNAs): currently potential bactericidal agents. Biomed Pharmacother. 2017;93:580–588. doi:10.1016/j.biopha.2017.06.092
61. Choudhury HG, Tong Z, Mathavan I, et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc Natl Acad Sci U S A. 2014;111(25):9145–9150. doi:10.1073/pnas.1320506111
62. Verhalen B, Dastvan R, Thangapandian S, et al. Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature. 2017;543(7647):738.
63. Hürlimann LM, Hohl M, Seeger MA. Split tasks of asymmetric nucleotide‐binding sites in the heterodimeric ABC exporter EfrCD. FEBS J. 2017;284(11):1672–1687. doi:10.1111/febs.14065
64. Cannon RD, Lamping E, Holmes AR, et al. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev. 2009;22(2):291–321.
65. Woebking B, Reuter G, Shilling RA, et al. Drug-lipid A interactions on the Escherichia coli ABC transporter MsbA. J Bacteriol. 2005;187(18):6363–6369. doi:10.1128/JB.187.18.6363-6369.2005
66. Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter inEscherichia coli. J Bacteriol. 2001;183(19):5639–5644. doi:10.1128/JB.183.19.5639-5644.2001
67. Locher KP. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23(6):487. doi:10.1038/nsmb.3216
68. Okada U, Yamashita E, Neuberger A, Morimoto M, van Veen HW, Murakami S. Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat Commun. 2017;8(1):1336. doi:10.1038/s41467-017-01399-2
69. Kuroda T, Tsuchiya TJ. Multidrug efflux transporters in the MATE family. Biochim Biophys Acta. 2009;1794(5):763–768. doi:10.1016/j.bbapap.2008.11.012
70. He X, Szewczyk P, Karyakin A, et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467(7318):991.
71. Su X-Z, Chen J, Mizushima T, Kuroda T, Tsuchiya TJ. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob Agents Chemother. 2005;49(10):4362–4364. doi:10.1128/AAC.49.10.4362-4364.2005
72. West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26(6):369–376. doi:10.1016/s0968-0004(01)01852-7
73. Baranova N, Nikaido HJ. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J Bacteriol. 2002;184(15):4168–4176. doi:10.1128/jb.184.15.4168-4176.2002
74. Perron K, Caille O, Rossier C, Van Delden C, Dumas J-L, Köhler TJ. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J Biol Chem. 2004;279(10):8761–8768. doi:10.1074/jbc.M312080200
75. Roberts MC. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19(1):1–24. doi:10.1111/j.1574-6976.1996.tb00251.x
76. Touzé T, Eswaran J, Bokma E, Koronakis E, Hughes C, Koronakis VJ. Interactions underlying assembly of the Escherichia coli AcrAB–TolC multidrug efflux system. Mol Microbiol. 2004;53(2):697–706. doi:10.1111/j.1365-2958.2004.04158.x
77. Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19(2):382–402. doi:10.1128/CMR.19.2.382-402.2006
78. Hassan KA, Liu Q, Elbourne LDH, et al. Pacing across the membrane: the novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res Microbiol. 2018;169(7–8):450–454. doi:10.1016/j.resmic.2018.01.001
79. Stavri M, Piddock LJ, Gibbons S. Bacterial efflux pump inhibitors from natural sources. J Antimicrob Chemother. 2006;59(6):1247–1260. doi:10.1093/jac/dkl460
80. Lorenzi V, Muselli A, Bernardini AF, et al. Geraniol restores antibiotic activities against multidrug-resistant isolates from gram-negative species. Antimicrob Agents Chemother. 2009;53(5):2209–2211. doi:10.1128/AAC.00919-08
81. Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler SJ. Bacterial persistence as a phenotypic switch. Science (New York, N.Y.). 2004;305(5690):1622–1625. doi:10.1126/science.1099390
82. Pu Y, Zhao Z, Li Y, et al. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol Cell. 2016;62(2):284–294.
83. Du D, Wang-Kan X, Neuberger A, et al. Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol. 2018;16:523–539. doi:10.1038/s41579-018-0048-6
84. Everett MJ, Jin YF, Ricci V, Piddock LJ. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996;40(10):2380–2386.
85. Zampieri M, Enke T, Chubukov V, Ricci V, Piddock L, Sauer UJ. Metabolic constraints on the evolution of antibiotic resistance. Mol Syst Biol. 2017;13(3):917.
86. Bailey AM, Ivens A, Kingsley R, Cottell JL, Wain J, Piddock LJV. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J Bacteriol. 2010;192(6):1607–1616. doi:10.1128/JB.01517-09
87. Yao H, Shen Z, Wang Y, et al. Emergence of a potent multidrug efflux pump variant that enhances campylobacter resistance to multiple antibiotics. mBio. 2016;7(5):e01543–16. doi:10.1128/mBio.01543-16
88. Ni W, Li Y, Guan J, et al. Effects of efflux pump inhibitors on colistin resistance in multidrug-resistant Gram-negative bacteria. Antimicrob Agents Chemother. 2016;60(5):3215–3218. doi:10.1128/AAC.00248-16
89. Aghamali M, Bialvaei AZ, Aghazadeh M, Asgharzadeh M, Kafil HS. Carbapenemase inhibitors. Rev Med Microbiol. 2017;28(3):104–113. doi:10.1097/MRM.0000000000000106
90. Pagès J-M, Masi M, Barbe J. Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol Med. 2005;11(8):382–389. doi:10.1016/j.molmed.2005.06.006
91. Yılmaz Ç, Özcengiz G. Antibiotics: pharmacokinetics, toxicity, resistance and multidrug efflux pumps. Biochem Pharmacol. 2017;133:43–62. doi:10.1016/j.bcp.2016.10.005
92. Su X-Z, Chen J, Mizushima T, Kuroda T, Tsuchiya T. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob Agents Chemother. 2005;49(10):4362–4364. doi:10.1128/AAC.49.10.4362-4364.2005
93. Li L, Hassan KA, Brown MH, Paulsen IT. Rapid multiplexed phenotypic screening identifies drug resistance functions for three novel efflux pumps in Acinetobacter baumannii. J Antimicrob Chemother. 2016;71(5):1223–1232. doi:10.1093/jac/dkv460
94. Roca I, Marti S, Espinal P, Martínez P, Gibert I, Vila J. CraA, a major facilitator superfamily efflux pump associated with chloramphenicol resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53(9):4013–4014. doi:10.1128/AAC.00584-09
95. Schneiders T, Findlay J, Amyes SGB. Efflux pumps in Acinetobacter baumannii. In: Bergogne-Bérézin E, Friedman H, Bendinelli M, editors. Acinetobacter Biology and Pathogenesis. New York, NY: Springer US; 2008:105–127.
96. Fernández-Alarcón C, Miranda CD, Singer RS, et al. Detection of the floR gene in a diversity of florfenicol resistant gram-negative bacilli from Freshwater Salmon farms in chile. Zoonoses Public Health. 2010;57(3):181–188. doi:10.1111/jvb.2010.57.issue-3
97. Rajamohan G, Srinivasan VB, Gebreyes WA. Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii. J Antimicrob Chemother. 2010;65(9):1919–1925. doi:10.1093/jac/dkq195
98. Hassan KA, Liu Q, Henderson PJF, Paulsen IT. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio. 2015;6(1):e01982–14. doi:10.1128/mBio.01982-14
99. Lin M-F, Lin Y-Y, Tu C-C, Lan C-Y. Distribution of different efflux pump genes in clinical isolates of multidrug-resistant Acinetobacter baumannii and their correlation with antimicrobial resistance. J Microbiol Immunol Infect. 2017;50(2):224–231. doi:10.1016/j.jmii.2015.04.004
100. Pérez-Varela M, Corral J, Aranda J, Barbé J. Functional characterization of AbaQ, a novel efflux pump mediating quinolone resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2018;62(9):e00906–18. doi:10.1128/AAC.00906-18
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
