Back to Journals » Drug Design, Development and Therapy » Volume 13
A novel mutation panel for predicting etoposide resistance in small-cell lung cancer
Authors Qiu Z, Lin A, Li K, Lin W , Wang Q, Wei T, Zhu W, Luo P , Zhang J
Received 16 February 2019
Accepted for publication 31 May 2019
Published 21 June 2019 Volume 2019:13 Pages 2021—2041
DOI https://doi.org/10.2147/DDDT.S205633
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Anastasios Lymperopoulos
Zhengang Qiu,1,2,* Anqi Lin,1,* Kun Li,1,* Weiyin Lin,1,* Qiongyao Wang,1 Ting Wei,1 Weiliang Zhu,1 Peng Luo,1 Jian Zhang1
1Department of Oncology, Zhujiang Hospital, Southern Medical University, Guangzhou 510282, People’s Republic of China; 2Department of Oncology, First Affiliated Hospital of Gannan Medical University, Ganzhou 341000, People’s Republic of China
*These authors contributed equally to this work
Purpose: Platinum-based chemotherapy, consisting of etoposide and cisplatin (EP), has been the cornerstone of therapy for extensive-stage small-cell lung cancer (ES-SCLC) for decades. Despite the marked initial sensitivity of SCLC to chemotherapy, EP regimens cannot avoid the emergence of drug resistance in clinical practice. With the rise of new chemotherapy regimens in recent years and the primary resistance or insensitivity of ES-SCLC to EP regimens, it is desirable to be able to identify patients with resistant or insensitive ES-SCLC.
Methods: The sequencing and drug sensitivity data of SCLC cell lines were provided by The Genomics of Drug Sensitivity in Cancer Project (GDSC). The data regarding sensitivity to etoposide of 54 SCLC cell lines were analyzed, and etoposide-sensitive cell lines and etoposide-resistant cell lines were differentiated according to the IC50 values defined by the GDSC. ROC curve analysis was performed on all mutations and combinations of mutations to select the optimal panel to predict resistance to etoposide.
Results: ROC analysis of etoposide resistance revealed that the most significant single gene mutation indicating resistance to etoposide was CSMD3, and the accuracy of predicting resistance to etoposide proved to be the highest when there was any mutation in CSMD3/PCLO/RYR1/EPB41L3, area under the curve =0.804 (95% confidence interval: 0.679–0.930,P<0.001).
Conclusion: This study found that a panel with four genes (CSMD3, EPB41L3, PCLO, and RYR1) can accurately predict sensitivity to etoposide. These findings provide new insights into the overall treatment for patients with ES-SCLC that is resistant or insensitive to etoposide.
Keywords: small-cell lung carcinoma, etoposide, EP regimens, IP regimens, gene mutation
Introduction
In recent years, humans have made significant progress in the early detection, early diagnosis, early treatment, and even prevention of cancer. However, lung cancer is the most commonly diagnosed cancer (11.6%) and the leading cause of cancer-related death (18.4%) worldwide.1 Currently, there are approximately 2.1 million lung cancer patients worldwide.1 Approximately 12–15% of new lung cancer patients are diagnosed with small-cell lung cancer (SCLC).2,3 According to the latest National Comprehensive Cancer Network (NCCN) Guidelines, an estimated 29,654 new cases of SCLC occurred in the United States in 2017.4,5 Studies have shown that the incidence of SCLC is attributable to cigarette smoking, and the smoking pack-years increases, so does the risk of SCLC. Ninety percent of patients with SCLC have been or are currently smokers, and smoking duration is positively associated with an increased risk of SCLC.6,7 In addition, SCLC is characterized by a high growth fraction, a high degree of malignancy, and the early development of widespread metastases.8,9 The 5-year survival rate in patients with SCLC is only 6.6%. Currently, SCLC is divided into limited-stage SCLC (LS-SCLC) and extensive-stage SCLC (ES-SCLC). Unfortunately, the 5-year survival rates are only 1.6% and 12.1% for patients with ES-SCLC (1/3) and ES-SCLC (2/3),8–11 respectively.
At present, surgery is one of the main methods of cancer treatment, but it is rarely used in the treatment of patients with SCLC. It is only suitable for a small number of stage I patients with SCLC (2%-5%) who do not have mediastinal lymph node metastasis. In the past few decades, a platinum compound in combination with the topoisomerase-II inhibitor etoposide beyond 4 to 6 cycles of chemotherapy (EP) has become the cornerstone of treatment for patients with ES-SCLC for palliative care.11–13 In recent years, the chemotherapy for ES-SCLC has mainly been irinotecan, cisplatin (IP) and EP regimens.14 Despite the substantial initial sensitivity of SCLC to chemotherapy in the early stages of treatment, more than 90% of patients eventually develop clinical drug resistance and die as a result of relapse.8,9 At present, there is a great deal of controversy about the therapeutic effect and safety tolerance of IP and EP in the treatment of ES-SCLC. In 2002, a randomized, multicenter, phase III trial (J9511) performed in Japan reported that patients with ES-SCLC who were treated with IP experienced a median survival of 12.8 months compared with 9.4 months for patients treated with EP (P=0.002). In addition, the 1-year survival rates were 58.4% vs 37.7% and the median progression-free survival (PFS) rates were 12.8 months vs 9.4 months in the IP and EP groups, respectively.15 Furthermore, Hermes et al studied 220 patients with ES-SCLC, and the results showed that the median overall survival (OS) was slightly higher in those receiving IP than in those receiving EP (8.5 months vs 7.1 months, P=0.04).16 However, it is surprising that there were no significant differences in the efficacy and survival of the IP and EP groups in 4 subsequent phase III trials.17–20 In a cohort study from Korea, the median OS and median PFS of patients with ES-SCLC treated with IP were 10.9 months and 6.5 months, respectively, whereas the median OS and PFS in the EP arm were 10.3 months (P=0.120) and 5.8 months (P=0.115), respectively. Similarly, no significant differences were observed in the 1- and 2-year survival rates in the IP versus EP groups. In the subgroup analysis, males, patients <65 years old and patients with Eastern Cooperative Oncology Group performance status (ECOG PS) ≤1 were treated with IP or EP, and the two groups had significant therapeutic differences. In addition, there was a significant difference in the objective response rate (ORR) between the IP group and the EP group (62.4% vs 48.2%, P=0.006).21
Currently, 4 to 6 cycles EP is the standard therapy widely used for a majority of SCLC in the clinic, with an ORR of 50%-80%.22 However, the median OS of patients with ES-SCLC is only 9 months, with only 2% of patients surviving after 5 years.14,23 Although SCLC usually responds well to chemotherapy regimens in the early stages of treatment, subsequent clinical drug resistance and disease recurrence occur in more than 90% of patients.8,9 This may be due to the existence of cancer stem cells that are relatively resistant to cytotoxic therapy. Chemotherapy cannot destroy residual tumor cells, leading to a high recurrence rate and a high drug resistance rate in SCLC.24 Primary resistance or acquired resistance to chemotherapy is a major factor in the poor prognosis of patients with lung cancer.25–27 In the drug sensitivity data from GDSC, we found that the IC50 of etoposide in the 54 SCLC cell lines ranged from 0.242 μM to 319 μM, and the drug resistance cut-off value provided by the website was 16 μM. In total, 65% of patients have SCLC that is sensitive to etoposide, which is close to the response rate for etoposide.28 Therefore, if we are able to select patients with ES-SLCL that is not sensitive to etoposide before treating them with standard chemotherapy, we could choose a different chemotherapy regimen to treat these patients, hopefully improving survival outcomes in those ES-SCLC patients. Survival time was significantly improved with the new chemotherapy compared with EP. However, there is currently no clinically relevant prediction factor and screening for appropriate means of insensitivity to etoposide.
To date, a growing number of studies have shown that the emergence of primary or acquired platinum and Topoisomerase Inhibitors resistance in EP is associated with certain gene expression changes or/and gene mutations.29 Chiu et al30 found that FBXL7 is a biomarker of poor prognosis in patients with ovarian cancer. A high expression level of FBXL7 is positively associated with a low survival rate in ovarian cancer patients, and the FBXL7 mRNA level and ovarian cancer cell line paclitaxel (PTX) IC50 values were positively correlated, leading to the speculation that the upregulation of FBXL7 expression results in resistant ovarian cancer cell lines. In addition, Chiu et al31 detected the transcriptional level of the shared gene in HCC38 (PTX-sensitive) and MDA-MB436 (PTX-resistant) TNBC cells posttreatment with paclitaxel. They found that the downregulation of miR-1180 may regulate OTUD7B, ultimately negatively regulating the NF-κB-Lin28 axis. This in turn triggers Let-7 microRNA-mediated caspase-3 downregulation, ultimately leading to resistance to PTX. Based on these findings, the sensitivity and drug resistance of tumor cells to chemotherapy can be predicted by gene expression levels. Thus, patients with ES-SLCL that is sensitive or insensitive to chemotherapy can be further distinguished. We hope that the sensitivity of ES-SCLC to etoposide can be predicted by gene mutation panels, allowing the selection of patients with ES-SCLC that is insensitive to etoposide before standard chemotherapy is administered and the development of personalized, precise chemotherapy to extend patients’ OS and improve their quality of life (QOL).
To this end, we analyzed the sequencing and drug sensitivity data for a SCLC cell line through the GDSC database to determine whether mutations can predict the primary resistance to etoposide and try to explain the potential underlying mechanism to provide first-line treatment recommendations for patients with ES-SCLC.
Methods
Drug response, gene expression and mutation data
The natural logarithm half maximal inhibitory concentration (IC50) of all selected erlotinib-related cell lines were obtained from the GDSC (
Screening of mutated resistance genes
There were 54 SCLC cell lines in the GDSC with drug sensitivity data for etoposide. The GDSC site defined etoposide-resistant cell lines as those with IC50 values ≥16 μM and etoposide-sensitive cell lines as those with IC50 values <16 μM. ROC curve analysis was performed for all mutations, and the cell lines with areas under the curve (AUCs) >0.5 were selected and randomly combined; then, resistance to etoposide was predicted by the combined mutation panels. The Youden Index values obtained by various combined ROC analyses were sorted to select the best combination.
Statistical analysis
The IC50 distribution for etoposide in various cell lines was obtained with the GDSC web tool. ROC analysis and mapping were performed with SPSS 21.0 (IBM SPSS Statistics, IBM Corporation); mutation and gene expression data were analyzed and mapped with the maftools32 and limma packages33 in R. In the differential analysis of the gene expression profiles, P<0.05 and FC>1.5 orFC<2/3 were considered to indicate significant differences. The survival analysis was with the log-rank test after the Kaplan-Meier analysis to investigate the predictive ability of a mutation panel with regard to survival. Gene Ontology (GO) annotation analysis and KEGG pathway enrichment analysis of the differentially expressed genes (DEGs) in this study were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (
Results
The sensitivity of cancer cell lines to drugs is mainly expressed as the IC50 value, which refers to the concentration of drug that kills half of the tumor cells in vitro. Because the drug concentration is diluted to 1/10 or 1/100, we used lnIC50 values to distinguish between resistant or sensitive cell lines. Based on the GDCS 7.0 database (updated on March 20, 2018), there are 64 SCLC cell lines, but only 54 of them have etoposide susceptibility data (drug sensitivity data), WES mutation data and RNA Seq data.
Using the GDSC website tools, we obtained the IC50 distribution for etoposide by tissue type (Figure 1A). We found that most of the tumors are sensitive to etoposide, and the IC50 values of most cell SCLC lines indicate that they are sensitive to etoposide. By analyzing the IC50 values of the 54 SCLC cell lines shown in Figure 1B, we found that there are 35 cell lines that are sensitive to etoposide, accounting for 64.8% of the total, and their median and mean IC50 values were 2.06 μM (range: 0.242–15.2 μM) and 4.02±4.07 μM, respectively. In total, 19 strains were resistant to etoposide, accounting for 35.2% of the total, and their median and mean IC50 values were 50.0 μM (range: 16.4–319.0 μM) and 71.9±71.8 μM, respectively. The raw data for the IC50 values of all cell lines with regard to etoposide can be found in Table S1.
 | Figure 1 (A) IC50 distribution for etoposide by tissue type. (B) The scatter plot of IC50 distribution for etoposide of 54 SCLC cell lines.Abbreviation: IC50, half maximal inhibitory concentration. |
After sorting the IC50 values for etoposide, we found that in the mutation landscape of the 54 SCLC cell lines (Figure 2), the genes with the highest mutation frequencies were TP53 (91%), TTN (78%) and Rb1 (70%). Among them, TP53 and TTN mutations were mainly missense mutations, while the Rb1 mutations were mainly nonsense and splice mutations.
 | Figure 2 Mutation landscape of 54 SCLC cell lines.Abbreviation: SCLC, small-cell lung cancer. |
We performed an ROC analysis of to predict etoposide resistance using all mutated genes (see Table S2). From the ROC curves, we found that the most significant single gene mutation associated with resistance to etoposide was CSMD3, with an AUC of 0.697 (P=0.016) (Table 1). By experimenting with different combinations, we found that when any mutations occurred in CSMD3/PCLO/RYR1/EPB41L3, the accuracy of predicting resistance to etoposide was the highest (AUC=0.804, 95% CI: 0.679–0.930, P<0.001) (Table 1). The ROC curve results of the panel composed of CSMD3/PCLO/RYR1/EPB41L3 and the individual genes are shown in Figure 3A.
 | Table 1 Receiver operator characteristic curve analysis for four-gene panel and four genes separately to etoposide resistance status in small-cell lung cancer cell lines |
 | Figure 3 (A) ROC curve of the panel and four mutations; (B) Kaplan–Meier overall survival analyses for the four-gene panel in clincal trial of SCLC.Abbreviation: SCLC, small-cell lung cancer. |
We performed a log-rank test with the Kaplan–Meier plots according to mutations and clinical follow-up data in 110 SCLCs published by George et al34 In addition, we found a significantly lower average survival time in patients with CLC with any mutation in CSMD3/PCLO/RYR1/EPB41L3 than in those with no mutations in all four genes (35.6±5.3 months vs 76.7±12.1 months, P=0.040) (Figure 3B). By analyzing significantly enriched KEGG pathways of DEGs, we found that there was a significant association between both CSMD3 and RYR1 mutations and MAPK signaling pathway (P=0.015 and P=0.023, respectively) (Table 2).
 | Table 2 Significantly enriched KEGG pathways of DEGs |
Discussion
EP has been the most common therapy for ES-SCLC for decades. As a standard treatment, it can inhibit tumor proliferation, relieve clinical symptoms, and achieve ideal results.13,34–37 We found that 19 (35.2%) of the 54 SCLC cell lines were insensitive to etoposide according to the data from the GDSC. Currently, the clinically accepted ORR of EP is 50–80%.23 Based on the above findings, the majority of patients with SCLC do not receive survival benefits from EP, indicating that screening for patients with primary resistance to etoposide is necessary. Therefore, this study further analyzed the mutation, gene expression and etoposide sensitivity data of 54 ES-SCLC cell lines obtained from the GDSC. We identified four genes, namely, CSMD3, EPB41L3, PCLO, and RYR1; mutations in these genes predict resistance to etoposide. The predictive sensitivity this four-gene panel for resistance to etoposide is as high as 85%, with 77.8% accuracy when screening for patients with primary etoposide resistance. In addition, the ROC showed an AUC of 0.804 (95% CI 0.679–0.930), and the model was considered to have a high degree of confidence.
Recently, a small phase III trial performed in Japan compared the efficacy of IP and EP in patients with ES-SCLC15. The trial results showed a higher median OS (12.8 months vs 9.4 months), 1-year survival rate (58.4% vs 37.7%) and 2-year survival rate (19.5% vs 5.2%) after IP than after EP. In addition, Hermes et al16 studied 220 patients with ES-SCLC, and the results showed a longer median OS resulting from the IP regimen compared with the EP regimen (8.5 months vs 7.1 months, P=0.04).
We analyzed the data and found that mutations in both CSMD3 and RYR1 can cause the activation of the downstream MAPK signaling pathway (Figure 4). In addition, Liu et al36 found that etoposide activates the MAPK/ERK signaling pathway, inhibits p53 expression and enhances c-Myc expression to decrease the sensitivity of gastric cancer cells to chemotherapy in. Therefore, we hypothesized that mutations in the CSMD3 and RYR1 genes may cause a significant resistance to etoposide in ES-SCLC via the downstream MAPK signaling pathway. It is well known that etoposide induces DNA double-strand breakage (DSB) and triggers the DNA damage response by activating the ataxia telangiectasia-mutated gene (ATM) DNA repair is a process of energy dissipation, and ATP-dependent chromatin remodeling complexes participate in DSB repair.37 In aerobic conditions, tumor cells preferentially perform glycolysis rather than providing energy for cell growth through the more efficient oxidative phosphorylation pathway and are therefore characterized by high glucose uptake, glycolysis activity levels and lactic acid content in the metabolites. Glycolysis consumes more glucose but produces less ATP.38 The PI3K/AKT signaling pathway promotes aerobic glycolysis by upregulating cell surface glucose transporters39 and glycolytic enzymes in tumor cells.40,41 Surprisingly, we found that the mutation of the EPB41L3 gene caused increased activity of the glucose metabolism pathway in tumor cells. Therefore, we speculate that mutations in EPB41L3 may reduce sensitivity to etoposide through DNA repair in tumor cells. In addition, AKT is involved in the repair of DNA damage caused by genotoxicity, mainly by the action of DNA-dependent protein kinase (DNA-PK), the kinase ATM/ATM and nonhomologous end joining (NHEJ) to repair DSB.42 Makinoshima et al43 found that PI3K/AKT/mTOR signaling inhibitors can effectively inhibit the expression of GLUT1 on the cell membrane. They used RNAi to interfere with the expression of GLUT1, ultimately reducing the aerobic glycolysis process and cell proliferation rate. Furthermore, our results suggest that PCLO mutations cause activation of the PI3K-Akt pathway, so we hypothesized that PCLO mutations may enhance glucose metabolism by activating the PI3K/Akt pathway, thereby enhance the ability of the tumor cell to repair DNA.
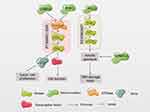 | Figure 4 Potential mechanism of the four-gene panel to predict the resistance of etoposide in SCLC.Abbreviation: SCLC, small-cell lung cancer. |
Identifying outpatients with ES-SCLC that is not sensitive to etoposide and treating them with another combination therapy are important steps in improving the survival of patients with SCLC. Screening for the sensitivity to etoposide in patients with SCLC who are receiving chemotherapy for the first time allows clinicians to use a different combination chemotherapy regimen (Table 3) in these patients to avoid treatment failure due to primary resistance to etoposide. Currently, alternative treatment options that are commonly used in clinical practice include IP protocols, platinum-based drugs plus paclitaxel, and IP plus sunitinib. A phase II clinical trial (NCT00454324) on the use of a platinum-based compound plus paclitaxel in patients with ES-SCLC has shown good efficacy.44 In a phase II clinical trial (NCT00695292),45 sunitinib combined with IP for patients with ES-SCLC showed potential clinical efficacy and safety, with an ORR of 59%, a one-year survival rate of 54% and a median PFS of 7.6 months. In recent years, combinations of various chemotherapy regimens have been shown to provide excellent survival advantages in patients with ES-SCLC. It may be possible to classify patients by adding inclusion criteria and then use a more specific new chemotherapy regimen as a clinical treatment to achieve individualized and precise treatment of ES-SCLC patients, overcoming the treatment bottleneck for patients with ES-SCLC that is resistant to EP and ultimately prolonging their survival time and improving their QOL.
 | Table 3 Completed/ongoing clinical trials of alternative treatment of etoposide in SCLC patients |
There were some limitations in this study. First, the most suitable alternative drug at present is irinotecan. GDSC does not provide data regarding the sensitivity to irinotecan, and the sensitivity of etoposide-resistant ES-SCLC to irinotecan is still unclear. Second, currently, there are no suitable large-sample clinical datasets that directly support our conclusions, and relevant clinical research needs to be further conducted to verify our hypothesis; moreover, we have initialed a clinical trial(NCT03162705) and hope this onging clincal trial could provide more direct evidence onni. Third, the accuracy of the model prediction is inadequate, and it may be necessary to expand the model to optimize it.
Conclusion
In conclusion, we analyzed the mutation and gene expression data from the GDSC of 54 ES-SCLC cell lines with regard to etoposide susceptibility and found that the panel including CSMD3, EPB41L3, PCLO, and RYR1 can likely predict the sensitivity of ES-SCLC to etoposide and, therefore, the clinical survival of patients with SCLC.
Acknowledgments
Work cited in this review was supported by the National Natural Science Foundation of China (grant numbers 81672267, 81772457, and 81871859) and the Province Natural Science Foundation of Guangdong (grant numbers 2016A030313632 and 2017A030313567).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
2. Haddadin S, Perry MC. History of small-cell lung cancer. Clin Lung Cancer. 2011;12(2):87–93. doi:10.1016/j.cllc.2011.03.002
3. Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clin Proc. 2008;83:355–367. doi:10.4065/83.3.355
4. Howlader N, Noone AM, Krapcho M, et al. SEER cancer statistics review, 1975–2014, National Cancer Institute. Bethesda, MD, based on November 2016 SEER data submission, posted to the SEER website, April 28, 2017. Available from:
5. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi:10.3322/caac.21442
6. Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi:10.1007/978-3-319-24223-1_1
7. Conen K, Hagmann R, Hess V, et al. Incidence and predictors of Bone metastases (BM) and Skeletal-related events (SREs) in Small cell lung cancer (SCLC): a Swiss patient cohort. J Cancer. 2016;7:2110–2116. doi:10.7150/jca.16211
8. Schiller JH, Adak S, Cella D, et al. Topotecan versus observation after cisplatin plus etoposide in extensive-stage small-cell lung cancer: E7593–a phase III trial of the eastern cooperative oncology group. J Clin Oncol. 2001;19:2114–2122. doi:10.1200/JCO.2001.19.8.2114
9. Jett JR, Schild SE, Kesler KA, et al. Treatment of small-cell lung cancer: diagnosis and management of lung cancer, 3rd ed: American college of chest physicians evidence-based clinical practice guidelines. Chest. 2013;143(5Suppl):e400S–e419S. doi:10.1378/chest.12-2363
10. SEER cancer statistics review. 1975–2009. Available from:
11. Stinchcombe TE, Gore EM. Limited-stage small-cell lung cancer: current chemoradiotherapy treatment paradigms. Oncologist. 2010;15:187–195. doi:10.1634/theoncologist.2009-0298
12. Morabito A, Carillio G, Daniele G, et al. Treatment of small-cell lung cancer. Crit Rev Oncol Hematol. 2014;9:257–270. doi:10.1016/j.critrevonc.2014.03.003
13. Früh M, De Ruysscher D, Popat S, et al. Small-cell lung cancer (SCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24:vi99–vi105. doi:10.1093/annonc/mdt178
14. Ogino H, Hanibuchi M, Kakiuchi S, et al. Analysis of the prognostic factors of extensive disease small-cell lung cancer patients intokushima university hospital. J Med Invest. 2016;63:286–293.
15. Noda K, Nishiwaki Y, Kawahara M, et al. Irinotecan plus cisplatin compared with etoposide plus cisplatin for extensive small-cell lung cancer. N Engl J Med. 2002;346:85–91. doi:10.1056/NEJMoa003034
16. Hermes A, Bergman B, Bremnes R, et al. Irinotecan plus carboplatin versus oral etoposide plus carboplatin in extensive small-cell lung cancer: a randomized phase III trial. J Clin Oncol. 2008;26:4261–4267. doi:10.1200/JCO.2007.15.7545
17. Hanna N, Bunn PA
18. Lara PN
19. Schmittel A, Sebastian M, Fischer von Weikersthal L, et al. A German multicenter, randomized phase III trial comparing irinotecan-carboplatin. Ann Oncol. 2011;22:1798–1804. doi:10.1093/annonc/mdq652
20. Zatloukal P, Cardenal F, Szczesna A, et al. A multicenter international randomized phase III study comparing cisplatin in combination with irinotecan or etoposide in previously untreated small-cell lung cancer patients with extensive disease. Ann Oncol. 2010;21(9):1810–1816. doi:10.1093/annonc/mdq036
21. Kim DW, Kim HG, Kim JH, et al. Randomized phase III trial of irinotecan plus cisplatin versus etoposide plus cisplatin in chemotherapy-naïve Korean patients with extensive-disease small-cell lung cancer. Cancer Res Treat. 2019;51:119–127. doi:10.4143/crt.2018.019
22. Rossi A, Di Maio M, Chiodini P, et al. Carboplatin- or cisplatin-based chemotherapy in first-line treatment of small-cell lung cancer: the COCIS meta-analysis of individual patient data. J Clin Oncol. 2012;30:1692–1698. doi:10.1200/JCO.2011.40.4905
23. Chute JP, Chen T, Feigal E, et al. Twenty years of phase III trials for patients with extensive-stage small-cell lung cancer: perceptible progress. J Clin Oncol. 1999;17:1794–1801. doi:10.1200/JCO.1999.17.6.1794
24. Goldie JH, Coldman AJ. A mathematic model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat Rep. 1979;63(11–12):1727–1733.
25. van Meerbeeck JP, Fennell DA, de Ruysscher DK. Small-cell lung cancer. Lancet. 2011;378(9804):1741–1755. doi:10.1016/S0140-6736(11)60984-7
26. Dingemans AM, Witlox MA, Stallaert RA, et al. Expression of DNA topoisomerase IIalpha and topoisomerase IIbeta genes predicts survival and response to chemotherapy in patients with small-cell lung cancer. Clin Cancer Res. 1999;5:2048–2058.
27. Viktorsson K, De Petris L, Lewensohn R. The role of p53 in treatment responses of lung cancer. Biochem Biophys Res Commun. 2005;331:868–880. doi:10.1016/j.bbrc.2005.03.192
28. Yang W, Soares J, Greninger P, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–61. doi:10.1093/nar/gks1111
29. Bonanno L, Favaretto A, Rugge M, et al. Role of genotyping in non-small-cell lung cancer treatment: current status. Drugs. 2011;71:2231–2246. doi:10.2165/11597700-000000000-00000
30. Chiu HW, Chang JS, Lin HY, et al. FBXL7 upregulation predicts a poor prognosis and associates with a possible mechanism for paclitaxel resistance in ovarian cancer. J Clin Med. 2018;7(10):330. doi:10.3390/jcm7100330
31. Chiu HW, Lin HY, Tseng IJ, et al. OTUD7B upregulation predicts a poor response to paclitaxel in patients with triple-negative breast cancer. Oncotarget. 2017;9:553–565. doi:10.18632/oncotarget.23074
32. Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi:10.1101/gr.239244.118
33. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi:10.1093/nar/gkv007
34. George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small-cell lung cancer. Nature. 2015;524(7563):47–53. doi:10.1038/nature14664
35. Liu SQ, Yu JP, Yu HG, et al. Activation of Akt and ERK signalling pathways induced by etoposide confer chemoresistance in gastric cancer cells. Dig Liver Dis. 2006;38:310–318. doi:10.1016/j.dld.2006.01.012
36. Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi:10.1038/nature01368
37. Lans H, Marteijn JA, Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin. 2012;5:4. doi:10.1186/1756-8935-5-4
38. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314.
39. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi:10.1016/j.cmet.2007.10.002
40. Majewski N, Nogueira V, Bhaskar P, et al. Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Mol Cell. 2004;16:819–830. doi:10.1016/j.molcel.2004.11.014
41. Buzzai M, Bauer DE, Jones RG, et al. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene. 2005;24:4165–4173. doi:10.1038/sj.onc.1208622
42. Xu N, Lao Y, Zhang Y, et al. Akt: a double-edged sword in cell proliferation and genome stability. J Oncol. 2012;2012:951724. doi:10.1155/2012/951724
43. Makinoshima H, Takita M, Saruwatari K, et al. Signaling through the Phosphatidylinositol 3-kinase (PI3K)/Mammalian target of Rapamycin(mTOR) axis is responsible for aerobic glycolysis mediated by glucose transporter in Epidermal growth factor receptor(EGFR) -mutated lung adenocarcinoma. J Biol Chem. 2015;290:17495–17504. doi:10.1074/jbc.M115.660498
44. Grilley-Olson JE, Keedy VL, Sandler A, et al. A randomized phase II study of carboplatin with weekly or every-3-week nanoparticle albumin-bound paclitaxel (abraxane) in patients with extensive-stage small-cell lung cancer. Oncologist. 2015;20:105–106. doi:10.1634/theoncologist.2014-0327
45. Spigel DR, Greco FA, Rubin MS, et al. Phase II study of maintenance sunitinib following irinotecan and carboplatin as first-line treatment for patients with extensive-stage small-cell lung cancer. Lung Cancer. 2012;77:359–364. doi:10.1016/j.lungcan.2012.03.009
Supplementary materials
 | Table S1 Etoposide IC50 values of 54 SCLC cell lines |
 |
 |  |  |  |
 |  |
 |  |
Table S2 ROC curve of all genes (mutation frequency >10%) |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
