Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
A Functional Variant rs2072915 is Associated with the Susceptibility and Mortality of Cervical Squamous Cell Carcinoma
Authors Li RL, Wu JH, Guo M, Sha LX, Xia SQ, Xu L
Received 10 March 2021
Accepted for publication 20 May 2021
Published 16 June 2021 Volume 2021:14 Pages 705—712
DOI https://doi.org/10.2147/PGPM.S310504
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Ren-Liang Li,1 Jiao-Hong Wu,1 Min Guo,1 Li-Xiao Sha,1 Shu-Qi Xia,1 Lian Xu2
1Department of Obstetrics and Gynecology, Wenzhou People’s Hospital, Wenzhou, Zhejiang, 325000, People’s Republic of China; 2Department of Pathology, West China Second University Hospital, Sichuan University, Chengdu, Sichuan, 610041, People’s Republic of China
Correspondence: Lian Xu Tel/Fax +86-57788306071
Email [email protected]
Purpose: Genetic variant has been demonstrated to be a risk factor for the occurrence and outcome of cervical squamous cell carcinoma (CSCC). From previous genome wide association studies, 6p21.32 has been identified as a susceptibility locus of CSCC. The purpose of this study was to investigate the association of a polymorphism rs2072915 located in 6p21.32 with the risk of CSCC and examine the potential mechanism of the rs2072915 in CSCC pathogenesis.
Patients and Methods: The rs2072915 was genotyped using polymerase chain reaction (PCR)-restriction fragment length polymorphism. miR-637 and RXRB mRNA expression levels in CSCC patients were examined using quantitative PCR. miR-637 target site was determined using the dual-luciferase reporter assay.
Results: The rs2072915 was associated with a significantly increased risk (AA vs TT: adjusted OR = 2.48, 95% CI, 1.57– 3.94, P < 0.001; AT/AA vs TT: adjusted OR = 1.38, 95% CI, 1.06– 1.80, P = 0.018; A vs T: adjusted OR = 1.49, 95% CI, 1.21– 1.84, P < 0.001, respectively) and shorter survival time of CSCC (P = 0.03). Patients with the rs2072915 AA genotype displayed lower levels of RXRB that is a target of miR-637.
Conclusion: These findings suggest that the rs2072915 T > A change might augment the binding energy of miR-637 to RXRB, result in lower levels of RXRB, and thus contribute to the risk of CSCC.
Keywords: miR-637, polymorphism, survival, cervical squamous cell carcinoma
Introduction
Cervical cancer is one of the most common malignancies in women worldwide, with about 570,000 newly diagnosed cases and 311,000 associated deaths annually (estimates for 2018). The incidence rates of cervical cancer are 7 to 10-fold greater in Sub-Saharan Africa and South-Eastern Asia than in North America, Australia, New Zealand, and Western Asia (Saudi Arabia and Iraq).1 In China, approximately 98,900 new cases and 30,500 deaths occurred in 2015, and a substantial increasing trend in cervical cancer incidence was observed.2 It is widely known that persistent infection with high-risk human papilloma virus (HPV) is a major reason for the tumorigenesis of cervical cancer.1,3–5 However, not all HPV carriers develop cervical cancer, suggesting that genetic susceptibility may be a risk factor for the occurrence of cervical cancer. In our previous work, we found that the risk-associated rs1041279 G allele in the promoter region of hepatocellular carcinoma upregulated long noncoding RNA may contribute to the risk of cervical squamous cell carcinoma (CSCC) via enhancing the transcriptional activity.6
microRNAs (miRNAs) are a class of non-coding RNAs with a length of 18–25 nucleotides.7 By binding to 3ʹ-untranslated region (3ʹ-UTR) of its target gene, miRNA can influence mRNA degradation and protein translation, playing a key role in tumorigenesis, chemoresistance and prognosis in a variety of cancers, including CSCC.7–11 Currently, several susceptibility factors of cervical cancer have been discovered, such as single nucleotide polymorphisms (SNPs) in the binding sites of miRNAs.12–16 The SNPs may affect miRNAs binding, alter the expression of miRNAs target genes, and finally modify the risk of CSCC.15,16 Moreover, genome wide association studies (GWAS) have identified 6p21.32 as a susceptibility locus of cervical cancer.17,18 After searching the region of 6p21.32 with an online database software,19 we found an SNP rs2072915 that is located in the 3ʹ-UTR of retinoid X receptor beta (RXRB) with a binding site of miR-637. miR-637 expression was down-regulated in cervical cancer and up-regulation of miR-637 can inhibit cell proliferation.20 Base on this background, we hypothesized that the rs2072915 may be a risk factor for the pathogenesis of CSCC. To test this hypothesis, we performed a case-control study to determine whether the rs2072915 affected CSCC risk in a Chinese population. We found that the rs2072915 AT/AA genotypes conferred to the susceptibility and outcome of CSCC partly by influencing the expression levels of RXRB. This study is helpful for identifying new susceptibility locus of CSCC through post-GWAS analysis.
Materials and Methods
Study Subjects
Using the Quanto software, we calculated the sample size of a power more than 80%. When we present the allele frequency of 10% and the relative risk of 1.6 under a dominant model, the minimum numbers of samples were 407. The study group is composed of 435 CSCC patients and 454 unrelated healthy controls, who were recruited from the Wenzhou People’s Hospital between April 2012 and December 2019. Patients were diagnosed by histological examination after surgical resection. The exclusion criteria of the patients were as follows: (a) patients receiving chemotherapy and/or radiotherapy before surgery; (b) non Han Chinese; (c) patients who had a family history of any malignancy. Detailed data of the patients’ characteristics were collected from medical records, such as age, race/ethnicity, living region, HPV infection information, differentiation degree, tumor size, regional lymph node and distal metastasis. Patients were followed by telephone call for 36 months, and overall survival was calculated from the date of surgery until death or last follow-up. At the same time, 454 female controls coming to the same hospital for physical examination were selected. The inclusion criteria were as follows: (a) healthy subjects who agreed to participate in the study; (b) Han Chinese living in Wenzhou or surrounding regions; (c) without a family history of any malignancy. This study was approved by the Ethics Committee of the Wenzhou People’s Hospital, and all subjects provided written informed consent according to the 1964 Helsinki Declaration.
SNP Selection
We searched a database of functional variants (http://m6avar.renlab.org/index.html)19 according to the following criteria: (a) SNP at 6p21.32 that is a susceptibility locus of cervical cancer;17,18 (b) SNP locating in the 3ʹ-UTR with a miRNA binding target; (c) minor allele frequency more than 10% in Han Chinese. Finally, only rs2072915 was identified, which is located in the 3ʹ-UTR of RXRB with a binding site of miR-637.
DNA Extraction and Genotyping
Approximately, 2–3 mL venous blood was taken from each participant. DNA was extracted from leukocytes using the genomic DNA isolation kit (Bioteke, Beijing, China) according to the manufacturer’s directions. The rs2072915 was genotyped using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). DNA fragments were amplified using the following primers: 5ʹ-TCTCATGTCCATCAGCTTGG-3ʹ and 5ʹ-CCATGATTTGGGGTGATTTC-3ʹ and digested with the Dpn II restriction enzyme (New England Biolabs, Ipswich, MA). The PCR reaction was performed in 10 μL of the final volume including the following materials: 1 μL template DNA, 10 μM of each primer, 10 X TransTaq HiFi buffer 1 μL, 2.5 mM dNTPs 0.8 μL, 10 X GC enhancer 1 μL and TransTaq HiFi DNA polymerase 0.1 μL (TransGen Biotech, Beijing, China). The annealing temperature of PCR was 58 °C. After amplification, PCR products were digested with Dpn II at 37 °C overnight, yielding a band of 183 bp (T allele) or two bands of 130 bp and 53 bp (A allele). Some samples were randomly selected for replicate genotyping and the results were identical.
RNA Isolation and Quantitative PCR (qPCR)
Primary CSCC tissues and adjacent normal tissues were collected from 82 patients who underwent tumor resection. The specimen was diagnosed by pathological evaluation. Total RNA was extracted from 50 mg tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) as described in our previous work.6 Reverse transcription was carried out using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Rockford, IL, USA) or the miDETECT A Track miRNA qRT-PCR Starter Kit (Ribobio, Guangzhou, China). qPCR assay was performed using SYBR Green Master mix from the miDETECT A Track miRNA qRT-PCR Starter Kit (Ribobio, Guangzhou, China) according to the manufacturer’s instruction. GAPDH or U6 was used as an internal control. The primer sequences of RXRB were as follows: 5ʹ-GGGACAACAAAGACTGCACA-3ʹ and 5ʹ-ATCCTGTCCACAGGCATCTC-3ʹ, and the primer sequences of GAPDH were as follows: 5ʹ-CTCTCTGCTCCTCCTGTTCGAC −3ʹ and 5ʹ-TGAGCGATGTGGCTCGGCT-3ʹ. Primer sets of miR-637 and U6 utilized in this study were purchased from Ribobio company (Guangzhou, China). Relative expression levels of RXRB mRNA and miR-637 in CSCC tissues were calculated using the 2 −ΔΔCt method.21
Plasmid Construction and Dual-Luciferase Reporter Assay
RXRB 3ʹ-UTR containing the rs2072915 A allele (WT) or some mismatch mutations including the rs2072915 T allele (mutant) was synthesized and inserted into pmirGLO vector. The inserted products were verified by Sanger sequencing. Human cervical cancer cell lines including HeLa and SiHa were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Hyclone, Utah, USA) and 1% penicillin-streptomycin. The cells were plated in 24-well plates and co-transfected with empty, WT or mutant vector and miR-637 mimic by Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA). At 48 h following transfection, cells were lysed and luciferase activities were examined by the Dual-Luciferase Reporter assay system (Promega, Madison, WI, USA) according to the manufacturer's manual. Relative luciferase activity was determined by the ratio of reporter (firefly) to control (Renilla) luciferase activity.
Statistical Analysis
Statistical Package for Social Sciences software version 13.0 (Chicago, IL, USA) was used for statistical analysis. The rs2072915 was tested for deviation from the Hardy–Weinberg equilibrium (HWE) using the χ2 test. Differences in characteristics of study population were compared using the χ2 test for categorical data and the Student’s t test for continuous data. Association between the rs2072915 and CSCC risk was estimated using the χ2 test by calculating odds ratios (ORs) and their 95% confidence intervals (CIs). RXRB mRNA levels in patients with different genotypes of the rs2072915 were analyzed by the Mann–Whitney U-test. Correlation between miR-637 and RXRB mRNA expression levels was analyzed by the Pearson correlation test. Survival curve was generated by the Kaplan–Meier method and difference of survival time was compared by the log rank test. P < 0.05 was considered statistically significant.
Results
Characteristics of Study Population
This study included 435 CSCC patients and 454 controls. Results from the Quanto software estimation revealed that the sample size included in this study provides a statistical power of 82.6%. The positive rate of HPV infection was 82.1 and 77.8% and the mean age was 45.7 and 44.8 years in cases and controls, respectively, with no statistical significance between the two groups. Among the patients, 43.4% was well to moderately differentiated, 46.0% was clinical stage I and 26.9% had lymph node metastasis (Table 1).
 |
Table 1 Characteristics of the Study Population |
Association Between the rs2072915 and Risk of CSCC
The genetic variant rs2072915 in controls maintained HWE (P = 0.10). The genotype and allele frequencies of the rs2072915 are shown in Table 2. The distributions of the rs2072915 homozygous genotype and dominant model in the case group were significantly different from those in the healthy control group (AA vs TT: adjusted OR = 2.48, 95% CI, 1.57–3.94, P < 0.001; AT/AA vs TT: adjusted OR = 1.38, 95% CI, 1.06–1.80, P = 0.018, respectively). Moreover, compared with the control group, the frequency of the rs2072915 A allele in the case group was significantly higher, with an adjusted OR of 1.49 and 95% CI of 1.21–1.84 (P < 0.001). However, the rs2072915 was not associated with clinical features of CSCC when stratified by differentiated status, clinical stage and lymph node metastasis (Table 3).
 |
Table 2 Association Between the rs2072915 and Risk of Cervical Squamous Cell Carcinoma |
 |
Table 3 Stratified Analysis of the rs2072915 with Clinical Features of Cervical Squamous Cell Carcinoma |
The rs2072915 AA Genotype Was Associated with Lower Levels of RXRB Expression
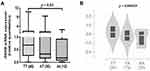
To study the correlation between the rs2072915 and RXRB mRNA expression, we analyzed the RXRB mRNA levels in 82 CSCC tissues using qPCR, and we also downloaded data from the Genotype-Tissue Expression (GTEx) project. As shown in Figure 1A and B, the rs2072915 AA genotype significantly correlated to elevated mRNA expression levels (P = 0.03 and 0.000029, respectively).
RXRB 3ʹ-UTR Containing the rs2072915 is a Target of miR-637
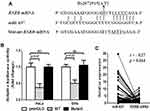
Through bioinformatics analysis using an online tool miRwalk,22 we found that RXRB 3ʹ-UTR containing the rs2072915 is a potential target of miR-637. To verify the accuracy of bioinformatics prediction, dual-luciferase reporter assay was performed in HeLa and SiHa cells. RXRB 3ʹ-UTR containing the rs2072915 A allele (WT) and some mismatch mutations containing the rs2072915 T allele (mutant) were inserted into pmirGLO vector (Figure 2A). As shown in Figure 2B, the luciferase activity of WT vector was significantly decreased in cells cotransfected with miR-637 mimic (P = 0.02), whereas the administration of mutant vector reversed the suppressive effect of miR-637 mimic. Additionally, the correlation between miR-637 and RXRB mRNA expression levels was negative in CSCC tissues (r = 0.27, P = 0.016) (Figure 2C).
The rs2072915 AT/AA Genotypes Carriers Had a Worse Survival Time
To determine whether the rs2072915 affects overall survival of CSCC, Log rank test was performed. Figure 3 showed the Kaplan–Meier curve for 36-month overall survival. The median survival time in patients with the rs2072915 AT/AA genotypes was significantly shorter than that in patients with the rs2072915 TT genotype (P = 0.03).
 |
Figure 3 Kaplan-Meier survival curves of rs2072915 in CSCC patients. |
Discussion
It is evident that about 70% of cervical cancer cases are caused by high-risk HPV infection. Persistent HPV infection may inactivate some tumor suppressors, such as p53 and members of the retinoblastoma family, resulting in a greater likelihood of acquiring cellular mutations, higher genomic instability and malignant progression.23,24 Recent evidence has shown that SNPs within miRNA binding sequences may affect the binding energy and expression of miRNA targets, and eventually contribute to the susceptibility of CSCC.12–16 For instance, an insertion/deletion rs3783553 polymorphism in the 3ʹ-UTR of interleukin-1A was associated with a decreased risk to develop CSCC.13 Similarly, the rs963918 C allele and rs963917 G allele in the 3ʹ-UTR of RAD51B conferred lower risks of cervical cancer.14 The rs4846048 G allele in the 3ʹ-UTR of methylenetetrahydrofolate reductase was associated with an increased risk of cervical cancer by enhancing the binding ability to miR-522.16 Through up-regulating the expression of caspase-3, the rs1049216 TT genotype in the miR-181a binding site conferred not only a significantly decreased risk but also the progression of cervical cancer.15 These findings provide a clue that genetic polymorphisms in the miRNA-target binding sites may be related to the susceptibility of cervical cancer.
Previously, genome wide association study (GWAS) has identified some susceptibility loci of cervical cancer, including SNPs at 6p21.32.17,18 In this study, we used an online database m6AVar19 and predicted that an m6A modification-associated SNP rs2072915 in the region of 6p21.32 may be a risk factor for the etiology of CSCC. Our findings confirmed this prediction. We found that carriers with the rs2072915 AT/AA genotypes had a 1.38-fold increased risk of CSCC and poor outcome. Although the exact mechanism by which the rs2072915 influencing CSCC risk is not well understood, we found that the rs2072915 was an expression quantitative trait locus of RXRB based on GTEx database’s annotation. Consistently, we observed a significantly lower RXRB mRNA expression in CSCC patients carrying the rs2072915 AA genotype compared to those carrying the rs2072915 TT genotype. RXRB methylation was observed in non-small cell lung cancer,25 having a 98% positive predictive value for malignant thyroid disease.26 Additionally, RXRB mRNA was down-regulated in CSCC tissues according to GEPIA annotation. Taken together, our finding of the rs2072915 AA increasing CSCC risk might be caused in part by decreasing RXRB expression.
Since we selected SNP locating in the 3ʹ-UTR that has a miRNA binding target, we predicted miRNA-RXRB binding using miRwalk22 and found that miR-637-RXRB binding sequence contains the rs2072915. Results from dual-luciferase reporter assay revealed that RXRB is a target of miR-637. miR-637, down-regulated in cervical cancer, could regulate cell proliferation by serving as a sponge.20 The observations above suggest that the rs2072915 T > A change might augment the binding energy of miR-637 to RXRB, result in lower levels of RXRB, and thus contribute to the risk of CSCC.
Although the sample size is not large enough that may be a limitation of this study, we have a 82.6% statistical power to detect the effect of the rs2072915 on CSCC occurrence. Additional studies with larger numbers are required for validating the association. Moreover, gene–gene interaction and gene-environment interaction were not performed in this study, which may interfere with the true association between the rs2072915 and CSCC risk. Further studies considering these factors are of great importance.
In conclusion, this is the first study reporting that the rs2072915 was associated with an increased risk of CSCC in the Chinese population partly by altering the expression of RXRB. Further studies are warranted to identify the association of the rs2072915 with CSCC risk in our and other ethnic groups, which constitutes a critical step towards a better understanding of the rs2072915 in the pathogenesis of CSCC.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval
This study was in accordance with the ethical standards of the Institutional Human Ethics Committee of the Wenzhou People’s Hospital and with the 1964 Helsinki declaration.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
All authors agree to publish the data.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
3. Munoz N, Bosch FX, de Sanjose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–527. doi:10.1056/NEJMoa021641
4. Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19. doi:10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F
5. Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55(4):244–265. doi:10.1136/jcp.55.4.244
6. Wang LY, Li RL, Guo M, Huang LX, Chen YM. Long noncoding RNA HULC promoter polymorphism rs1041279 is associated with an increased risk of cervical squamous cell carcinoma. Reprod Sci. 2020;27(1):93–99. doi:10.1007/s43032-019-00013-8
7. Mathonnet G, Fabian MR, Svitkin YV, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317(5845):1764–1767. doi:10.1126/science.1146067
8. Li JH, Xiao X, Zhang YN, et al. MicroRNA miR-886-5p inhibits apoptosis by down-regulating Bax expression in human cervical carcinoma cells. Gynecol Oncol. 2011;120(1):145–151. doi:10.1016/j.ygyno.2010.09.009
9. Ke G, Liang L, Yang JM, et al. MiR-181a confers resistance of cervical cancer to radiation therapy through targeting the pro-apoptotic PRKCD gene. Oncogene. 2013;32(25):3019–3027. doi:10.1038/onc.2012.323
10. Wu DW, Chuang CY, Lin WL, Sung WW, Cheng YW, Lee H. Paxillin promotes tumor progression and predicts survival and relapse in oral cavity squamous cell carcinoma by microRNA-218 targeting. Carcinogenesis. 2014;35(8):1823–1829. doi:10.1093/carcin/bgu102
11. Wang F, Liu M, Li X, Tang H. MiR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett. 2013;587(5):488–495. doi:10.1016/j.febslet.2013.01.016
12. Yu A, Zhang J, Mei Y, Zhong H, Chen S, Song Q. Correlation between single nucleotide polymorphisms of an miRNA binding site in the 3ʹUTR of PTEN and risk of cervical cancer among the Han Chinese. Genet Test Mol Biomarkers. 2020;24(7):381–389. doi:10.1089/gtmb.2019.0269
13. Huang J, Ni S, Li D, He Y. An insertion/deletion polymorphism at miRNA-122 binding site in the IL1A is associated with a reduced risk of cervical squamous cell carcinoma. Genet Test Mol Biomarkers. 2015;19(6):331–334. doi:10.1089/gtmb.2015.0015
14. Hang D, Zhou W, Jia M, et al. Genetic variants within microRNA-binding site of RAD51B are associated with risk of cervical cancer in Chinese women. Cancer Med. 2016;5(9):2596–2601. doi:10.1002/cam4.797
15. Guo X, Dong Z, Yamada S, et al. Association of Casp3 microRNA target site (1049216) SNP with the risk and progress of cervical squamous cell carcinoma. Int J Gynecol Cancer. 2017;27(2):206–213. doi:10.1097/IGC.0000000000000881
16. Zhou X, Shan L, Na J, Li Y, Wang J. The SNP rs4846048 of MTHFR enhances the cervical cancer risk through association with miR-522: a preliminary report. Mol Genet Genom Med. 2020;8(1):e1055. doi:10.1002/mgg3.1055
17. Shi Y, Li L, Hu Z, et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat Genet. 2013;45(8):918–922. doi:10.1038/ng.2687
18. Miura K, Mishima H, Yasunami M, et al. A significant association between rs8067378 at 17q12 and invasive cervical cancer originally identified by a genome-wide association study in Han Chinese is replicated in a Japanese population. J Hum Genet. 2016;61(9):793–796. doi:10.1038/jhg.2016.50
19. Zheng Y, Nie P, Peng D, et al. m6AVar: a database of functional variants involved in m6A modification. Nucleic Acids Res. 2018;46(D1):D139–D145. doi:10.1093/nar/gkx895
20. Rui X, Xu Y, Jiang X, Ye W, Huang Y, Jiang J. Long non-coding RNA C5orf66-AS1 promotes cell proliferation in cervical cancer by targeting miR-637/RING1 axis. Cell Death Dis. 2018;9(12):1175. doi:10.1038/s41419-018-1228-z
21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
22. Sticht C, De la torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018;13(10):e0206239. doi:10.1371/journal.pone.0206239
23. Schiffman M, Wentzensen N, Wacholder S, Kinney W, Gage JC, Castle PE. Human papillomavirus testing in the prevention of cervical cancer. J Natl Cancer Inst. 2011;103(5):368–383. doi:10.1093/jnci/djq562
24. Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10(8):550–560. doi:10.1038/nrc2886
25. Lee SM, Lee JY, Choi JE, Lee SY, Park JY, Kim DS. Epigenetic inactivation of retinoid X receptor genes in non-small cell lung cancer and the relationship with clinicopathologic features. Cancer Genet Cytogenet. 2010;197(1):39–45. doi:10.1016/j.cancergencyto.2009.10.008
26. Hoftijzer HC, Liu YY, Morreau H, et al. Retinoic acid receptor and retinoid X receptor subtype expression for the differential diagnosis of thyroid neoplasms. Eur J Endocrinol. 2009;160(4):631–638. doi:10.1530/EJE-08-0812
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.