Back to Journals » Infection and Drug Resistance » Volume 13
A Comparative Study of Fluoroquinolone-Resistant Escherichia coli Lineages Portrays Indistinguishable Pathogenicity- and Survivability-Associated Phenotypic Characteristics Between ST1193 and ST131
Authors Huang J, Zhang S, Zhang S, Zhao Z , Cao Y, Chen M, Li B
Received 19 August 2020
Accepted for publication 19 October 2020
Published 20 November 2020 Volume 2020:13 Pages 4167—4175
DOI https://doi.org/10.2147/IDR.S277681
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Jiangqing Huang,1 Shengcen Zhang,1 Shuyu Zhang,2 Zhichang Zhao,3 Yingping Cao,1 Min Chen,2 Bin Li1
1Department of Clinical Laboratory, Fujian Medical University Union Hospital, Fuzhou, Fujian 350001, People’s Republic of China; 2Department of Laboratory Medicine, Fujian Medical University, Fuzhou, Fujian 350001, People’s Republic of China; 3Department of Pharmacy, Fujian Medical University Union Hospital, Fuzhou, Fujian 350001, People’s Republic of China
Correspondence: Bin Li
Department of Clinical Laboratory, Fujian Medical University Union Hospital, 29 Xinquan Road, Fuzhou, Fujian 350001, People’s Republic of China
Email [email protected]
Background: Sequence type 1193 is a new such lineage among fluoroquinolone-resistant Escherichia coli, which has risen dramatically within the last several years. However, reasons for rapid emergence and successful spread of E. coli ST1193 remain unclear. The aim of this study was to compare the pathogenicity and survivability features of E. coli ST1193 with global epidemic lineage, ST131.
Methods: A total of 30 E. coli were used in this study. Isolates were divided into two groups, ST1193 (n=15) and ST131 (n=15). Adhesion and invasion to T24 cells and resistance to serum were quantified and compared among two groups. Biofilm formation capacity was assessed by crystal violet assay. Macrocolony formation was assessed on macrocolony formation plates. Resistance to hydrogen peroxide was performed by broth microdilution. RAW264.7 cells were used to assess the anti-phagocytic function of different isolates.
Results: Adhesion and invasion assays revealed that E. coli ST1193 could adhere and invade T24 cells (p < 0.05). 93.3% of E. coli ST1193 could form biofilms. The majority of E. coli ST1193 (66.7%) possessed no curli/no cellulose on macrocolony formation plates. E. coli ST1193 showed significant growth in serum and hydrogen peroxide and illustrated higher anti-phagocytic function to RAW264.7 cells (p < 0.05). Group analysis showed that E. coli ST1193 was similar to ST131 in pathogenicity- and survivability-associated phenotypic characteristics (p > 0.05).
Conclusion: Our study provided more insights into pathogenicity and survivability features of E. coli ST1193, which was similar to ST131. Our study could be of great importance in understanding the emergence of global spread E. coli ST1193. Strategic and continued surveillance should be carried out to prevent the infections caused by E. coli ST1193.
Keywords: ST1193, ST131, pathogenicity, survivability
Introduction
Escherichia coli (E. coli) is usually found in the gut flora of humans and animals and can cause many kinds of extraintestinal infections, such as septicemias, meningitis and urinary tract infections.1 E. coli is a bacterial species with high genetic diversity.2,3 Multilocus sequence typing (MLST) was usually used for typing E. coli due to its detection of the genetic relatedness among strains.3,4 Meanwhile, MLST has a high discriminatory capability to identify a specific lineage in various bacterial pathogens.5,6 Therefore, it was usually regarded as a powerful tool for global and long-term surveillance.7
Many studies have reported that E. coli has a high degree of diversity in multilocus sequence typing, such as ST131, ST69, ST73 and ST95 and nearly half of clinical isolates belong to those ST types.8–10 Among them, E. coli ST131 was currently recognized as one such predominant global epidemic clonal group and multidrug-resistant lineage.11 A study based on E. coli ST131 has proved that the enhanced pathogenicity and better survivability in intestinal and extraintestinal sites contributed to ST131 epidemiological success.12
Recently, the proportion of E. coli ST1193 has risen dramatically among fluoroquinolone-resistant (FQr) E. coli.13,14 E. coli ST1193 shares many common features with ST131, including fluoroquinolone resistance, lactose non-fermenting and phylogenetic group B2.13 Meanwhile, E. coli ST1193 has conserved virulence gene profiles, which suggested that this lineage has potential pathogenicity in human.15 However, reasons for the rapid emergence and successful spread of E. coli ST1193 remain unclear. This has taken widespread concern and deserves our attention to clarify the biological characteristics of this lineage. The aim of this study was to compare the pathogenicity and survivability features of FQr E. coli ST1193 with global epidemic ST131, which might help to understand the factors of ST1193 epidemiological success.
Materials and Methods
Bacterial Isolates
A total of 30 E. coli isolates were used in this study and were divided into two groups, group ST1193 (n=15) and group ST131 (n=15). Those isolates were collected at Fujian Medical University Union Hospital (Fuzhou, Fujian province, China) between August 2014 and August 2015 which were reported in our previous studies.13,16 And those isolates were selected at random.
Pathogenicity-Associated Phenotypic Assays
Adhesion and Invasion Assay
The interaction of E. coli with the human bladder cancer cells (T24 cells) (Anchorage-dependent cell, FH0171, FuHeng Cell Center, Shanghai, China) was performed as previously described.17 Briefly, for adhesion assay, T24 cells were grown in 24-well plates at 37°C in a CO2 incubator. The T24 cells were infected with different strains at a multiplicity of infection (MOI) of 10 in triplicates. After 3 hours of incubation, T24 cells were washed with 1× phosphate-buffered saline (PBS) thrice and lysed using 0.1% Triton X-100 (Beyotime Institute of Biotechnology, China) for 10 minutes. The lysates were collected and diluted serially. Then the diluted lysates were plated on MH-ager plates. CFU counts were determined for all strains after incubated overnight at 37°C. E.coli strain EC505 and E. coli DH5α were served as positive control and negative control.
For invasion assay, similar to adhesion assay, an additional step was carried out. The medium was replaced with fresh medium containing 100 µg/mL of gentamicin (Yeasen Biotech, China) which incubated for 1.5 hours to kill all extracellular strains after 3 hours of incubation with T24 cells. After incubation, the cells were washed with 1×PBS thrice and lysed using 0.1% Triton X-100 for 10 minutes. The lysates were collected and diluted serially. The diluted lysates were then plated on MH-ager plates. CFU counts were determined for all strains after incubated overnight at 37°C. E.coli strain EC505 and E. coli DH5α were served as positive control and negative control.
Biofilm Formation
Biofilm formation capacity was assessed in 96-well plates by crystal violet assay as described previously and the optical density was measured at 595 nm in ELISA reader (Thermo Scientific, China).18 Each assay was repeated in three replicates and the results were averaged. Medium alone served as a negative control (ODc) and Enterococcus faecalis (E. faecalis) ATCC 29,212 was used as positive control.19,20 The degree of biofilm production was scored as follows: absent (OD<ODc), weak (ODc<OD<1.5×ODc), moderate (1.5×ODc<OD<2×ODc) and strong (OD>2× ODc).18
Macrocolony Formation
Five microliters of overnight cultures grown in LB broth medium at 37°C were dropped on NaCl-free LB plates with 0.04% congo red (CR) (Solarbio, China) and 0.02% coomassie brilliant blue G (CB) (Solarbio, China). Plates were incubated at 28°C for up to 5 days. The results were recorded by a stereomicroscope (8mm, 66 Vision Tech Co., Ltd, China) and classified into four categories: curli+cellulose, curli only, cellulose only and no curli/no cellulose.21 E. coli strains EC720 (curli +cellulose), E. coli K-12 (curli only) and EC505 (cellulose only) were included as positive controls. Strain EC700 (no curli/no cellulose) was used as negative control. Strains EC720, EC505 and EC700 were collected in our laboratory.
Survivability-Associated Phenotypic Assays
Serum Resistance Assay
Serum resistance assay was tested using 50% human serum for a 3-hour incubation as a previous study.17 The growth in the serum of isolate was detected as the total number of CFU/mL recovered per well.17 E. coli strain EC505 and E. coli DH5α were used as the positive and negative control.
Hydrogen Peroxide Resistance Assay
The hydrogen peroxide resistance assay was performed by broth microdilution according to the Clinical and Laboratory Standards Institute (CLSI, 2020) standards.22 The hydrogen peroxide (AR, 7722–84-1, Wokai Biotechnology Co., LTD, China) was used in this study and stored according to the manufacturers’ instructions. The results were observed in terms of MIC and MBC.23 E. coli ATCC 25,922 was used as quality control.
Anti-Phagocytic Function Assay
Anti-phagocytic function assay was performed as described previously.24 The RAW264.7 cells (Anchorage-dependent cell, FH0328, FuHeng Cell Center, Shanghai, China) were used in this study. 2×105 CFU/mL RAW264.7 cells were incubated with 2×106 CFU/mL bacterial at 37°C for 1.5 hours. The RAW264.7 cells were infected with different strains at a multiplicity of infection (MOI) of 10. And trypan blue staining cell viability assay kit (Beyotime, China) was used to determine the cell viability of RAW264.7 cells. The phagocytosed E.coli presenting inside the RAW264.7 cells were counted under a microscope by a person blinded to the experimental design. All experiments were repeated in three replicates. The anti-phagocytosis rate (PR) was calculated as following: PR = (total numbers of cells harbored the phagocytosed E.coli in 200 RAW264.7 cells)/200×100%. E. coli DH5α and E. coli ATCC 25,922 were used as negative and positive controls, respectively.
Statistical Analysis
SAS 9.4 was used for statistical analysis. The chi-square test or Fisher’s exact test (two-tailed) or Mann–Whitney test was performed for data comparison. Only p <0.05 was considered statistically significant.
Results
In this study, we explored the pathogenicity- and survivability-associated characteristics of FQr E. coli ST1193, and compared these phenotypic characteristics between ST1193 and pandemic ST131. Among them, adhesion capability, invasion capability and biofilm formation capability were used to assess the pathogenicity of strains. Serum resistance, hydrogen peroxide resistance and anti-phagocytic function were used to assess the survivability of strains.
Pathogenicity-Associated Phenotypic Characteristics
Adhesion and Invasion Capabilities
E. coli ST1193 could adhere and invade the bladder epithelial cell line T24 compared to E. coli DH5α (Figure 1, p <0.05). The adhesion and invasion abilities of ST1193 were compared with pandemic ST131. The results showed that E. coli group ST1193 was comparable with group ST131 lacking any significant difference (Figure 1, p >0.05).
Biofilm Formation Capability
As quantified by crystal violet staining, we found 93.3% of E. coli ST1193 could form biofilms. However, 66.7% of E. coli ST1193 showed weak biofilm-forming ability. No significant difference between two groups (E. coli ST1193 and ST131) was observed (p >0.05). The result of biofilm formation is shown in Table 1.
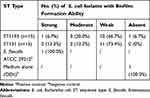 |
Table 1 Prevalence of Biofilm Formation Among Different E. coli Isolates |
Macrocolony Formation
To further explore the roles (curli and cellulose) of specific matrix components in shaping biofilm architecture, macrocolony formation assay was performed in E. coli ST1193 and ST131. The results of the positive and negative controls and E. coli group ST1193 and group ST131 are shown in Figure 2. The majority of E. coli ST1193 (66.7%) possessed no curli/no cellulose. Compared to ST131, E. coli ST1193 exhibited a statistical difference in curli only (p <0.05). We did not find a significant difference between these two groups in cellulose curli+cellulose, cellulose only and no curli/no cellulose (p >0.05).
Survivability-Associated Phenotypic Characteristics
Serum Resistance Assay
In our study, all strains were tested in 50% human serum to investigate their capacity to resist the serum bactericidal activity. E. coli ST1193 showed significant growth compared to E. coli DH5α (Figure 3A, p <0.05). Group analysis showed that E. coli group ST1193 had a similar capability compared to group ST131 (p >0.05).
Hydrogen Peroxide Resistance Assay
In this study, MIC of hydrogen peroxide for all E. coli group ST1193 was 16 μg/mL or higher and MBC of hydrogen peroxide for 46.7% of ST1193 strains was 32 μg/mL or higher. E. coli ATCC 25,922 had a MIC of 8 μg/mL and an MBC of 8 μg/mL, respectively. Both MIC and MBC results showed a significant difference (p <0.05) compared to E. coli ATCC 25,922. No significant difference in susceptibility was observed between the mean hydrogen peroxide resistance of E. coli group ST1193 and group ST131 at MIC level (Figure 3B) and MBC level (Figure 3C).
Anti-Phagocytic Function Assay
The results of anti-phagocytic function assay are shown in Figure 3D. The levels of phagocytic activity in the RAW264.7 cells against the E. coli ST1193 were statistically lower than E. coli DH5α (p <0.05). Meanwhile, E. coli group ST1193 had no significant difference in anti-phagocytic function compared to group ST131 (p >0.05).
Discussion
E. coli ST1193 has risen dramatically in the last decade and has made a significant contribution to the population of FQr E.coli around the world.13,14 E. coli ST1193 exhibits a large diversity of virulence gene profiles, indicating that ST1193 harbors potential pathogenicity.15 The epidemiological success of this bacteria is associated with its enhanced pathogenicity and better survivability of strains. In this study, we performed comparative analysis of pathogenicity- and survivability-associated characteristics in E. coli ST1193 and pandemic ST131 clones, which could provide new support for the epidemiological success of ST1193.
Firstly, we assessed the pathogenicity-associated characteristics of E. coli ST1193 and pandemic ST131, including adhesion capability, invasion capability and biofilm formation capacity. Adhesion and invasion capabilities seemed to be necessary for bacterial persistence and pathogenesis.25 Effective cellular adhesion and followed by invasion played a key role in the pathogenicity of strains.26 In our study, E. coli ST1193 exhibited strong adhesion and invasion ability to the human bladder cancer cells (Figure 1), which indicated that ST1193 could efficiently colonize and subsequently establish infections when they entered into the tissue sites of humans. These findings might prove the evidence to the fact that E. coliST1193 usually causes serious urinary tract infections.15 It should be noted that better adhesion and invasion capabilities of E. coli ST1193 might be important for its dissemination. One significant finding in our study was that E. coli ST1193 exhibited similar adhesion and invasion abilities compared to pandemic ST131 (Figure 1, p >0.05). Thus, we supposed that those capabilities might contribute to ST1193 epidemiological success. Additionally, the genes associated with adherence and invasion, such as ibeA and ibeB, were usually found in E. coli ST131 genomes.17 However, there are no reports regarding those genes in E. coli ST1193. Future studies need to be carried out to investigate this situation in E. coli ST1193.
Biofilm formation ability of bacteria is related with causing infections.27 Furthermore, bacteria in biofilm become tolerant and resistant to antibiotics, increasing the difficulties for the clinical treatment of microbial infections as the antimicrobial was hard to penetrate the biofilm.28 In our study, we found that most of E. coli ST1193 could form biofilms, which was similar to previous study.29 These findings suggested that refractory infections caused by E. coli ST1193 were related to its biofilm formation ability. Moreover, E. coli ST1193 and ST131 showed no significant difference in their specific biofilm-forming capacities (p >0.05), demonstrating similar pathogenicity in terms of their biofilm forming capability between the two groups. To further elucidate the roles of specific matrix components (curil and cellulose) in shaping biofilm architecture, we performed phenotypic macrocolony formation assay for those two groups (Figure 2). Curli promotes biofilm adhesion to abiotic surfaces or cells and contributes to persistence in hosts.30 Cellulose promotes delayed bacterial clearance.31 We found that the majority of E. coli ST1193 and ST131 possessed no curli/no cellulose in this study. This finding was different from a previous study, which might be due to regional disparity.32 Interestingly, E. coli ST1193 exhibited a statistical difference in curli only when compares to ST131 (p <0.05). Our results suggested that curli expression may increase the virulence of E. coli ST1193 by the participation of biofilm formation. Furthermore, whether the specific matrix component (curli) is a feature of E. coli ST1193 should be further investigated in the future.
Better survivability of strains was connected with its epidemiological success. Resistance to the bactericidal effect of serum and anti-phagocytic function were recognized as important survivability determinants in vivo for bacteria.17,33 Additionally, hydrogen peroxide is usually used as cleaning and disinfecting agents for controlling the growth of or killing bacteria in vitro.34 In this study, we found that E. coli ST1193 showed significant growth in serum and illustrated higher anti-phagocytic function (Figure 3, p <0.05). Serum resistance is one of the major survival mechanisms adopted by pathogenic E. coli to survive in the bloodstream of the host.17 Our study indicated that E. coli ST1193 has the potential to cause bloodstream infection and subsequently lead to septicemia. Anti-phagocytic function plays a significant role in the pathogenesis in extraintestinal infections.35 Enhanced anti-phagocytic function might protect the E. coli ST1193 strains from phagocytosis and promote them survival in vivo which conduced to cause infections. In addition, capsule, lipopolysaccharide, mannose receptor, ClpX protease and carnitine metabolism have been demonstrated to mediate anti-phagocytosis in bacteria cells and phagocyte.36–38 However, the specific factors of anti-phagocytosis function in E. coli ST1193 are unclear up to now and they need to be explored in the future. These results demonstrated that E. coli ST1193 could be resistant to immediate bactericidal effect of human immune system, then causing infections promptly. Furthermore, resistance to biocides seemed to be linked to incidence of bacterial infections in hospitals.23 E. coli ST1193 showed significant growth in hydrogen peroxide (Figure 3, p <0.05) in our study, suggesting that hydrogen peroxide resistance might be a contributory factor in the incidence of E. coli ST1193 infections. Moreover, E. coli ST1193 and ST131 were indistinguishable in terms of survivability-associated phenotypic characteristics. These results suggested E. coli ST1193 had similar survivability to predominant global epidemic clonal groups, ST131.
There are several limitations to our study. First, we have investigated the biofilm formation ability of E.coli ST1193 and ST131 lineages in this study. The majority of isolates E. coli ST1193 (66.7%) and ST131 (73.4%) presented weak biofilm formation ability. Similar results were found in previous study about E. coli ST131 linage, indicating that weak biofilm formation ability might be a feature of ST1193 and ST131 lineages.39 However, it is unclear whether the phenotype features of their biofilm formation ability were sufficient for its participation in the infection process and its refraction based on current experiments in this study. Second, we did not perform further experiments to verify the bacterial survival in RAW 264.7 cells which may help to understand the pathogenesis in vivo. Future study will be carried out to assess them.
Conclusions
Our study provided more insights into pathogenicity and survivability features of E. coli ST1193, which was similar to global epidemic lineages, ST131. Our report could be of great importance in understanding the emergence of global spread ST1193. This study rendered much needed support and reinforcement to the observations of virulence gene profiles among E. coli ST1193. Strategic and continued surveillance should be carried out to prevent the infections caused by E. coli ST1193.
Abbreviations
MLST, multilocus sequence typing; ST, sequence type; MIC, minimum inhibitory concentration; MBC, minimum bactericidal concentration; PR, anti-phagocytosis rate.
Ethics Approval and Informed Consent
All procedures of this study involving humans (individuals, medical records, human samples, clinical isolates and human cell lines) were reviewed and approved by the Medical Ethics Committee of Fujian Medical University Union Hospital (2020KY0121). All the patients participating in this study signed informed consent, while the guardians of children aged less than 18 years signed on behalf of them. We confirm that this study was conducted in accordance with the Declaration of Helsinki.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Joint Funds for the innovation of science and Technology, Fujian province [Grant number: 2017Y9049] and the Educational and Scientific Research Project for Young and Middle-Aged Teachers of Fujian Province (Grant number: JAT190191).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin Microbiol Rev. 2019;32(3):e00135–18. doi:10.1128/CMR.00135-18
2. Touchon M, Hoede C, Tenaillon O, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5(1):e1000344. doi:10.1371/journal.pgen.1000344
3. Chaudhuri RR, Henderson IR. The evolution of the Escherichia coli phylogeny. Infection, Genetics Evolution. 2012;12(2):214–226. doi:10.1016/j.meegid.2012.01.005
4. Riley LW. Pandemic lineages of extraintestinal pathogenic. Escherichia Coli Clinical Microbiology Infection. 2014;20(5):380–390. doi:10.1111/1469-0691.12646
5. Maiden MC, Jansen van Rensburg MJ, Bray JE, et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013;11(10):728–736. doi:10.1038/nrmicro3093
6. Pérez-Losada M, Cabezas P, Castro-Nallar E, Crandall KA. Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infection, Genetics Evolution. 2013;16:38–53. doi:10.1016/j.meegid.2013.01.009
7. Feijao P, Yao HT, Fornika D, et al. MentaLiST – a fast MLST caller for large MLST schemes. Microbial Genomics. 2018;4(2):e000146. doi:10.1099/mgen.0.000146
8. Kallonen T, Brodrick HJ, Harris SR, et al. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017;27(8):1437–1449. doi:10.1101/gr.216606.116
9. Johnson JR, Porter S, Thuras P, Castanheira M. Epidemic emergence in the United States of Escherichia coli sequence type 131-H30 (ST131-H30), 2000 to 2009. Antimicrob Agents Chemother. 2017;61(8):e00732–17. doi:10.1128/AAC.00732-17
10. Fibke CD, Croxen MA, Geum HM, et al. Genomic epidemiology of major extraintestinal pathogenic Escherichia coli lineages causing urinary tract infections in young women across Canada. Open Forum Infectious Diseases. 2019;6(11):ofz431. doi:10.1093/ofid/ofz431
11. Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27(3):543–574. doi: 10.1128/CMR.00125-13.
12. Ranjan A, Shaik S, Hussain A, et al. Genomic and functional portrait of a highly virulent, CTX-M-15-producing H30-Rx subclone of Escherichia coli sequence type 131. Antimicrob Agents Chemother. 2015;59(10):6087–6095. doi:10.1128/AAC.01447-15
13. Wu J, Lan F, Lu Y, He Q, Li B. Molecular characteristics of ST1193 clone among phylogenetic group B2 non-ST131 fluoroquinolone-resistant Escherichia coli. Front Microbiol. 2017;8:2294. doi:10.3389/fmicb.2017.02294
14. Tchesnokova V, Radey M, Chattopadhyay S, et al. Pandemic fluoroquinolone resistant Escherichia coli clone ST1193 emerged via simultaneous homologous recombinations in 11 gene loci. Proc Natl Acad Sci U S A. 2019;116(29):14740–14748. doi:10.1073/pnas.1903002116
15. Johnson TJ, Elnekave E, Miller EA, et al. Phylogenomic analysis of extraintestinal pathogenic Escherichia coli sequence type 1193, an emerging multidrug-resistant clonal group. Antimicrob Agents Chemother. 2019;63(1):e01913–18. doi: 10.1128/AAC.01913-18.
16. Li B, Lu Y, Lan F, He Q, Li C, Cao Y. Prevalence and characteristics of ST131 clone among unselected clinical Escherichia coli in a Chinese university hospital. Antimicrob Resist Infect Control. 2017;6:118. doi:10.1186/s13756-017-0274-0
17. Shaik S, Ranjan A, Tiwari SK, et al. Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal pathogenic Escherichia coli (ExPEC) lineages. mBio. 2017;8(5):e01596–17. doi:10.1128/mBio.01596-17
18. Wang Y, Yi L, Wang Y, et al. Isolation, phylogenetic group, drug resistance, biofilm formation, and adherence genes of Escherichia coli from poultry in central China. Poult Sci. 2016;95(12):2895–2901. doi:10.3382/ps/pew252
19. Tsikrikonis G, Maniatis AN, Labrou M, et al. Differences in biofilm formation and virulence factors between clinical and fecal enterococcal isolates of human and animal origin. Microb Pathog. 2012;52(6):336–343. doi:10.1016/j.micpath.2012.03.003
20. Cieśla J, Stępień-Pyśniak D, Nawrocka A, et al. Surface properties of Enterococcus faecalis cells isolated from chicken hearts determine their low ability to form biofilms. Biofouling. 2018;34(2):149–161. doi:10.1080/08927014.2017.1416105
21. Serra DO, Hengge R. Experimental detection and visualization of the extracellular matrix in macrocolony biofilms. Methods Molecular Biology. 2017;1657:133–145. doi: 10.1007/978-1-4939-7240-1_11.
22. CLSI. M100-S28. Performance Standards for Antimicrobial Susceptibility Testing: 30th Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
23. Alotaibi SMI, Ayibiekea A, Pedersen AF, et al. Susceptibility of vancomycin-resistant and -sensitive Enterococcus faecium obtained from Danish hospitals to benzalkonium chloride, chlorhexidine and hydrogen peroxide biocides. J Med Microbiol. 2017;66(12):1744–1751. doi: 10.1099/jmm.0.000642.
24. Zhu H, Yan L, Gu J, Hao W, Kv CJ. 1.3 channel blockade enhances the phagocytic function of RAW264.7 macrophages. Sci China Life Sci. 2015;58(9):867–875. doi:10.1007/s11427-015-4915-3
25. Chakroun I, Cordero H, Mahdhi A, et al. Adhesion, invasion, cytotoxic effect and cytokine production in response to atypical Salmonella Typhimurium infection. Microb Pathog. 2017;106:40–49. doi:10.1016/j.micpath.2016.11.004
26. Hussain A, Shaik S, Ranjan A, et al. Genomic and functional characterization of poultry Escherichia coli from India revealed diverse extended-spectrum β-lactamase-producing lineages with shared virulence profiles. Front Microbiol. 2019;10:2766. doi:10.3389/fmicb.2019.02766
27. Beebout CJ, Eberly AR, Werby SH, et al. Respiratory heterogeneity shapes biofilm formation and host colonization in uropathogenic. Escherichia Coli mBio. 2019;10(2):e02400–18.
28. Verderosa AD, Totsika M, Fairfull-Smith KE. Bacterial biofilm eradication agents: a current review. Frontierschem. 2019;7:824. doi:10.3389/fchem.2019.00824
29. Crémet L, Caroff N, Giraudeau C, Reynaud A, Caillon J, Corvec S. Detection of clonally related Escherichia coli isolates producing different CMY β-lactamases from a cystic fibrosis patient. J Antimicrob Chemother. 2013;68(5):1032–1035. doi:10.1093/jac/dks520
30. Somorin YM, Vollmerhausen T, Waters N, et al. Absence of curli in soil-persistent Escherichia coli is mediated by a C-di-GMP signaling defect and suggests evidence of biofilm-independent niche specialization. Front Microbiol. 2018;9:1340. doi:10.3389/fmicb.2018.01340
31. Slettengren M, Mohanty S, Kamolvit W, van der Linden J, Brauner A. Making medical devices safer–impact of plastic and silicone oil on microbial biofilm formation. J Hosp Infect. 2020;106(1):155–162. doi:10.1016/j.jhin.2020.07.011
32. Schaufler K, Semmler T, Wieler LH, et al. Genomic and functional analysis of emerging virulent and multidrug-resistant Escherichia coli lineage sequence type 648. Antimicrob Agents Chemother. 2019;63(6):e00243–19. doi: 10.1128/AAC.00243-19.
33. Xu B, Zhang P, Zhou H, Sun Y, Tang J, Fan H. Identification of novel genes associated with anti-phagocytic functions in Streptococcus equi subsp. zooepidemicus. Vet Microbiol. 2019;233:28–38. doi:10.1016/j.vetmic.2019.04.023
34. Ríos-Castillo AG, González-Rivas F, Rodríguez-Jerez JJ. Bactericidal efficacy of hydrogen peroxide-based disinfectants against gram-positive and gram-negative bacteria on stainless steel surfaces. J Food Sci. 2017;82(10):2351–2356. doi:10.1111/1750-3841.13790
35. Mu X, Gao R, Xiao W, et al. EntE, EntS and TolC synergistically contributed to the pathogenesis of APEC strain E058. Microb Pathog. 2020;141:103990. doi:10.1016/j.micpath.2020.103990
36. March C, Cano V, Moranta D, et al. Role of bacterial surface structures on the interaction of Klebsiella pneumoniae with phagocytes. PLoS One. 2013;8(2):e56847. doi:10.1371/journal.pone.0056847
37. Pan Y-J, Lin T-L, Hsu C-R, Wang J-T. Use of a Dictyostelium model for isolation of genetic loci associated with phagocytosis and virulence in Klebsiella Pneumoniae. Infection and Immunity. 2011;79(3):997–1006. doi: 10.1128/IAI.00906-10.
38. Gyorfy Z, Duda E, Vizler C. Interactions between LPS moieties and macrophage pattern recognition receptors. Vet Immunol Immunopathol. 2013;152(1–2):28–36. doi:10.1016/j.vetimm.2012.09.020
39. Flament-Simon SC, Duprilot M, Mayer N, et al. Association between kinetics of early biofilm formation and clonal lineage in Escherichia coli. Front Microbiol. 2019;10:1183. doi:10.3389/fmicb.2019.01183
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



