Back to Journals » OncoTargets and Therapy » Volume 8
A common genetic variation in CEBPE and acute lymphoblastic leukemia: a meta-analysis of the available evidence
Authors Zhang X, Du Y, Zhai Y, Gao F, Yang Y, Ma X , Lu J , Zheng J
Received 31 May 2015
Accepted for publication 5 August 2015
Published 7 September 2015 Volume 2015:8 Pages 2443—2451
DOI https://doi.org/10.2147/OTT.S89661
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Xiao-Xia Zhang,1,* Yue-Feng Du,2,* Ya-Jing Zhai,1 Fan Gao,3 Yu-Juan Yang,4 Xian-Cang Ma,3 Jun Lu,3 Jie Zheng3
1Department of Pharmacy, The First Affiliated Hospital, Xi’an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China; 2Department of Urology, The First Affiliated Hospital, Xi’an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China; 3Clinical Research Center, The First Affiliated Hospital, Xi’an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China; 4The Third Department of Cardiology, Shaanxi Provincial People’s Hospital, Xi’an, Shaanxi, People’s Republic of China
*These authors contributed equally to this work
Abstract: Acute lymphoblastic leukemia (ALL) has been studied intensively for decades, but the details of its etiology and underlying mechanisms have yet to be fully elucidated. It is now generally acknowledged that genetic factors contribute greatly to the development of this disease. The gene encoding CCAAT/enhancer-binding protein ε (CEBPE) is involved in the development of leukemia, and in particular the rs2239633 single nucleotide polymorphism (SNP) of CEBPE. The association between rs2239633 and risk of ALL has been well studied, but remains unclear. Therefore, a meta-analysis was performed in this study to establish a more precise estimation of that relationship. A comprehensive literature search of the PubMed electronic database was conducted, and relevant studies published up to February 20, 2015 were selected for analysis. The references of the retrieved articles were also screened. The extracted data were analyzed statistically, and pooled odds ratios with 95% confidence intervals were calculated using Review Manager (version 5.2) to estimate the association strength. Finally, eleven studies were included in the meta-analysis. The pooled analyses revealed that rs2239633 was associated with an increased risk of childhood ALL in Caucasians under any contrast models (P<0.01). However, this SNP did not affect the risk of ALL in adulthood among Caucasians, or in childhood among East Asians. In conclusion, these findings confirm that the CEBPE rs2239633 SNP could be considered a good marker of pediatric ALL risk in Caucasians, but not in East Asians; it is not a good marker of adult ALL risk in Caucasians.
Keywords: CEBPE, rs2239633, ALL, risk, meta-analysis
Background
Acute lymphoblastic leukemia (ALL) is the most common malignancy in childhood, accounting for 30% of all childhood cancers.1 The etiology of childhood ALL is mostly unknown, although infections in the first years of life and some environmental factors, such as ionizing radiation and parental alcohol and tobacco use, are thought to play a causative role.2
ALL is known to result from an accumulation of mutations in tumor suppressor genes and oncogenes, and genetic alterations affecting several chromosomes.2 Thus, the genetic variations may be important factors in its development. This finding has recently led to the performance of several genome-wide association studies to identify common single nucleotide polymorphisms (SNPs) affecting the susceptibility to childhood ALL.2–6 One of the most widely investigated SNPs identified in these studies is the common rs2239633 SNP of CEBPE, the gene encoding CCAAT/enhancer-binding protein ε (CEBPE), which plays an important role in the regulation of myelopoiesis.7 Papaemmanuil et al4 also found that CEBPE is a suppressor of myeloid leukemogenesis, and may be involved in B-cell precursor ALL development. The populations studied in the aforementioned genome-wide association studies were limited, and the many subsequent association studies that have been conducted with a view to verifying the described effects of rs2239633 on susceptibility to ALL in childhood in different populations have yielded varying results.8–13
A meta-analysis was performed in the present study to review and compare all relevant studies, enabling more accurate conclusions to be drawn. In addition, since the genetic background of ALL differs between the adult and childhood forms, some studies were also performed to make clear if this SNP was a risk factor for adult ALL.14,15 In this study, those two relevant studies were also compared to examine the differential roles of rs2239633 in children and adults.
Materials and methods
Literature search strategy
Two of the authors (Zhang and Du) each performed the literature search of the PubMed database independently. The process was supervised by the third author (Zhai), and any disagreement was resolved by consensus. The search strategy was to identify all possible studies that involved the use of the following test words: (14q11.2 “or” CEBPE “or” rs2239633) “and” (leukemia). The language of publication was restricted to English. The reference lists of retrieved articles were curated manually. The literature search was last updated on February 20, 2015. This study was judged exempt regarding ethics.
Selection criteria
All studies with the aim of investigating the association between presence of the rs2239633 SNP and ALL risk were potentially included. The following inclusion criteria were applied:
- Published articles on human genetics (full texts or abstracts) without racial restriction.
- If articles contained more than one geographic or other clinical characteristic subgroup, each subgroup was considered separately.
- If multiple studies were derived from the same population, only the study with the largest sample was analyzed to avoid overlapping data.
- Presentation of sufficient data on the distribution of rs2239633 in cases and in control groups or the data necessary to calculate those distributions. If the data were deficient or presented in an inappropriate form, the original authors were contacted and asked for the raw data.
Extracted information
A standard data-collection procedure was used, in line with the aforementioned inclusion criteria. Two of the authors (Zhang and Du) independently extracted the variables from the individual eligible studies in duplicate and made the characteristics compatible in a pooled database. Any encountered disagreements were resolved by discussion in order to reach a consensus. The following information was extracted from all eligible studies: name of the first author, year of publication, ethnicities of the study population, age and number of cases and controls, and main finding. The distribution of the genotypes among the controls was also tested for conformity to Hardy–Weinberg equilibrium (HWE).
Statistical analysis
The meta-analysis examined the overall association for allele contrasts, homozygote contrasts, and recessive and dominant models. The presence of heterogeneity was assessed using Cochran’s Q-statistic (with a significance cutoff of P<0.1) and quantified using the I2 statistic, which is proportional to the degree of heterogeneity; the value of I2 statistic lies in the range 0%–100%, with a value of 0% indicating homogeneity, and a value above 50% indicating the presence of a very high degree of heterogeneity.16
The raw data for genotype distribution were used to calculate the study-specific estimates of odds ratio (OR) and 95% confidence interval (CI). The overall pooled OR and 95% CI were estimated using the Mantel–Haenszel method, with a fixed-effects model when no significant clinical or statistical heterogeneity was present; when substantial heterogeneity was present, the random-effects model was used. The significance of the polled OR was determined by a test. The cutoff for statistical significance was set at P<0.05.
Potential publication bias was estimated by constructing funnel plots. If most of the data appear at the top of a funnel plot and are distributed roughly symmetrically, this suggests the absence of obvious publication bias, and vice versa.13 There was no need to construct funnel plots when there were too few analyzed studies (ie, n<5). All of these statistical analyses were performed using Review Manager (version 5.2) (Cochrane, London, UK).
The conformity of the rs2239633 SNP to HWE was tested among the controls in each individual study via the chi-square test or Fisher’s exact test using SPSS software (SPSS Inc., Chicago, IL, USA). Sensitivity analyses were performed by excluding those studies for which the data deviated from HWE (P<0.05), omitting the highest weight or changing the model.
Results
Literature search and characteristics of the retrieved studies
Figure 1 presents a flow diagram summarizing the process of study selection. The final search generated 65 potentially relevant publications. After reading the titles and abstracts, 44 articles were found to be irrelevant to this study in that the research did not cover the association between rs2239633 and susceptibility to ALL, and were thus excluded from the analysis. Of the remaining 21 publications, 3 were case reports, 2 were reviews, and 1 was a conference proceedings; these were therefore also excluded. Finally, 15 publications were identified, in which the effect of rs2239633 on the risk of both adult and childhood ALL was evaluated. Of these, four did not supply enough data, two were about adults,14,15 and nine were about children.4,8–13,17,18 These studies, providing enough data, were conducted in various populations of different ethnicities: eight involved Caucasians,4,10,11,13–15,17,18 two involved East Asians,9,12 and one involved multiple races.8 The basic data for every eligible study were extracted and are listed in Table 1. Although the authors of some of the publications were contacted to obtain the raw genotype data, some of the data could not be obtained.
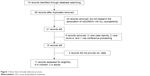  | Figure 1 Flow chart of study selection process. |
  | Table 1 Study characteristics |
Meta-analysis
rs2239633 SNP and ALL risk in childhood
As stated, nine papers studied the association between rs2239633 and risk of ALL in childhood and provided data about genotype distribution. The data from these studies were pooled and subjected to a meta-analysis. Overall, this SNP was found to be significantly associated with risk of ALL in childhood under the dominant model (Figure 2A, P<0.00001, OR =1.27, 95% CI =1.15–1.39), recessive model (Figure 2B, P<0.00001, OR =1.23, 95% CI =1.14–1.33), and allele contrast (Figure 2C, P<0.00001, OR =1.18, 95% CI =1.12–1.24). However, the presence of heterogeneity was found in the recessive model (Figure 2B, I2=56%) and allele contrast (Figure 2C, I2=57%), indicating the requirement for a subanalysis. Therefore, a submeta-analysis based on races was conducted. Among the included studies, the subjects in six were Caucasians,4,10,11,13,17,18 those in two were East Asians,9,12 and those in the remaining study were a mixed ethnic population.8 The submeta-analysis revealed that under the three contrast models, rs2239633 was significantly associated with childhood ALL in Caucasians (Figure 2A, P<0.00001, OR =1.31, 95% CI =1.18–1.46 for dominant model; Figure 2B, P<0.00001, OR =1.32, 95% CI =1.21–1.44 for recessive model; Figure 2C, P<0.00001, OR =1.23, 95% CI =1.16–1.30 for allele contrast), but not in East Asians (Figure 2). The degree of heterogeneity in both the Caucasian and East Asian subgroups was relatively small (Figure 2, I2=28%, 29%, 36% for Caucasian under the dominant, recessive, and allele contrast models, respectively; I2=0 for East Asian under the dominant, recessive, and allele contrast models).
rs2239633 and ALL risk in adults
Two of the studies investigated the relationship between the rs2239633 SNP and risk of ALL in adults, and provided the genotype data.14,15 The subjects in both of those studies were Caucasians. After pooling these data, further analysis revealed that this SNP was not associated with risk of ALL in adults in any model of comparison (Figure 3A, P=0.18, OR =1.21, 95% CI =0.91–1.61 for dominant model; Figure 3B, P=0.79, OR =1.03, 95% CI =0.81–1.32 for recessive model; Figure 3C, P=0.35, OR =1.08, 95% CI =0.92–1.26 for allele contrast). There was no heterogeneity in any of the three contrast models (Figure 3, I2=0).
Publication bias and sensitivity analysis
The publication bias for the research with pediatric subjects was assessed. Overall, no obvious publication bias was found under the recessive model, and the funnel plot exhibited good symmetry under the dominant model if the study of Vijayakrishnan et al12 was excluded. The result did not change after excluding this article. The symmetry of the funnel plot was poor under the allele contrast model, which suggested obvious publication bias (Figure 4).
  | Figure 4 The publication bias for researches of childhood. |
The stability of the results involving Caucasians was evaluated by performing a sensitivity analysis. Neither omitting Prasad et al’s13 study (highest weight) nor changing the model (random-effects model) made a significant difference to the pooled effects of the three contrast models, suggesting that the meta-analysis findings were highly stable.
Discussion
CEBPE is involved in terminal differentiation and functional maturation of neutrophils and macrophages, and is critical for transcription of most granule proteins localized to specific and gelatinase granules as well as for azurophil granule proteins expressed in the late promyelocyte stage.19,20 It has also been reported that CEBPE is targeted by recurrent immunoglobulin heavy-chain (locating on chromosome band 14q32) chromosomal translocations in B-cell precursor ALL, and mutations in CEBPE lead to neutrophil-specific granule deficiency.9,19,20 All of these findings suggested that CEBPE plays a role in the development of ALL. The studies conducted thus far on the relationship between rs2239633 and risk of ALL have yielded a relatively small amount of data and have produced conflicting results. Those data were evaluated in the present meta-analysis. The primary findings of this investigation are the increased OR of childhood ALL susceptibility associated with presence of the rs2239633 SNP in a fixed-effects model, and the lack of an association between this SNP and risk of ALL in adults.
Subgroup analysis for childhood ALL according to race revealed that this SNP impacted Caucasians, and not Asians. This finding was extremely robust, remaining significant irrespective of the different model assumptions (fixed-effects versus random-effects models), genetic models, and additional sensitivity analysis. This disparity between races may reflect the existence of a true population-specific disease variant, but may also be attributable to differences in genomic structure at these loci between populations. Wang et al9 determined the linkage disequilibrium (LD) patterns at the locus of rs2239633 in European and Chinese populations from the HapMap database, and found that rs2239633 is located in different LD blocks in the two populations. Therefore, we speculate that the pathogenic locus might not be rs2239633, but rather other SNPs in the LD block. Other factors contributing to the differential results for the two populations include differences in allele frequency and differences in genetic and environmental backgrounds that interact with the variants.12
It has been proposed that genetics plays a role in adult cancer susceptibility, although this has been difficult to evaluate because of confounders, such as environmental factors, the immune system, and exposure to pathogens. To the best of our knowledge, no predisposing genetic marker has been proven for adult ALL. No association between rs2239633 and risk of adult ALL was found in this study. Although the failure to detect any weak association may be affected by the relatively small sample, these data suggest that the genetic impact of polymorphisms at these loci is at least weaker for adult ALL than it is for pediatric ALL and highlights genetic differences between ALL occurring in adults versus children.15
Based on a meta-analysis, Wang et al21 concluded that rs2239633 confers an increased risk of childhood ALL, especially among Caucasians and Hispanics. The main findings of the present meta-analysis are consistent with that conclusion. While, the adults’ ALL risk related with rs2239633 was also evaluated, three different genetic models were used in our study, which showed more comprehensive evaluation of the impact of rs2239633 on ALL. In addition, we believe that the article of Wang et al21 contained some data errors. The research data – and in particular the OR and 95% CI values – of Chokkalingam et al22 and Walsh et al23 that were used in the meta-analysis performed by Wang et al21 were calculated using logistic regression. We speculate that since the OR and 95% CI values generally differed before and after data adjustment, Wang et al21 did not use the raw data from the studies of Chokkalingam et al22 and Walsh et al23 thus potentially causing bias. It is not possible to derive raw data using the adjusted data. Before completing our study, we attempted to contact the authors of those two articles to obtain the raw genotype data,22,23 but were unsuccessful. Therefore, these two articles were excluded from the analysis, which we believe was the most appropriate action.
It is worth noting that there were no differences in the results among the three contrast models, although the funnel figure obtained under the allele contrast model exhibited poor symmetry, and a clear outlier appeared in that obtained under the dominant model, indicating that the impact of genetic models on the results is not large.
Our study demonstrated that genotype testing is very important, although just a statistical association was found. Based on genotype testing, the susceptible population will be able to take the necessary protective measures, which is particularly important for those with long-term exposure to environmental pollutants.
This study had some limitations. First, the effect of gene–gene and gene–environmental interactions was not addressed in the meta-analysis due to lack of data. Second, the meta-analysis was based on unadjusted estimates, and the confounding factors could not be controlled for because most studies did not provide these data. Third, for publication bias, the best symmetry was observed for the funnel figure produced under the recessive genetic model, but not under the dominant model and allele contrast. Finally, the conclusions regarding risk of ALL among adults and Asian children are based on relatively few cases, and so the statistical power thereof is not strong.
Conclusion
In summary, the findings of the present meta-analysis support a positive association between CEBPE rs2239633 and childhood risk of ALL in Caucasians, but there was no such association for risk of ALL in Caucasian adults or East Asian children. Further large-scale studies assessing gene–gene and gene–environment interactions are required.
Acknowledgment
This work was supported by the Natural Science Foundation of Shaanxi Province (number 2015JM8415) and the Fundamental Research Funds for the Central Universities of China (number 2011jdhz55).
Disclosure
The authors report no conflicts of interest in this work.
References
Karathanasis NV, Choumerianou DM, Kalmanti M. Gene polymorphism sin childhood ALL. Pediatr Blood Cancer. 2009;52(3):318–323. | ||
Han S, Lee KM, Park SK, et al. Genome-wide association study of childhood acute lymphoblastic leukemia in Korea. Leuk Res. 2010;34(10):1271–1274. | ||
Borst L, Wesolowska A, Joshi T, et al. Genome-wide analysis of cytogenetic aberrations in ETV6/RUNX1-positive childhood acute lymphoblastic leukemia. Br J Haematol. 2012;157(4):476–482. | ||
Papaemmanuil E, Hosking FJ, Vijayakrishnan J, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1006–1010. | ||
Trevino LR, Yang W, French D, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1001–1005. | ||
Yang JJ, Cheng C, Yang W, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301(4):393–403. | ||
Akasaka T, Balasas T, Russell LJ, et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Blood. 2007;109(8):3451–3461. | ||
Emerenciano M, Barbosa TC, Lopes BA, et al. ARID5B polymorphism confers an increased risk to acquire specific MLL rearrangements in early childhood leukemia. BMC Cancer. 2014;14:127. | ||
Wang Y, Chen J, Li J, et al. Association of three polymorphisms in ARID5B, IKZF1 and CEBPE with the risk of childhood acute lymphoblastic leukemia in a Chinese population. Gene. 2013;524(2):203–207. | ||
Ross JA, Linabery AM, Blommer CN, et al. Genetic variants modify susceptibility to leukemia in infants: a Children’s Oncology Group report. Pediatr Blood Cancer. 2013;60(1):31–34. | ||
Pastorczak A, Gorniak P, Sherborne A, et al. Role of 657del5 NBN mutation and 7p12.2 (IKZF1), 9p21 (CDKN2A), 10q21.2 (ARID5B) and 14q11.2 (CEBPE) variation and risk of childhood ALL in the Polish population. Leuk Res. 2011;35(11):1534–1536. | ||
Vijayakrishnan J, Sherborne AL, Sawangpanich R, Hongeng S, Houlston RS, Pakakasama S. Variation at 7p12.2 and 10q21.2 influences childhood acute lymphoblastic leukemia risk in the Thai population and may contribute to racial differences in leukemia incidence. Leuk Lymphoma. 2010;51(10):1870–1874. | ||
Prasad RB, Hosking FJ, Vijayakrishnan J, et al. Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood. 2010;115(9):1765–1767. | ||
Burmeister T, Bartels G, Groger D, et al. Germline variants in IKZF1, ARID5B, and CEBPE as risk factors for adult-onset acute lymphoblastic leukemia: an analysis from the GMALL study group. Haematologica. 2014;99(2):e23–e25. | ||
Peyrouze P, Guihard S, Grardel N, et al. Genetic polymorphisms in ARID5B, CEBPE, IKZF1 and CDKN2A in relation with risk of acute lymphoblastic leukaemia in adults: a Group for Research on Adult Acute Lymphoblastic Leukaemia (GRAALL) study. Br J Haematol. 2012;159(5):599–602. | ||
Wu Z, Lou Y, Qiu X, et al. Association of cholesteryl ester transfer protein (CETP) gene polymorphism, high density lipoprotein cholesterol and risk of coronary artery disease: a meta-analysis using a Mendelian randomization approach. BMC Med Genet. 2014;15(1):118. | ||
Lautner-Csorba O, Gezsi A, Semsei AF, et al. Candidate gene association study in pediatric acute lymphoblastic leukemia evaluated by Bayesian network based Bayesian multilevel analysis of relevance. BMC Med Genomics. 2012;5:42. | ||
Orsi L, Rudant J, Bonaventure A, et al. Genetic polymorphisms and childhood acute lymphoblastic leukemia: GWAS of the ESCALE study (SFCE). Leukemia. 2012;26(12):2561–2564. | ||
Akagi T, Thoennissen NH, George A, et al. In vivo deficiency of both C/EBPbeta and C/EBPepsilon results in highly defective myeloid differentiation and lack of cytokine response. PLoS One. 2010;5(11):e15419. | ||
Gombart AF, Shiohara M, Kwok SH, Agematsu K, Komiyama A, Koeffler HP. Neutrophil-specific granule deficiency: homozygous recessive inheritance of a frameshift mutation in the gene encoding transcription factor CCAAT/enhancer binding protein – epsilon. Blood. 2001;97(9):2561–2567. | ||
Wang C, Chen J, Sun H, Sun L, Liu Y. CEBPE polymorphism confers an increased risk of childhood acute lymphoblastic leukemia: a meta-analysis of 11 case-control studies with 5,639 cases and 10,036 controls. Ann Hematol. 2015;94(2):181–185. | ||
Chokkalingam AP, Hsu LI, Metayer C, et al. Genetic variants in ARID5B and CEBPE are childhood ALL susceptibility loci in Hispanics. Cancer Causes Control. 2013;24(10):1789–1795. | ||
Walsh KM, Chokkalingam AP, Hsu LI, et al. Associations between genome-wide native American ancestry, known risk alleles and B-cell ALL risk in Hispanic children. Leukemia. 2013;27(12):2416–2419. | ||
Enciso-Mora V, Hosking FJ, Sheridan E, et al. Common genetic variation contributes significantly to the risk of childhood B-cell precursor acute lymphoblastic leukemia. Leukemia. 2012;26(10):2212–2215. | ||
Migliorini G, Fiege B, Hosking FJ, et al. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood. 2013;122(19):3298–3307. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


