Back to Journals » International Journal of Nanomedicine » Volume 12
The influence of surface charge on serum protein interaction and cellular uptake: studies with dendritic polyglycerols and dendritic polyglycerol-coated gold nanoparticles
Authors Bewersdorff T , Vonnemann J, Kanik A, Haag R, Haase A
Received 10 October 2016
Accepted for publication 10 January 2017
Published 14 March 2017 Volume 2017:12 Pages 2001—2019
DOI https://doi.org/10.2147/IJN.S124295
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Tony Bewersdorff,1 Jonathan Vonnemann,2 Asiye Kanik,1 Rainer Haag,2 Andrea Haase1
1Department of Chemical and Product Safety, German Federal Institute for Risk Assessment (BfR), Berlin, Germany; 2Institute of Chemistry and Biochemistry, Freie Universität Berlin, Berlin, Germany
Abstract: Nanoparticles (NPs) have gained huge interest in the medical field, in particular for drug delivery purposes. However, binding of proteins often leads to fast NP uptake and rapid clearance, thereby hampering medical applications. Thus, it is essential to determine and control the bio–nano interface. This study investigated the serum protein interactions of dendritic polyglycerols (dPGs), which are promising drug delivery candidates by means of two dimensional gel electrophoresis (2DE) in combination with mass spectrometry. In order to investigate the influence of surface charge, sulfated (sulfated dendritic polyglycerol [dPGS]) and non-sulfated (dPGOH) surfaces were applied, which were synthesized on a gold core allowing for easier separation from unbound biomolecules through centrifugation. Furthermore, two different sizes for dPGS were included. Although size had only a minor influence, considerable differences were detected in protein affinity for dPGS versus dPGOH surfaces, with dPGOH binding much less proteins. Cellular uptake into human CD14+ monocytes was analyzed by flow cytometry, and dPGOH was taken up to a much lower extent compared to dPGS. By using a pull-down approach, possible cellular interaction partners of serum pre-incubated dPGS-Au20 NPs from the membrane fraction of THP-1 cells could be identified such as for instance the transferrin receptor or an integrin. Clathrin-mediated endocytosis was further investigated using chlorpromazine as an inhibitor, which resulted in a 50% decrease of the cellular uptake of dPGS. This study could confirm the influence of surface charge on protein interactions and cellular uptake of dPGS. Furthermore, the approach allowed for the identification of possible uptake receptors and insights into the uptake mechanism.
Keywords: sulfated dendritic polyglycerols, protein corona, cellular uptake, uptake receptors, clathrin-mediated endocytosis
Introduction
Nanotechnology is often regarded as one of the key enabling technologies of the century. Nanoparticles (NPs) are already used for various diverse purposes including medical applications such as drug delivery.1 NP-based drug delivery systems offer several advantages. In particular, they are used with difficult-to-formulate chemicals, can protect drugs from degradation, enhance drug half-lives, and finally can enable drug targeting as NPs may reach and accumulate at specific sites in the body, where they may also be used as local drug depots. Thus, often for nano-formulated drugs, lower doses can be applied compared to conventional formulations leading to significantly reduced side effects.2 Some NP drug carrier systems are already approved, mostly for tumor therapy such as DaunoXome®, a liposome-based formulation of daunorubicin being used to treat human immunodeficiency virus-related Kaposi’s sarcoma.3 However, not only liposome-based formulations but also dendrimers seem to be useful candidates for drug delivery purposes. They allow packaging of larger molecules and are in particular useful to solubilize poorly water-soluble drugs.4,5
Often the use of NPs in medicine is hampered by the fast binding of plasma proteins to their surfaces (opsonization), leading to quick clearance of the circulating NPs by the reticuloendothelial system, which is also called the mononuclear phagocytic system.6–8 This may decrease the injected dose by as much as 50% within a few hours.9 Thus, for medical applications, it is often desired to synthesize NPs that are rather repellent to plasma protein interaction. Polyethylene glycol as a surface modifier for example was shown to enhance blood circulation and half-life of superparamagnetic iron oxide nanoparticles in vivo in mice fivefold.10 On the other hand, in order to achieve site-specific targeting of a drug specific interactions of a NP with selected serum proteins may be desired. One example is poly(n-butyl-2-cyanoacrylate) NP, which upon coating with polysorbate 80 was able to mediate transport across the blood brain barrier in vivo.11–13 This was shown to be dependent on recruiting apolipoprotein E from blood plasma onto the NP surface.14,15 Thus, in nanomedicine, it is necessary to understand the behavior of NPs in physiological environments and to analyze the NP protein corona. The protein corona formation depends on several physico-chemical properties of NPs such as size, shape, surface charge, or hydrophobicity and can influence cellular uptake in vitro.16–23 Therefore, the protein corona can also influence NP functionality and their biological effects.24,25 For example, transferrin coating was shown to enhance the endocytosis of lipid-coated NPs in A549 cells in vitro and furthermore to have the best inhibitory effect on tumor growth in vivo compared to the other used NPs.26
Within this study, we focused on dendritic polyglycerols (dPGs), which are interesting carrier systems for medical applications. They are highly biocompatible and can be synthesized with different functionalities on their surface offering a high variability and allowing modulation of the binding affinity.27–29 Sulfated dendritic polyglycerol (dPGS) has been shown to possess anti-inflammatory properties in vivo and in vitro with the relevant cellular targets being L- and P-selectins.28,30,31 The aim of this study was to investigate the influence of size and charge of dPGs on the interactions with serum proteins and to study the influence of the protein corona on cellular uptake in human CD14+ monocytes by flow cytometry. Furthermore, this study aimed to elucidate cellular interaction partners being possibly involved in cellular uptake by using a pull-down approach with purified cell membrane proteins of THP-1 cells and to get first insights into the uptake mechanisms.
Materials and methods
Synthesis of the NPs
The synthesis of the native citrate-functionalized gold NPs as well as their functionalization with TA (thioctic acid)-dPGS-10 kDa and TA-PGOH-10 kDa is described in detail elsewhere.32 Briefly, native citrate-functionalized gold NPs were synthesized by the reduction of tetrachloroauric acid at elevated temperatures in an aqueous solution.33 Larger-sized gold NPs (50 nm) were obtained by a seed-mediated growth procedure according to a protocol established by Ziegler and Eychmüller.34 The functionalization of the gold NPs with TA-PGOH-10 kDa and TA-dPGS-10 kDa, respectively, was performed according to an own established salt aging procedure to maximize the ligand packing on the NPs.32 Briefly, the citrate-stabilized gold NPs were incubated with TA-dPGS-10 kDa and TA-PGOH-10 kDa, respectively, under rapid stirring. After 30 min, the ionic strength of the solution was increased from 0 to 1.5 M over 30 min by the drop-wise addition of a NaCl (5 M) stem solution. The particles were incubated for 1 h under rapid stirring with 2 s sonification every 20 min. For purification, the colloidal dispersion was purified 6 times by centrifugation (15,000× g, 20 min, room temperature [RT]) for the removal of the supernatant and redispersion of the pellet in water. The concentration of gold NPs in the solutions was calculated by the ratio of the mass concentration of gold in the colloidal solution as determined by atomic adsorption spectroscopy and the mass of one gold NP as calculated by the diameter obtained from dynamic light scattering (DLS) and the assumption of a perfect sphere.32,35
The syntheses of rhodamine B-labeled dendritic polyglycerol (dPGOH-RB) and rhodamine B-labeled dendritic polyglycerol sulfate (dPGS-RB) are explained and depicted schematically in Figures S1 and S2, complete details are described elsewhere.36
Characterization of the NPs
Transmission electron microscopy (TEM)
Droplets of the corresponding sample solutions (5 μL) were placed for 30 s on hydrophilized (60 s plasma treatment at 8 W using a BAL-TEC MED020 device [Leica Microsystems, Wetzlar, Germany]) carbon-coated copper grid. Excess fluid was removed with a filter paper, and the samples were analyzed in a CM-12 by FEITM with an accelerating voltage of 60 kV.
DLS and zeta potential measurement
NPs were diluted 1:200 either in ultra-pure water (Milli-Q [MQ] reference ultrapure water purification system; Merck Millipore, Darmstadt, Germany), phosphate-buffered saline (PBS; PAN Biotech GmbH, Aidenbach, Germany) or Dulbecco’s Modified Eagle’s Medium (DMEM, without phenol red and L-glutamine; PAN Biotech GmbH) containing 10% non-heat-inactivated fetal bovine serum (FBS gold, PAA Laboratories GmbH, Cölbe, Germany). The NPs (dPGOH-Au50 with a final concentration of 24.6 μg/mL, dPGS-Au50 with 20 μg/mL, and dPGS-Au20 with 10.08 μg/mL) were dispersed at RT under constant stirring (700 rounds per minute, rpm) with a magnetic stirring device (HMC Cyclone; HMC Europe, Tüssling, Germany). These concentrations are in line with the ones used for studying cellular uptake. After 120 min, samples were taken and measured using a Zetasizer Nano ZS (Malvern Instruments GmbH, Herrenberg, Germany) equipped with a red laser (He-Ne-Laser 633 nm, maximum 4 mW). Pre-equilibration time for the samples of 60 s was used before the measurement (5 runs with 10 measurement points [equals n=50] for each sample) at 25°C. The attenuator and voltage calibration were selected automatically during the automatic measurements.
In order to measure zeta potential, NP dispersions were measured at 1 mg/mL in 10 mM NaCl dissolved in ddH2O (Merck Millipore) at 25°C using Zetasizer Nano ZS. A pre-equilibration time of 2 min was also used, attenuator and voltage were selected automatically, and three runs of 30 measurements were used.
Nanoparticle tracking analysis (NTA)
The NanoSight LM20 system (638 nm visible radiation, maximum output 40 mW, Malvern Instruments GmbH) was used for NTA. Sizes were calculated from videos (60 s, 30 frames per second, camera gain =1, and background extract on) by the NTA software (Version 2.3 Build 0033). Particles were diluted using filtrated MQ water (filtered through a 0.2 μm filter) to final concentrations of dPGOH-Au50 2.46 μg/mL, dPGS-Au50 2 μg/mL, and dPGS-Au20 1.008 μg/mL and measured at 25°C.
Dispersion of the NPs for protein corona analysis
For analysis of the protein corona, the concentrations of all NPs (dPGS-Au20, dPGS-Au50, and dPGOH-Au50) were adjusted such that protein to surface ratio was kept constant (total available NP surface was adjusted to approximately 66 cm2. Ratio was kept at approximately 300 microgram protein/cm2.). The total available surface for protein binding was calculated by assuming a perfect spherical shape. NPs were dispersed in DMEM containing 10% FBS gold in a total volume of 5 mL in sterile glass vials containing magnetic stir bar by stirring (700 rpm for 2 h at RT) with a magnetic stirring device (HMC Cyclone; HMC Europe).
Protein elution and analysis of protein content in eluate
For the separation of the NPs with adsorbed corona from unbound proteins, the NP dispersions (5 mL) were loaded on top of 2 M sucrose (4.5 mL), and the NPs were pelleted through ultracentrifugation (17,900× g, 2.5 h at RT). DMEM with 10% FBS gold without NPs served as control and was treated similarly. Proteins in the NP pellets were eluted in 500 μL two dimensional (2D) lysis buffer containing 7 M urea, 2 M thiourea, 4% chaps, 2% pharmalyte pH 4–7, 1% dithiothreitol (DTT), and protease inhibitors for 2DE analysis while rotating for 1 h at RT. Samples for 2DE analysis were centrifuged (40,000× g for 1 h at 15°C) and total protein content was measured using the 2D Quant kit (GE Healthcare, Berlin, Germany) according to manufacturer’s instructions.
2D-Polyacrylamide gel electrophoresis (PAGE) analysis of the protein corona
For the isoelectric focusing (IEF), samples (500 μL) were loaded on nonlinear IPG strips (24 cm Immobiline™ DrySrip pH 4–7 [NL]; GE Healthcare) and equilibrated in equilibration buffer EB1 containing 360 mg/mL urea (Carl Roth GmbH, Karlsruhe, Germany), 24 mg/mL sodium dodecyl sulfate (SDS; Carl Roth GmbH) and 50.4 mM/mL tris(hydroxymethyl)aminomethane-hydrogen chloride (Tris-HCl), pH 8.6 (Carl Roth GmbH) for 1 h. Active rehydration and focusing were performed (15 h at 30 V, 1.5 h at 200 V, 1 h at 500 V, 13.5 h gradient 500–1,000 V, 3 h gradient 1,000–8,000 V, and 6 h at 8,000 V) with the GE Ettan IPGphor 3 (GE Healthcare). After the IEF proteins were first reduced using DTT (1% DTT in EB1; Carl Roth GmbH) and then alkylated using iodacetamide (4% IAA in EB1; SERVA, Heidelberg, Germany) 15 min each. For the second dimension, strips were transferred onto 12.5% SDS-polyacrylamide gels. Electrophoresis was carried out with the GE Ettan DALTtwelve System Seperation Unit (GE Healthcare). Later, the gels were fixed using 30% ethanol (Sigma-Aldrich, Munich, Germany) and 10% acetic acid (Carl Roth GmbH) in water and thereafter stained with ruthenium II tris (bathophenanthroline disulfate) chelate (0.4 μM RU-II and 20% ET in MQ).37 After scanning of the gels with the FLA 9,500 (GE Healthcare, excitation λ=473 nm and detection at λ=610 nm), they were analyzed with Delta2D version 4.4 (Decodon, Greifswald, Germany). Each sample was analyzed in three independent replicates. Spots with an increased intensity compared with control gels without NPs (>1.5 fold increase, n=3, p<0.05) were further analyzed by mass spectrometry to identify the respective proteins.
Matrix-assisted laser desorption ionization (MALDI)-time of flight (TOF)/TOF
Spots of interest were excised using a spot picker (Proteome Factory, Berlin, Germany), destained in 200 mM ammonium bicarbonate (ABC; Sigma-Aldrich) containing 50% acetonitrile (ACN; Sigma-Aldrich) and digested overnight with trypsin using a standard protocol (50 mM ABC and 5% ACN, 1.2 ng/μL trypsin). Peptides were extracted from the gel piece with 60% ACN and then with 100% ACN. Collected and combined supernatants were dried using a speedvac. Dry peptide pellets were resuspended in 0.1% TFA and purified using a C18 ZipTip (Merck Millipore). Samples were spotted with an α-cyano-4-hydroxy-cinnamic acid (HCCA; Sigma-Aldrich) matrix on AnchorChip targets (383/800; Bruker, Bremen, Germany) and measured using a UltrafleXtreme MALDI-TOF/TOF (Bruker) device equipped with FlexControl (version 3.3) and FlexAnalysis (version 3.3) software packages. For analysis, ProteinScape (version 4.0) and MASCOT (version 2.3.02) were used based on search in Swissprot database (MS tolerance 50 ppm, MS/MS tolerance 0.8 Da, Carbamidomethyl [Cys] and Oxidation [Met] as variable modifications, 1 partial cleavage).
Cell culture THP-1 cells
The human monocytic cell line THP-1 was obtained from DSMZ (German Collection of Microorganisms and Cell Cultures, Germany) and was cultivated in RPMI-1640 complete medium containing 10% FBS, 2 mM L-glutamine, 100 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid, 100 units/mL penicillin, 0.1 mg/mL streptomycin in an incubator (37°C, 5% CO2). Cell culture medium, FBS, and additives were obtained from PAN Biotech GmbH. Cell viability and quantity were determined through a cell counter Casy TTC (Roche Diagnostics GmbH, Mannheim, Germany).
Isolation of the CD14+ cells through CD14 MicroBeads
Primary human CD14+ monocytes were isolated from blood samples (buffy coats) obtained from Deutsche Rote Kreuz (Berlin, Germany). Samples were diluted with PBS, layered on Ficoll-Paque™ Plus (GE Healthcare) solution and centrifuged (400× g for 35 min at RT) to obtain peripheral blood mononuclear cells (PBMCs). PBMCs were washed with PBS and finally dispersed in auto MACS™ Running buffer (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) (40 μL per 107 cells). CD14+ MicroBeads (Miltenyi Biotec GmbH) were added (10 μL per 107 cells). After 15 min of incubation, CD14+ cells were separated using positive selective LS columns (Miltenyi Biotec GmbH). Figure S3 depicts quality control through flow cytometry before and after CD14 MicroBeads purification. For analysis 1×105 cells (PBMC or CD14+ monocytes) were washed (300 g, 5 min) with buffer (PBS, 1% FBS and 2 mM ethylenediaminetetraacetate [EDTA]) and stained in 50 μL buffer with a-CD14-PE (TÜK4) as a marker for CD14+ monocytes, with a-DC-Sign-APC (DCN47.5) as a marker for dendritic cells and with a-CD86-Fitc (FM95) as an activation marker (all 1:50; Miltenyi Biotec GmbH) for 30 min at 4°C. After washing, the samples were measured using the BD FACSAria III with Diva 6.0 software (BD Biosciences, Heidelberg, Germany).
Investigation of the uptake of NPs through flow cytometry
CD14+ cells (105 cells per well in 1 mL RPMI-1640 medium containing 10% FBS) were seeded in 12-well plates (TPP, Trasadingen, Switzerland). After 24 h, the medium was replaced with either RPMI-1640 medium containing 10% FBS or cell culture RPMI-1640 medium containing 3 mg/mL bovine serum albumin (BSA) or RPMI-1640 medium without any proteins. Rhodamine B-labeled dPG NPs were diluted in the respective cell culture medium (final concentration dPGOH-RB 50 μg/mL and dPGS-RB 20 μg/mL) and pre-incubated for 1 h (at 37°C) such that a protein corona can form. Cell culture medium from the monocytes was then replaced with the respective cell culture medium containing the NPs and incubated for 1 or 6 h, respectively. Later, the cells were rinsed with PBS and detached using trypsin (PAN Biotech GmbH), washed again using PBS and fixed using 100 μL 1× CellFix solution (BD Biosciences). Uptake was analyzed using an FACS Aria III (BD Biosciences), equipped with a blue laser (488 nm) and fluorescence detector (585/42 nm) using a 70 μm nozzle.
Purification of cell membrane protein fraction
Biotinylation of the THP-1 cells
Prior to the treatment with sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate (Biotin; Antibodies-online GmbH, Aachen, Germany), THP-1 cells were washed three times with PBS (500 g for 10 min, 4°C). After incubation with Biotin for 10 min at RT (0.05 mg/mL biotin diluted in PBS per 5×106 THP-1 cells) Tris-HCl (pH 7.5 with 50 mM) was added followed by incubation for another 10 min at RT. Thereafter cells were washed with PBS five times (4°C at 500 g for 10 min) and lyzed by 30 min incubation on ice in modified RIPA buffer (0.05 M Tris-HCl [pH 7.4], 0.15 M NaCl, 0.001% EDTA, 1% Igepal CA-630, 0.25% sodiumdeoxycholate) followed by centrifugation (14,000× g for 20 min at 4°C). All chemicals were obtained from Sigma-Aldrich. Total protein content was determined in supernatants using a Bradford assay (BioRad, Munich, Germany) according to the manufacturer’s protocol.
Purification of the biotinylated membrane proteins through avidin-agarose beads
Biotinylated proteins were isolated using avidin-agarose beads (15 μL of a 50% w/w slurry per 25 mg lysate), which were incubated with the cell lysates (three runs, each 15 min incubation). After binding, beads were washed with PBS (five times) and proteins eluted by adding DTT (100 mM in PBS, 1 h at RT). Concentration of the eluate was determined by Bradford assay according to manufacturer instructions. In total, 615 μg of protein was obtained using this protocol from 250×106 cells.
Pull-down experiment with dPG NPs
The NPs dPGS-Au50 and dPGOH-Au50 (final concentrations 100 μg/mL) were washed with PBS three times and incubated for 2 h at RT with the relevant medium (RPMI-1640 medium ±10% FBS), later washed again with PBS three times. Then, NPs with or without serum pre-treatment were incubated (2 h at RT) with the purified membrane protein fraction (115 μg total protein per 100 μg NP) and later washed with PBS three times and pelleted by centrifugation. Proteins from NP pellets were eluted using Laemmli buffer (0.25 M Tris-HCL pH 6.8, 4% SDS, 20% glycerol [Sigma-Aldrich] with 10% β-mercaptoethanol [Merck Millipore]) at 95°C for 10 min. Eluted proteins were separated through 10% SDS-PAGE and stained with Coomassie brilliant blue R 250 (AppliChem GmbH, Darmstadt, Germany). Each lane was cut into 10 bands, proteins were digested, peptides were eluted, dried, and redissolved in ACN/0.1% TFA (ratio 7:3). Peptides were then analyzed by nano high performance liquid chromatography (nanoHPLC) coupled offline with MALDI-MS/MS.
Analysis of pulled down proteins through nanoHPLC-MALDI-MS/MS
For the chromatographic separation of the samples, a reversed-phase HPLC UltiMate3000 (Thermo Scientific) with gradient elution was used. The nonpolar stationary phase (Acclaim® PepMap RSLC, 75 μm ×50 cm nano Viber, C18, 2 μm, 100 Å) consists of C18-modified silica. Equilibration was done with solvent A (0.1% TFA) for 5 min, and the separation of the samples was done by increasing the concentration of the nonpolar solvent B (0.08% TFA/80% TFA, reaching following concentrations at the given time points: 1 min 20%, 11 min 25%, 95 min 50%, 105 min 80%, and after 115 min 95% concentration of solvent B). Ultraviolet spectrum was measured at 214 nm. Eluates from nanoHPLC were automatically spotted with matrix solution (HCCA) at a target (AnchorChip 1536 TF, Bruker) and measured through an UltrafleXtreme MALDI-TOF/TOF (Bruker).
Investigation of possible uptake pathways of dPGS-RB in THP-1 cells
THP-1 cells were seeded in 96-well plates (TPP) at 75,000 cells per well in 100 μL cell culture medium (CCM, RPMI-1640 medium containing 10% FBS) 24 h before treatment. To investigate energy-dependent uptake, cells were then either transferred to 4°C or were kept at 37°C (control) for the rest of the treatment. After 4 h at the respective temperature conditions, 20 μg/mL of pre-incubated dPGS-RB NPs (2 h in CCM, RT) were added for another 2 or 4 h. To investigate endocytosis, cell culture medium was replaced with serum-free RPMI-1640 medium (control) or serum-free RPMI-1640 medium containing 5 or 10 μg/mL chlorpromazine (CP) to investigate clathrin-mediated endocytosis (Sigma-Aldrich) or 200 μM genistein (Sigma-Aldrich) and incubated for 2 h. Then, 20 μg/mL of pre-incubated dPGS-RB NPs (in CCM) were added for another 2 h. Thereafter in both cases, cells were washed twice with ice cold PBS (300× g, 5 min). Life/Dead Fixable Near-IR Dead cell Stain Kit (ThermoFisher Scientific, Darmstadt, Germany) diluted 1:1,000 in ice cold PBS was added for 30 min (constant shaking, 4°C). Samples were washed twice with PBS (300× g, 5 min) and fixed (2 h, 4°C) using 100 μL 1× CellFix solution (BD Biosciences). Uptake was analyzed using a FACS Aria III (BD Biosciences), equipped with a blue laser (488 nm) and fluorescence detector (585/42 nm) using a 70 μm nozzle.
Results
Characterization of the NPs
This study used gold NPs functionalized with either sulfated or non-sulfated dPGs with a molecular weight of 10 kDa, dPGS-10 kDa or dPGOH-10 kDa, respectively. Figure 1 schematically depicts the chemical structures of dPGS-10 kDa and dPGOH-10 kDa as well as the NPs used in this study.
Both, dPGS-10 kDa and dPGOH-10 kDa were functionalized with ~1 TA anchor moiety, resulting in TA-dPGS-10 kDa and TA-dPGOH-10 kDa, respectively, to permit the binding to the gold NP surface through stable sulfur–gold linkage.38 The dPGOH- and dPGS-coated gold NPs (dPGS-Au and dPGOH-Au) were synthesized by ligand exchange from citrate-coated gold NPs according to a previously established salt-aging procedure, which permits a maximization of the ligand density on the NPs.32 This leads to a slight increase in NP radii compared with the original citrate-coated gold NPs (Figure 2). The dPGS-Au was synthesized in two different sizes, using 20 and 50 nm gold cores, herein referred to as dPGS-Au20 and dPGS-Au50, respectively. In the case of dPGOH-Au, a 50 nm gold core was used for the synthesis, resulting in dPGOH-Au50. NPs were fully characterized with respect to size and size distribution using TEM, DLS, and NTA. The DLS and NTA results are summarized in Table 1, and TEM images and DLS data are depicted in Figure 2.
NPs were dispersed in different biological dispersion media, that is, PBS or complete CCM, consisting of DMEM supplemented with 10% FBS. NPs seem to be stably dispersed in PBS and CCM over the time course of the experiment, that is, for up to 2 h. However, a shift in NP hydrodynamic radii was observed over time in the protein containing CCM dispersion medium compared with PBS, in particular for the dPGS-Au NPs. This may indicate the formation of a protein corona (Table 1). For dPGS-Au50, the increase in hydrodynamic radius in CCM accounts for 25 nm, which is consistent with the reported changes that occurred because of the formation of a protein corona of ~50 nm NPs as reviewed in Walkey and Chan.39 In addition, the zeta potential in 10 mM NaCl in MQ water was also determined for dPGS (−25.7 mV) and dPGOH (+12.1 mV) particles. For studying cellular uptake, rhodamine-labeled dPGS-RB and dPGOH-RB were used, which were also fully characterized (Table S1).
NP protein corona
The NPs were incubated in CCM for 2 h such that they could form a stable protein corona. As of course the total amount of bound protein would depend on available NP surface, the NP concentrations for this experiment were adjusted such that similar ratios of total protein to NP surface were obtained.
A detailed analysis of protein coronas was performed using 2D gel electrophoresis (2DE) in combination with MALDI-TOF/TOF. 2DE analysis revealed qualitative and quantitative differences between the coronas of all NPs. Each NP displayed an individual spot pattern. It becomes obvious that dPGOH surface binds considerable less protein compared with dPGS surfaces (Figure 3). Significant quantitative and qualitative differences were observed in protein coronas for the different surface functionalized NPs, that is, dPGOH-Au50 versus dPGS-Au50, whereas the differences for the two sizes of dPGS, that is, dPGS-Au20 versus dPGS-Au50, appeared marginal only (Figure S4).
In order to identify relevant proteins, spot patterns of all gels were analyzed and compared with control gels. Protein spots with significantly increased intensity compared with controls (ie, intensity increased >1.5, p<0.05, n=3) were excised and analyzed by MALDI-TOF/TOF (Figure S5).
Clearly, dPGS surfaces bind more proteins compared with dPGOH (ie, compare dPGS-Au50 versus dPGOH-Au50, Figure 3). The corona of dPGOH-Au50 consisted only of 36 protein spots, whereas 169 were detected on dPGS-Au50 and 114 on dPGS-Au20. Similarities and differences in the protein coronas are visualized in a Venn diagram based on the spot numbers (Figure 3C). Comparing the proteins in the different coronas as identified by mass spectroscopy, it was found that most proteins detected on dPGOH-Au50 were also found on dPGS-Au50 (for instance complement factor H and serum albumin). However, several of them such as for instance serotransferrin and vitronectin (VN) were unique for the dPGS NP coronas (Table 2).
Influence of the protein corona on cellular uptake
Next, the study aimed to analyze whether the differences in protein binding would correlate with differences in cellular uptake. For this analysis, we used rhodamine B-labeled dPGS and dPGOH NPs without a gold core, herein referred to as dPGS-RB and dPGOH-RB (refer to Figures S1 and S2 for a summary of their syntheses and to Table S1 for their physico-chemical characterization). The fluorescence label allowed for easy quantification of cellular uptake using flow cytometry. NPs were pre-incubated for 2 h in different biological media. DMEM cell culture medium was used without serum (ie, no proteins were present), CCM with serum such that a complete protein corona can form or cell culture medium containing BSA only, which was considered a rather unspecific binder with moderate affinity.
Figure 4 shows that dPGS-RB was taken up much more efficiently by primary human monocytes compared with dPGOH-RB, regardless of the kind of pre-incubation. Using confocal laser scanning microscopy, it could be confirmed that dPGS-RB NPs (Figure S6) and also dPGOH-RB NPs (Figure S7) were taken up into monocytes. However, amounts of dPGOH-RB taken up seemed to be very low.
Comparing different conditions of pre-incubation for dPGS, it was found that without any protein pre-incubation, dPGS was taken up most efficiently (Figures 4 and S6). Thus, clearly the presence or absence of a protein corona influences the cellular uptake of dPGS. In addition, the composition of the corona seems to affect cellular uptake as well (compare uptake in the presence of BSA alone, which is a rather unspecific binder compared to uptake in the presence of complete serum).
Identification of putative cellular interaction partners
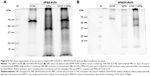
This prompted us to further analyze the interactions between protein-coated NPs and cellular surfaces in more detail. For this purpose, the cell membrane proteins of THP-1 cells were purified after biotinylation by affinity chromatography. Purified membrane fraction was used in pull-down experiments with dPGS-Au50 (Figure 5A) and dPGOH-Au50 as control (Figure 5B), both of which have been pre-incubated in CCM. Later, pulled down proteins were identified using a nanoHPLC-MALDI-TOF/TOF approach.
A total of 138 proteins could be identified in the pull-down experiment with CCM-pre-incubated dPGS-Au50, out of which 22 were membrane or membrane-associated proteins. A few selected identified membrane proteins, that is, those for which possible protein interaction partners may be suggested from the identified proteins in the protein corona of dPGS, are listed in Table 3, and all of them are listed in Table S2. In particular, transferrin receptor protein 1 and the integrin βII might be involved in cellular recognition of the dPGS.
Investigation of possible uptake pathways
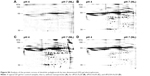
In the pull-down experiment, the transferrin receptor was identified as a possible cellular interaction partner, which indicates that clathrin-mediated endocytosis is involved in the cellular uptake of dPGS as the transferrin receptor is internalized through this pathway.40 To further look into this, this study first aimed to analyze whether the uptake process is energy dependent, which may be expected for endocytotic processes and then whether it may be inhibited by CP, which is a known inhibitor for clathrin-mediated endocytosis.41 When transferring the cells to 4°C and thereby blocking energy-dependent uptake processes, it could be shown that the uptake of dPGS-RB NPs is indeed significantly decreased, that is, ~79% after 2 h and ~66% after 4 h (Figure 6A). When pretreating THP-1 cells for 2 h with CP, the uptake was decreased by ~25% after 2 h using 5 μg/mL and by ~50% using 10 μg/mL (Figure 6B). Caveolin-mediated endocytosis was also considered, and cellular uptake could be blocked by ~20% after 2 h and by ~30% after 4 h incubation with genistein (Figure 6C). Therefore, it is concluded that clathrin-mediated endocytosis seems to be the major uptake pathway for dPGS particles. However, other pathways, in particular caveolin-mediated endocytosis also contribute but to a lesser extent. Also energy-independent processes seem to exist and can also contribute to cellular uptake of dPGS.
Discussion
The aim of this study was to investigate the influence of size and surface chemistry for dPGs on interactions with serum proteins as they may be widely used in medical applications. A reduced protein affinity was clearly observed for the non-sulfated variant dPGOH compared with dPGS, which indicates that serum proteins preferably interact with negatively charged NP surfaces, as also shown by others.28 Differences in protein binding with respect to NP size were also observed. However, the influence of size on the protein composition of the NP protein corona seems to be minor compared with the influence of the surface charge. The effect of size on the composition of the protein corona may be explained by the different curvature of NPs, which could affect the binding of differently shaped proteins to different extents. Often, globular proteins absorbed on small particles seem to retain their native conformation with increasing curvature as shown for BSA on monodisperse silica spheres.42 In contrast, fibrillar proteins may lose their native conformation with decreasing particle size as shown for fibrinogen.42
Here we could identify different serum proteins as components of the protein corona for each NP, among them constituents of the adaptive immune response, cell-adhesion (-migration, -motility, and -proliferation), activators, and regulators of the complement system. Some of these proteins may play a role in the cellular interaction of the NPs and thus could be interesting with respect to NP targeting.
In order to further analyze the interactions between NP protein corona and cell membranes, pull-down experiments were performed to identify possible interaction partners for dPGS. Through this approach, the transferrin receptor protein, HLA class 1 histocompatibility antigens, and a member of the integrin receptor family, which could be potentially involved in cellular recognition and uptake of the dPGS NPs, could be identified.
Others have already performed intensive research on the role of transferrin in NP uptake. Transferrin has been frequently identified in the protein coronas of several NPs, for example, silver, iron oxide, and silica.43–45 Here we could identify serotransferrin in the corona of dPGS but not in the corona of dPGOH NPs. Serotransferrin can interact with the transferrin receptor and mediate an endocytotic uptake through clathrin-mediated endocytosis.46 It was shown that the presence of transferrin enhanced the endocytosis of lipid-coated NPs in A549 cells in vitro.26 In this study, the transferrin receptor could be identified as a possible cellular interaction partner of CCM pre-incubated dPGS NPs using a pull-down approach. Furthermore, by using CP, a known inhibitor of clathrin-mediated endocytosis, cellular uptake of dPGS was decreased by 50% in this study. Therefore, clathrin-mediated endocytosis plays a major role in cellular uptake of dPGS. However, other pathways, in particular caveolin-mediated endocytosis seem to contribute as well.
In addition, a membrane protein of the integrin family was identified, which can also be involved in mediating cellular uptake of dPGS NPs. Integrins have the ability to recognize multiple ligands and may bind to interaction partners with an RGD sequence (Arg-Gly-Asp), an acidic motif termed “LDV,” or a collagenous GFOGER motif.47–53 VN, which was identified as a component of the serum corona for dPGS but not for dPGOH NPs, can interact with αvβ3 integrin and could thus possibly be important for the uptake of dPGS NPs as well.54–56 For FBS-treated TiO2 NPs, it was already shown that cellular uptake into human lung carcinoma cells (A549) was reduced when cells were pre-treated with anti-VN antibodies.57
Taken together, several lines of evidence that the protein coronas of dPG NPs can alter the NP cell interactions and influence cellular uptake are presented. These data indicate that clathrin-mediated endocytosis is the major cellular uptake pathway for dPGS, whereas other pathways such as caveolin-mediated endocytosis contribute as well. Therefore, most likely different proteins and different pathways are involved in parallel in cellular uptake of dPGS NPs. Indeed, for the first time, a few possible cellular uptake receptors for dPGS were identified. As shown by others, different proteins in the protein corona can be involved in the cellular uptake of NPs. A study performed by Ritz et al showed that binding of ApoA4 and ApoC3 to four polymeric NPs was increasing the uptake while the binding of ApoH was decreasing it.58 Therefore, as NPs are typically covered by a protein corona consisting of plenty of different proteins, there seems to be a complex interplay of different corona components. Different proteins may influence cellular uptake through different pathways.
Conclusion
In this study, different dPG-coated gold NPs were synthesized, and it could be shown that the surface charge had a large influence on the protein corona compared to size, which only had a minor effect on corona composition in this study. Sulfated dPG NPs (dPGS) showed a higher tendency to interact with serum proteins compared to the non-sulfated dPG NPs (dPGOH). The different protein affinities were well reflected by different rates of cellular uptake with dPGS but not dPGOH being strongly taken up by human monocytes. A pull-down approach allowed us to identify possible cellular interaction partners for dPGS in human monocytes such as the transferrin receptor or an integrin, which correlated well with the presence of serotransferrin or VN, respectively, in the corona of dPGS. Thus, this approach allows for the first time to get insights into the role of specific proteins of the dPGS corona in cellular uptake and thereby to better understand possible uptake mechanisms. Furthermore, these data indicate that clathrin-mediated endocytosis is the major though not the only cellular uptake pathway for cellular uptake. It may be concluded that most likely different pathways involving different receptors exist for mediating cellular uptake of dPGS NPs.
Acknowledgments
The authors thank their institutions for supporting this project. The authors also would like to thank Sabine Reimann (FU) for providing materials. In addition, the Helmholtz Virtual Institute on Functional Biomaterials should be acknowledged for financial support.
Disclosure
The authors have no affiliation or financial involvement with any organization or entity with a financial interest. This paper was presented at the 2nd NanoSafety Forum for Young Scientists in Visby, Gotland, Sweden in September 2016 as an oral presentation with interim findings. The authors report no other conflicts of interest in this work.
References
Torchilin VP. Targeted pharmaceutical nanocarriers for cancer therapy and imaging. AAPS J. 2007;9(2):E128–E147. | ||
Arachchige MC, Reshetnyak YK, Andreev OA. Advanced targeted nanomedicine. J Biotechnol. 2015;202:88–97. | ||
Petre CE, Dittmer DP. Liposomal daunorubicin as treatment for Kaposi’s sarcoma. Int J Nanomedicine. 2007;2(3):277–288. | ||
Caminade A-M, Turrin C-O. Dendrimers for drug delivery. J Mater Chem B. 2014;2(26):4055–4066. | ||
Madaan K, Kumar S, Poonia N, Lather V, Pandita D. Dendrimers in drug delivery and targeting: drug-dendrimer interactions and toxicity issues. J Pharm Bioallied Sci. 2014;6(3):139–150. | ||
Owens DEr, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307(1):93–102. | ||
Oh N, Park JH. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int J Nanomedicine. 2014;9(Suppl 1):51–63. | ||
Jones SW, Roberts RA, Robbins GR, et al. Nanoparticle clearance is governed by Th1/Th2 immunity and strain background. J Clin Invest. 2013;123(7):3061–3073. | ||
Li SD, Huang L. Stealth nanoparticles: high density but sheddable PEG is a key for tumor targeting. J Control Release. 2010;145(3):178–181. | ||
Allard-Vannier E, Cohen-Jonathan S, Gautier J, et al. Pegylated magnetic nanocarriers for doxorubicin delivery: a quantitative determination of stealthiness in vitro and in vivo. Eur J Pharm Biopharm. 2012;81(3):498–505. | ||
Kreuter J, Ramge P, Petrov V, et al. Direct evidence that polysorbate-80-coated poly(butylcyanoacrylate) nanoparticles deliver drugs to the CNS via specific mechanisms requiring prior binding of drug to the nanoparticles. Pharm Res. 2003;20(3):409–416. | ||
Kurakhmaeva KB, Djindjikhashvili IA, Petrov VE, et al. Brain targeting of nerve growth factor using poly(butyl cyanoacrylate) nanoparticles. J Drug Target. 2009;17(8):564–574. | ||
Lin Y, Pan Y, Shi Y, Huang X, Jia N, Jiang JY. Delivery of large molecules via poly(butyl cyanoacrylate) nanoparticles into the injured rat brain. Nanotechnology. 2012;23(16):165101. | ||
Kreuter J. Nanoparticulate systems for brain delivery of drugs. Adv Drug Deliv Rev. 2001;47(1):65–81. | ||
Mulik RS, Monkkonen J, Juvonen RO, Mahadik KR, Paradkar AR. ApoE3 mediated polymeric nanoparticles containing curcumin: apoptosis induced in vitro anticancer activity against neuroblastoma cells. Int J Pharm. 2012;437(1–2):29–41. | ||
Jo DH, Kim JH, Lee TG, Kim JH. Size, surface charge, and shape determine therapeutic effects of nanoparticles on brain and retinal diseases. Nanomedicine. 2015;11(7):1603–1611. | ||
Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. What the cell “sees” in bionanoscience. J Am Chem Soc. 2010;132(16):5761–5768. | ||
Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3(1–2):40–47. | ||
Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Protein-nanoparticle interactions: opportunities and challenges. Chem Rev. 2011;111(9):5610–5637. | ||
Monopoli MP, Walczyk D, Campbell A, et al. Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J Am Chem Soc. 2011;133(8):2525–2534. | ||
Zanganeh S, Spitler R, Erfanzadeh M, Alkilany AM, Mahmoudi M. Protein corona: Opportunities and challenges. Int J Biochem Cell Biol. 2016;75:143–147. | ||
Cheng X, Tian X, Wu A, et al. Protein corona influences cellular uptake of gold nanoparticles by phagocytic and nonphagocytic cells in a size-dependent manner. ACS Appl Mater Interfaces. 2015;7(37):20568–20575. | ||
Ritz S, Schottler S, Kotman N, et al. Protein corona of nanoparticles: distinct proteins regulate the cellular uptake. Biomacromolecules. 2015;16(4):1311–1321. | ||
Salvati A, Pitek AS, Monopoli MP, et al. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nature Nanotechnol. 2013;8(2):137–143. | ||
Mirshafiee V, Mahmoudi M, Lou K, Cheng J, Kraft ML. Protein corona significantly reduces active targeting yield. Chem Commun. 2013;49(25):2557–2559. | ||
Guo Y, Wang L, Lv P, Zhang P. Transferrin-conjugated doxorubicin-loaded lipid-coated nanoparticles for the targeting and therapy of lung cancer. Oncol Lett. 2015;9(3):1065–1072. | ||
Kainthan RK, Janzen J, Levin E, Devine DV, Brooks DE. Biocompatibility testing of branched and linear polyglycidol. Biomacromolecules. 2006;7(3):703–709. | ||
Dernedde J, Rausch A, Weinhart M, et al. Dendritic polyglycerol sulfates as multivalent inhibitors of inflammation. Proc Natl Acad Sci U S A. 2010;107(46):19679–19684. | ||
Khandare J, Calderon M, Dagia NM, Haag R. Multifunctional dendritic polymers in nanomedicine: opportunities and challenges. Chem Soc Rev. 2012;41(7):2824–2848. | ||
Weinhart M, Groger D, Enders S, et al. The role of dimension in multivalent binding events: structure-activity relationship of dendritic polyglycerol sulfate binding to L-selectin in correlation with size and surface charge density. Macromol Biosci. 2011;11(8):1088–1098. | ||
Weinhart M, Groger D, Enders S, Dernedde J, Haag R. Synthesis of dendritic polyglycerol anions and their efficiency toward L-selectin inhibition. Biomacromolecules. 2011;12(7):2502–2511. | ||
Vonnemann J, Liese S, Kuehne C, et al. Size dependence of steric shielding and multivalency effects for globular binding inhibitors. J Am Chem Soc. 2015;137(7):2572–2579. | ||
Lou X, Wang C, He L. Core-shell Au nanoparticle formation with DNA-polymer hybrid coatings using aqueous ATRP. Biomacromolecules. 2007;8(5):1385–1390. | ||
Ziegler C, Eychmüller A. Seeded growth synthesis of uniform gold nanoparticles with diameters of 15−300 nm. J Phys Chem C. 2011;115(11):4502–4506. | ||
Vonnemann J, Sieben C, Wolff C, et al. Virus inhibition induced by polyvalent nanoparticles of different sizes. Nanoscale. 2014;6(4):2353–2560. | ||
Groger D, Kerschnitzki M, Weinhart M, et al. Selectivity in bone targeting with multivalent dendritic polyanion dye conjugates. Adv Healthcare Mater. 2014;3(3):375–385. | ||
Rabilloud T, Strub J-M, Luche S, Girardet J, van Dorsselaer A, Lunardi J. Ruthenium II tris (bathophenanthroline disulfonate), a powerful fluorescent stain for detection of proteins in gel with minimal interference in subsequent mass spectrometry analysis. Proteome. 2000;1(1):1–14. | ||
Vonnemann J, Beziere N, Bottcher C, et al. Polyglycerolsulfate functionalized gold nanorods as optoacoustic signal nanoamplifiers for in vivo bioimaging of rheumatoid arthritis. Theranostics. 2014;4(6):629–641. | ||
Walkey CD, Chan WC. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem Soc Rev. 2012;41(7):2780–2799. | ||
Le Roy C, Wrana JL. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol. 2005;6(2):112–126. | ||
Wang LH, Rothberg KG, Anderson RG. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol. 1993;123(5):1107–1117. | ||
Roach P, Farrar D, Perry CC. Surface tailoring for controlled protein adsorption: effect of topography at the nanometer scale and chemistry. J Am Chem Soc. 2006;128(12):3939–3945. | ||
Shannahan JH, Lai X, Ke PC, Podila R, Brown JM, Witzmann FA. Silver nanoparticle protein corona composition in cell culture media. PLoS One. 2013;8(9):e74001. | ||
Sakulkhu U, Mahmoudi M, Maurizi L, Salaklang J, Hofmann H. Protein corona composition of superparamagnetic iron oxide nanoparticles with various physico-chemical properties and coatings. Sci Rep. 2014;4:5020. | ||
Tenzer S, Docter D, Rosfa S, et al. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS Nano. 2011;5(9):7155–7167. | ||
McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12(8):517–533. | ||
Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. Ligand binding to integrins. J Biol Chem. 2000;275(29):21785–21788. | ||
Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. | ||
Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011;3(3):a004994. | ||
Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin alpha2beta1. Cell. 2000;101(1):47–56. | ||
Arosio D, Casagrande C. Advancement in integrin facilitated drug delivery. Adv Drug Deliv Rev. 2016;97:111–143. | ||
Raj A, Saraf P, Javali NM, Li X, Jasti B. Binding and uptake of novel RGD micelles to the alphavbeta3 integrin receptor for targeted drug delivery. J Drug Target. 2014;22(6):518–527. | ||
Lahti M, Heino J, Kapyla J. Leukocyte integrins alphaLbeta2, alphaMbeta2 and alphaXbeta2 as collagen receptors – receptor activation and recognition of GFOGER motif. Int J Biochem Cell Biol. 2013;45(7):1204–1211. | ||
Horton MA. The alpha v beta 3 integrin “vitronectin receptor”. Int J Biochem Cell Biol. 1997;29(5):721–725. | ||
Pytela R, Pierschbacher MD, Ruoslahti E. A 125/115-kDa cell surface receptor specific for vitronectin interacts with the arginine-glycine-aspartic acid adhesion sequence derived from fibronectin. Proc Natl Acad Sci U S A. 1985;82(17):5766–5770. | ||
Li R, Luo M, Ren M, et al. Vitronectin regulation of vascular endothelial growth factor-mediated angiogenesis. J Vasc Res. 2014;51(2):110–117. | ||
Tedja R, Lim M, Amal R, Marquis C. Effects of serum adsorption on cellular uptake profile and consequent impact of titanium dioxide nanoparticles on human lung cell lines. ACS Nano. 2012;6(5):4083–4093. | ||
Ritz S, Schöttler S, Kotman N, et al. Protein corona of nanoparticles: distinct proteins regulate cellular uptake. Biomacromolecules. 2015;16:1311–1321. |
Supplementary materials
  | Figure S1 Schematic illustration of the synthesis of rhodamine-B-labeled dendritic polyglycerol (dPGOH-RB). |
  | Figure S2 Schematic illustration of the synthesis of rhodamine-B-labeled dendritic polyglycerol sulfate (dPGS-RB). |
Reference
Groger D, Kerschnitzki M, Weinhart M, et al. Selectivity in bone targeting with multivalent dendritic polyanion dye conjugates. Adv Healthcare Mater. 2014;3(3):375–385. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.