Back to Journals » Clinical Ophthalmology » Volume 13
The Association Between Interleukin 1 Beta Promoter Polymorphisms And Keratoconus Incidence And Severity In An Egyptian Population
Authors Nabil KM , Elhady GM , Morsy H
Received 25 June 2019
Accepted for publication 27 August 2019
Published 14 November 2019 Volume 2019:13 Pages 2217—2223
DOI https://doi.org/10.2147/OPTH.S220723
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Karim Mahmoud Nabil,1 Ghada Mohamed Elhady,2 Heba Morsy2
1Department of Ophthalmology, Faculty of Medicine, University of Alexandria, Alexandria, Egypt; 2Department of Human Genetics, Medical Research Institute, University of Alexandria, Alexandria, Egypt
Correspondence: Karim Mahmoud Nabil
Department of Ophthalmology, Faculty of Medicine, University of Alexandria, Sultan Hussein Street, Khartoum Square, Alexandria 21523, Egypt
Tel +2034869545
Fax +2034255781
Email [email protected]
Purpose: In the present study, we investigated whether interleukin 1 beta (IL1B) promoter polymorphisms are associated with keratoconus in an Egyptian population and their association with disease severity.
Methods: A total of 95 Egyptian keratoconus patients and 126 Egyptian healthy controls were enrolled in the study. Two IL1B single nucleotide polymorphisms (SNPs) (rs1143627 and rs16944) were genotyped using Taqman real-time PCR to compare haplotype, genotype, and allele frequencies between cases and controls (primary outcome) and their association with disease severity (secondary outcome).
Results: Statistically significant association was observed for rs1143627 and rs16944; the T allele of rs1143627 and the G allele of rs16944 were associated with an increased risk of keratoconus (p < 0.001, odds ratio = 3.313, 4.770, respectively). The TT genotype of rs1143627 and the GG genotype of rs16944 were strongly associated with an increased risk of keratoconus (p < 0.001, odds ratio = 5.631, 11.478, respectively). The G allele of rs16944 was associated with an increased curvature of the flattest corneal meridian Kf in keratoconus (p = 0.041). The GG genotype of rs16944 was associated with an increased curvature of the flattest corneal meridian Kf, steepest corneal meridian Ks and average corneal curvature Kavg in keratoconus (p = 0.01, 0.046, 0.023, respectively).
Conclusion: IL1B is suspected to play a crucial role, both in development and severity of keratoconus in Egyptian population.
Keywords: interleukin 1 beta, promoter polymorphisms, keratoconus, Egypt
Introduction
Keratoconus (OMIM 148300) is a corneal pathology showing progressive noninflammatory corneal thinning, which can result in profound visual disability. Keratoconus prevalence rates show marked variation through studies, geographic areas and ethnic groups, falling in the range between 0.0002% and 2.34%.1 The definitive etiological mechanism of keratoconus remains unclear, but the pathogenesis is believed to be multifactorial, involving the interplay of variable genetic and environmental factors. Twin and family studies supply profound evidence of the genetic role in keratoconus development. Keratoconus prevalence among keratoconus cases relatives is higher than the general population,2,3 and the concordance rate for keratoconus is high in monozygotic twins.4
Linkage analysis allowed to identify the association between keratoconus and several genetic loci, with special emphasis on 20p11.21, harboring the visual system homeobox 1 (VSX1) gene. Although multiple studies recognized VSX1 mutations as a possible candidate gene for keratoconus in various ethnic groups,5–12 numerous studies found VSX1 mutations to be nonspecific for the disease,13–17 suggesting that VSX1 mutations may play a minor effect, with other genetic factors having a more powerful role in keratoconus development. Besides VSX1, multiple candidate genes were investigated, including secreted protein acidic and rich in cysteine abbreviation of the genes, superoxide dismutase 1 (SOD1),11,18 human leukocyte antigen (HLA),19 mitochondrial complex I genes,20 lipoxygenase,21 transforming growth factor, beta 1 (TGF-β1),22 and collagen type IV, alpha 3 and collagen type IV, alpha four.23 However, different population and ethnic groups' studies failed to identify associations with these candidate genes.11–13,24
Interleukin (IL)-1 is a proinflammatory cytokine inducing chemokines and cytokines production and playing a key role in the inflammatory processes. IL-1 contributes to multiple cellular activities, including cell differentiation, proliferation, and cellular apoptosis. IL-1 comprises IL-1 receptor antagonist (IL-1Ra) and two proinflammatory cytokines (IL-1α and IL-1β), encoded by IL1RN, IL1A, and IL1B, respectively, comprising a cluster that spans 360 kb on chromosome 2q14. Recently, Takenori et al studied the correlation between IL1A and IL1B polymorphisms with keratoconus in a Japanese population and stated that IL1B promoter polymorphisms rs1143627 and rs16944 showed statistically significant association with keratoconus development risk.25 However, replication studies are lacking in groups of different ethnicities.
The aim of the current study was to explore whether IL1B promoter polymorphisms are associated with keratoconus in an Egyptian population and their correlation with disease severity.
Methods
The study adhered to the Declaration of Helsinki guidelines and was approved by the Ethics committees of the Faculty of Medicine and the Medical Research Institute, University of Alexandria. Details of the current study were highlighted to all participants and written informed consents were collected before the start of the study.
We enrolled 95 unrelated Egyptian keratoconus patients and 126 unrelated healthy Egyptian, age- and sex-matched controls without any control disease, at Department of Ophthalmology, Faculty of Medicine, University of Alexandria, Egypt. In case of contact lens wear, both cases and controls were instructed to discontinue their contact lenses at least 1 week before undergoing topography tests. Keratoconus diagnosis relied on biomicroscopic signs detected by slit lamp examination (BQ 900 slit lamp Haag‑Streit AG), such as stromal corneal thinning, Fleischer’s ring, Munson’s sign or striae of Vogt. Corneal topographic patterns ensuring keratoconus diagnosis, detected by Topographic Modeling System TMS‑5 (Tomey Corporation, Nagoya, Japan) and Allegro Oculyzer (Wavelight, GmbH, Erlangen, Germany), included corneal inferior steepening, corneal inferocentral thinning, or bowtie of asymmetric pattern and radial axes skewing. The controls were sex- and age-matched patients. Multiple pathologies have been linked to IL1B polymorphisms including Alzheimer's disease, osteomyelitis, end-stage diabetic kidney failure, stomach carcinoma, inflammatory bowel syndrome, and Parkinsonian disease. These disease states were excluded from our cases and controls.
Genomic DNA was extracted from peripheral blood samples using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) per manufacturer's protocol. Methodology was undertaken under standard conditions to avoid DNA quality alteration. Two SNPs reportedly linked with keratoconus, 1143627 and rs16944, in IL1B were investigated.25 SNPs were genotyped applying TaqMan 5′ exonuclease assay with TaqMan primer-probe validated sets provided by Thermo Scientific (USA). Quantitative polymerase chain reaction (PCR) was undertaken in a reaction mixture of 20 μL sum volume containing 1X TaqMan Genotyping PCR Master Mix (Thermo Scientific, USA), 1 μL TaqMan genotyping assay mix (20✕) and 5 ng genomic DNA. PCR conditions were: 95°C for 10 mins, then 40 denaturation cycles at 92°C for 15 s and annealing/extension at 60°C for 1 min. The probe fluorescence signal detection was performed by Eco illumine real-time PCR (Illumina, USA).
Statistical Analysis
The frequencies of alleles and genotypes were counted directly. Hardy–Weinberg equilibrium (HWE), differences in allele and genotype frequencies between cases and controls (primary outcome) and genotype–phenotype association in patients (secondary outcome) were assessed with Chi-square, Mann–Whitney and Kruskal–Wallis tests.
Results
The present study involved 95 unrelated Egyptian patients with keratoconus and 126 unrelated healthy Egyptian age- and sex-matched controls as shown in Table 1.
 |
Table 1 Demographic And Clinical Criteria Of Keratoconus Cases And Healthy Controls |
Hardy–Weinberg equilibrium and allele and genotype frequencies: The two SNPs were in HWE in patient and control groups. The T allele of rs1143627 and the G allele of rs16944 were associated with an increased risk of keratoconus (p < 0.001, odds ratio = 3.313, 4.770, respectively). The TT genotype of rs1143627 and the GG genotype of rs16944 showed strong association with increased keratoconus risk (p < 0.001, odds ratio = 5.631, 11.478, respectively) as shown in Table 2.
Genotype–phenotype correlation: The G allele of rs16944 was associated with an increase of curvature of the flattest corneal meridian Kf in keratoconus (p = 0.041) as shown in Table 3.
 |
Table 3 Clinical Association Of Allele Frequencies Of The Studied SNPs In Keratoconus Cases, Showing Increased Curvature Of The Flattest Corneal Meridian Associated With The G Allele |
The GG genotype of rs16944 was associated with an increase of curvature of the flattest corneal meridian (Kf), steepest corneal meridian (Ks) and average corneal curvature (Kavg) in keratoconus (p = 0.01, 0.046, 0.023, respectively) as shown in Table 4 and Figure 1.
 |
Table 4 Clinical Association Of Genotype Frequencies Of The Studied SNPs In Keratoconus Cases, Showing Increased Corneal Curvature Associated With GG Genotype |
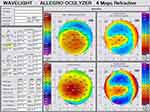 |
Figure 1 Scheimpflug tomography of a keratoconus patient (Krumeich class 4), with GG rs16944 genotype. |
Discussion
The purpose of the current study was to investigate whether IL1B polymorphisms affect the development and severity of keratoconus in an Egyptian population. Therefore, two promoter region polymorphisms were genotyped, outlining a statistically significant association between IL1B promoter polymorphism rs1143627 and rs16944 and keratoconus in Egyptian population, suggesting that the IL1B promoter polymorphisms play a crucial role in the development of keratoconus in Egyptian patients.
The SNPs rs1143627 and rs16944 lie at IL1B promoter region. Since promoter sequences represent possible polymorphism sources altering genetic expression, they could profoundly contribute in IL1B genetic expression, playing a confound role in keratoconus development. This theory is assisted by research demonstrating that IL1B promoter polymorphisms can influence IL1B expression.26 The IL1-B protein was detected in human corneal endothelial, stromal fibroblast, and epithelial cells,27 and this protein expression has been proven to be enhanced in keratoconus corneas in comparison to normal corneas.28 In keratoconus corneas, keratocyte apoptosis has been speculated to play a role in the corneal thinning process,29 suggesting that the enhanced IL1B expression induced by the promoter polymorphism can result in IL1-B protein overexpression, leading to increased apoptotic activity in corneal tissue documented in keratoconus patients. Considering the noninflammatory nature in definition of keratoconus, these findings revealed that the inflammation has a role in keratoconus induction and progression, thus the definition goes under question suggesting possible revision for keratoconus definition.
This hypothesis supports the findings highlighted in the present study, where the G allele of rs16944 was linked with an increase of curvature of the flattest corneal meridian (Kf) in keratoconus (p = 0.041) and the GG genotype was associated with an increased curvature of the flattest corneal meridian (Kf), steepest corneal meridian (Ks) and average corneal curvature (Kavg) in keratoconus (p = 0.01, 0.046, 0.023, respectively).
Takenori et al studied the correlation between IL1A and IL1B polymorphisms with keratoconus in a Japanese population and stated that IL1B promoter polymorphisms rs1143627 and rs16944 showed statistically significant association with keratoconus development risk.25 The T allele of rs1143627 was associated with increased keratoconus risk. The C allele of rs16944 in the IL1B promoter region had a 1.33-fold increased keratoconus risk. The TT genotype of rs1143627 was weakly associated with an increased risk of keratoconus. However, no significant differences were found in the allele and genotype frequencies between the cases and controls for rs2071376 in IL1A. Regarding haplotypic diversity, the haplotype created by the T allele of rs1143627 and C allele of rs16944 was associated with a 1.72-fold increased risk of keratoconus.25
Kim SH et al investigated the genetic association between unrelated Korean keratoconus patients and IL1A and IL1B. The C allele of rs16944 and the T allele of rs1143627 were associated with a significantly increased risk of keratoconus in Korean patients.30 However, replication studies are lacking in groups of different ethnicities.
Since keratoconus is genetically heterogeneous, detecting susceptibility genes can supply valuable information considering the etiological mechanism of this imperfectly understood pathology. In the current work, IL1B promoter polymorphism, rs1143627 and rs16944, showed statistically significant association with keratoconus in the Egyptian population. These findings are in concordance with previous studies in Korean and Japanese populations, highlighting that the IL1B promoter polymorphism plays a crucial role in keratoconus development. To our knowledge, this is the first study to demonstrate IL1B promoter polymorphism crucial role, both in development and severity of keratoconus.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gordon-Shaag A, Millodot M, Shneor E. The epidemiology and etiology of keratoconus. Int J Keratoco Ectatic Corneal Dis. 2012;1:7–15. doi:10.5005/jp-journals-10025-1002
2. Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi:10.1016/S0039-6257(97)00119-7
3. Wang Y, Rabinowitz YS, Rotter JI, Yang H. Genetic epidemiological study of keratoconus: evidence for major gene determination. Am J Med Genet. 2000;93:403–409. doi:10.1002/(ISSN)1096-8628
4. Tuft SJ, Hassan H, George S, Frazer DG, Willoughby CE, Liskova P. Keratoconus in 18 pairs of twins. Acta Ophthalmol (Copenh). 2012;90:e4826. doi:10.1111/aos.2012.90.issue-6
5. Nielsen K, Hjortdal J, Pihlmann M, Corydon TJ. Update on the keratoconus genetics. Acta Ophthalmol (Copenh). 2013;91:106–113. doi:10.1111/aos.2013.91.issue-2
6. Eran P, Almogit A, David Z, et al. The D144E substitution in the VSX1 gene: a non-pathogenic variant or a disease causing mutation? Ophthalmic Genet. 2008;29:53–59. doi:10.1080/13816810802008242
7. Mok JW, Baek SJ, Joo CK. VSX1 gene variants are associated with keratoconus in unrelated Korean patients. J Hum Genet. 2008;53:842–849. doi:10.1007/s10038-008-0319-6
8. Paliwal P, Singh A, Tandon R, Titiyal JS, Sharma A. A novel VSX1 mutation identified in an individual with keratoconus in India. Mol Vis. 2009;15:2475–2479.
9. Dash DP, George S, O’Prey D, et al. Mutational screening of VSX1 in keratoconus patients from the European population. Eye (Lond). 2010;24:1085–1092. doi:10.1038/eye.2009.217
10. Paliwal P, Tandon R, Dube D, Kaur P, Sharma A. Familial segregation of a VSX1 mutation adds a new dimension to its role in the causation of keratoconus. Mol Vis. 2011;17:481–485.
11. De Bonis P, Laborante A, Pizzicoli C, et al. Mutational screening of VSX1, SPARC, SOD1, LOX, and TIMP3 in keratoconus. Mol Vis. 2011;17:2482–2494.
12. Saee-Rad S, Hashemi H, Miraftab M, et al. Mutation analysis of VSX1 and SOD1 in Iranian patients with keratoconus. Mol Vis. 2011;17:3128–3136.
13. Stabuc-Silih M, Strazisar M, Hawlina M, Glavac D. Absence of pathogenic mutations in VSX1 and SOD1 genes in patients with keratoconus. Cornea. 2010;29:172–176. doi:10.1097/ICO.0b013e3181aebf7a
14. Stabuc-Silih M, Strazisar M, Ravnik-Glavac M, Hawlina M, Glavac D. Genetics and clinical characteristics of keratoconus. Acta Dermatovenerol Alp Panonica Adriat. 2010;19:3–10.
15. Tanwar M, Kumar M, Nayak B, et al. VSX1 gene analysis in keratoconus. Mol Vis. 2010;16:2395–2401.
16. Abu-Amero KK, Kalantan H, Al-Muammar AM. Analysis of the VSX1 gene in keratoconus patients from Saudi Arabia. Mol Vis. 2011;17:667–672.
17. Jeoung JW, Kim MK, Park SS, et al. VSX1 gene and keratoconus: genetic analysis in Korean patients. Cornea. 2012;31:746–750. doi:10.1097/ICO.0b013e3181e16dd0
18. Udar N, Atilano SR, Brown DJ, et al. SOD1: a candidate gene for keratoconus. Invest Ophthalmol Vis Sci. 2006;47:3345–3351. doi:10.1167/iovs.05-1500
19. Adachi W, Mitsuishi Y, Terai K, et al. The association of HLA with young-onset keratoconus in Japan. Am J Ophthalmol. 2002;133:557–559. doi:10.1016/S0002-9394(01)01368-X
20. Pathak D, Nayak B, Singh M, et al. Mitochondrial complex 1 gene analysis in keratoconus. Mol Vis. 2011;17:1514–1525.
21. Bykhovskaya Y, Li X, Epifantseva I, et al. Variation in the lysyl oxidase (LOX) gene is associated with keratoconus in family-based and case-control studies. Invest Ophthalmol Vis Sci. 2012;53:4152–4157. doi:10.1167/iovs.11-9268
22. Guan T, Liu C, Ma Z, Ding S. The point mutation and polymorphism in keratoconus candidate gene TGFBI in Chinese population. Gene. 2012;503:137–139. doi:10.1016/j.gene.2012.04.061
23. Stabuc-Silih M, Ravnik-Glavac M, Glavac D, Hawlina M, Strazisar M. Polymorphisms in COL4A3 and COL4A4 genes associated with keratoconus. Mol Vis. 2009;15:2848–2860.
24. Udar N, Kenney MC, Chalukya M, et al. Keratoconus – no association with the transforming growth factor beta-induced gene in a cohort of American patients. Cornea. 2004;23:13–17. doi:10.1097/00003226-200401000-00003
25. Takenori M, Akira M, Takeshi T, et al. Interleukin 1 beta promoter polymorphism is associated with keratoconus in a Japanese population. Mol Vis. 2013;19:845–851.
26. Chen H, Wilkins LM, Aziz N, et al. Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum Mol Genet. 2006;15:519–529.
27. Weng J, Mohan RR, Li Q, Wilson SE. IL-1 upregulates keratinocyte growth factor and hepatocyte growth factor mRNA and protein production by cultured stromal fibroblast cells: interleukin-1 beta expression in the cornea. Cornea. 1997;16:465–471. doi:10.1097/00003226-199707000-00015
28. Zhou L, Yue BY, Twining SS, Sugar J, Feder RS. Expression of wound healing and stress-related proteins in keratoconus corneas. Curr Eye Res. 1996;15:1124–1131. doi:10.3109/02713689608995144
29. Kaldawy RM, Wagner J, Ching S, Seigel GM. Evidence of apoptotic cell death in keratoconus. Cornea. 2002;21:206–209. doi:10.1097/00003226-200203000-00017
30. Kim SH, Mok JW, Kim HS, Joo CK. Association of −31T>C and −511 C>T polymorphisms in the interleukin 1 beta (IL1B) promoter in Korean keratoconus patients. Mol Vis. 2008;14:2109–2116.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

