Back to Journals » International Journal of Nanomedicine » Volume 11
Nanoparticle-based strategy for personalized B-cell lymphoma therapy
Authors Martucci NM, Migliaccio N, Ruggiero I, Albano F, Calì G, Romano S, Terracciano M, Rea I, Arcari P, Lamberti A
Received 1 August 2016
Accepted for publication 29 September 2016
Published 16 November 2016 Volume 2016:11 Pages 6089—6101
DOI https://doi.org/10.2147/IJN.S118661
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Nicola M Martucci,1,* Nunzia Migliaccio,1,* Immacolata Ruggiero,1,* Francesco Albano,2 Gaetano Calì,3 Simona Romano,1 Monica Terracciano,4 Ilaria Rea,4 Paolo Arcari,1 Annalisa Lamberti1
1Department of Molecular Medicine and Medical Biotechnology, University Federico II of Naples, Naples, 2Department of Experimental and Clinical Medicine, University of Catanzaro “Magna Graecia”, Catanzaro, 3Institute of Endocrinology and Molecular Oncology, 4Institute for Microelectronics and Microsystems, National Research Council, Naples, Italy
*These authors contributed equally to this work
Abstract: B-cell lymphoma is associated with incomplete response to treatment, and the development of effective strategies targeting this disease remains challenging. A new personalized B-cell lymphoma therapy, based on a site-specific receptor-mediated drug delivery system, was developed in this study. Specifically, natural silica-based nanoparticles (diatomite) were modified to actively target the antiapoptotic factor B-cell lymphoma/leukemia 2 (Bcl2) with small interfering RNA (siRNA). An idiotype-specific peptide (Id-peptide) specifically recognized by the hypervariable region of surface immunoglobulin B-cell receptor was exploited as a homing device to ensure specific targeting of lymphoma cells. Specific nanoparticle uptake, driven by the Id-peptide, was evaluated by flow cytometry and confocal microscopy and was increased by approximately threefold in target cells compared with nonspecific myeloma cells and when a random control peptide was used instead of Id-peptide. The specific internalization efficiency was increased by fourfold when siRNA was also added to the modified nanoparticles. The modified diatomite particles were not cytotoxic and their effectiveness in downregulation of gene expression was explored using siRNA targeting Bcl2 and evaluated by quantitative real-time polymerase chain reaction and Western blot analyses. The resulting gene silencing observed is of significant biological importance and opens new possibilities for the personalized treatment of lymphomas.
Keywords: diatomite, active targeting, Bcl2, small interfering RNA, personalized therapy
Introduction
Cancer is one of the leading causes of death worldwide. Currently, cancer treatments, including radiations and chemotherapeutics, present a range of critical drawbacks, including low treatment efficiency and serious side effects, due to non-specific impairment of normal tissues and cells. Targeted drug delivery and release to tumor sites is believed to enhance therapeutic efficiency and limit side effects. Subsequent to the discovery of the first drug delivery system based on liposomes in the 1960s, nanomedicine has made significant advances in the pharmaceutical and biotechnology fields.1–10
Gene therapy using small interfering RNA (siRNA) to downregulate the expression of cancer-associated genes represents a promising tool in cancer treatment.11–13 However, naked siRNA is susceptible to enzymatic degradation and is thus not able to survive in biological fluids for extended periods of time. Moreover, the ability of siRNA molecules to penetrate the cell membrane is limited by their strong negative charge and high molecular weight. Thus, successful RNA interference therapy is considerably reliant on the development of efficient gene delivery systems.
A wide range of nanomaterials have been explored for siRNA delivery, and, among them, inorganic systems based on silica are the most attractive bio-interfaces, since they couple interesting properties with reduced toxicity.14–16 In addition, silica is one of the few inorganic materials recognized as “safe” by the US Food and Drug Administration. In this study, diatomite-based nanoparticles were employed as a delivery platform to specifically vehiculate siRNA to lymphoma cancer cells.17–20
B-cell lymphoma is a clonal expansion of neoplastic cells that can have a fatal outcome. Tumorigenic B-cell lymphomas are sensitive to conventional anticancer treatments, including chemotherapy, radiation, and corticosteroids; however, the disease is associated with incomplete response to clinical treatment, resulting in a minimal residual disease, where some undetected cancerous cells supply the cancer cell reservoir in vivo.21 Despite considerable efforts in lymphoma therapy, the development of an effective strategy to target the anatomic homing of tumorigenic B cells still remains challenging.
B-cell lymphoma/leukemia 2 (Bcl2) is an apoptosis-regulating protein, and its overexpression is associated with an abnormal apoptotic pathway conferring a protective effect toward death signals on malignant cells. Impairment of Bcl2 has been identified as contributing to the onset of a large number of cancers, including lymphoma.22 Bcl2 overexpression results in chemotherapy-resistant disease, an aggressive clinical course and poor survival in patients with B-cell lymphoproliferative disorders.23–25 A successfully developed Bcl2-targeted drug has the potential to enhance cell killing, when used in combination with traditional cytotoxic agents.26 Attempts to downregulate Bcl2 by antisense therapy have been pursued as an antitumor strategy.27,28
In this study, an active targeting strategy for Bcl2 siRNA delivery was developed to specifically target extremely aggressive murine A20 lymphoma cells, using as ligand an idiotype-specific peptide (Id-peptide) endowed with high affinity toward B-cell receptor (BCR, homing device).29,30 The idiotype determinants of the immunoglobulin B-cell receptor (Ig-BCR) of B-cell malignancies are unique for a given clonal population and function as patient-specific tumor antigen that may be exploited for active therapeutic targeting in personalized cancer therapy.30 The choice of Id-peptides was based on the following properties: 1) binding to tumor target cells with high specificity and sensitivity, both in vitro and in vivo, and 2) internalization into target tumor cells by BCR-mediated endocytosis, which overcomes drug resistance.29–31 Based on these considerations, diatomite nanoparticles (DNPs) conjugated with an Id-peptide and loaded with siRNA directed against Bcl2, highly expressed in most B-cell lymphomas, were explored as a ligand-mediated delivery platform.
Materials and methods
Reagents
Calcined diatomite was obtained from Deref Spa (Genova, Italy); 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), (3-aminopropyl)triethoxysilane (APTES), N-hydroxysuccinimide (NHS), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), MES buffer, propidium iodide (PI) and H2SO4 were purchased from Sigma-Aldrich (St Louis, MO, USA). Phosphate-buffered saline (PBS) was purchased from Gibco (Grand Island, NY, USA). ON-TARGET plus smart pool mouse Bcl2 (siRNA Bcl2), ON-TARGET plus control pool nontargeting pool (siRNA scramble, siRNASCR) and fluorescent (Dy547) siGLO RISC-Free siRNA (siRNA*) were from Thermo Fisher Scientific (Waltham, MA, USA). Synthesis of poly D-Arg peptide (sequence RRRRRRRRC) was performed by ProteoGenix (Schiltigheim, France). HCl was purchased from ROMIL (Cambridge, UK). A20 target-specific peptide (pA2036) (sequence EYVNCDNLVGNCVI), pA2036-9R [sequence EYVNCDNLVGNCVI-(L-Arg)8(D-Arg)], pA2036-fluorescein isothiocyanate (FITC) (pA2036*) random peptide (pRND) (sequence SSAYGSCKGPCSSGVHSI), pRND-9R [sequence SSAYGSCKGPCSSGVHSI(L-Arg)8(D-Arg)], and pRND-FITC (pRND*) were synthetized by CASLO Laboratory, Lyngby, Denmark. Cell-Titer GLO was purchased from Promega (Madison, WI, USA).
Preparation of modified DNPs
The diatomite nanopowder was obtained from calcined diatomite as already reported and purified with hot strong acids to remove organic and inorganic contaminants.17 After purification treatments, DNPs were aminosilanized by 1 h incubation at room temperature (RT) with a 5% (v/v) APTES solution in absolute ethanol to form a thin silane film on their surface.32,33 The amino groups of silanized DNPs were covalently conjugated to the carboxyl groups of peptides alone and/or complexed with siRNA via NHS/EDC chemistry; briefly, DNPs (~1.0 mg) were incubated with 150 μM peptide in the presence of EDC (20 mM)/NHS (12 mM) solution in 10 mM MES buffer pH 5.5 to promote the reaction (ON at RT, under stirring). Peptide/siRNA complex was previously obtained by mixing 150 μM peptide with 7.5 μM siRNA (molar ratio 20:1) for 2 h at RT in PBS pH 7.4. After the reaction, diatomite suspension was centrifuged and the pellet was collected and washed twice with PBS. The siRNA loaded was ~0.5 pmol per 1.0 μg of diatomite, as determined by UV reading at 260 nm.
Size and morphology of DNPs
Morphology of bare DNPs was investigated by atomic force microscopy (AFM). A XE-100 AFM (Park Systems) was used for the imaging of DNPs deposited on silicon substrate. Surface imaging was obtained in noncontact mode using silicon/aluminum-coated cantilevers (PPP-NCHR 10 M; Park Systems) 125 μm long with a resonance frequency of 200–400 kHz and a nominal force constant of 42 N/m. The scan frequency was typically 1 Hz per line.
The size and surface charge measurements of DNPs dispersed in water (pH 7) were investigated by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments, UK) equipped with a He–Ne laser (633 nm, fixed scattering angle of 173°C, 25°C).
Cells and reagents
A20 is a murine cell line derived from a spontaneously arising tumor in an aged BALB/c mouse. It pathologically mimics the characteristics of human diffuse large B-cell lymphoma.29,34 The 5T33 murine myeloma (5T33MM) cells were used as control. The cell lines were kindly provided by Prof C Palmieri (University of Catanzaro, Italy). The cell lines were grown in suspension culture with Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco), supplemented with 10% fetal bovine serum (FBS; Gibco), 50 units/mL penicillin, 50 μg/mL streptomycin and 2 mM L-glutamine at 37°C in a 5% CO2 atmosphere. Human peripheral blood mononuclear cells (PBMCs) from healthy donors were obtained from heparinized peripheral blood samples by centrifugation (400× g for 30 min) over a Ficoll-Hipaque (Lonza, MD, USA) density gradient. Cells were resuspended in RPMI 1640 medium supplemented with 10% FBS.
Cytofluorimetric analysis
Cell lines (2.0×106) were incubated with pA2036- or pRND FITC-functionalized DNPs (DNPs•pA2036*/pRND*) at 50 μg/mL for 2 h at 37°C. After three washes in PBS, cells were divided into two aliquots; the first aliquot was resuspended in PBS and the other aliquot was permeabilized using 2% paraformaldehyde at RT for 30 min, washed twice with PBS and incubated with 0.1% Triton X-100 in PBS on ice for 1–2 min. After washing, the cells were resuspended in PBS, and both aliquots were treated with 0.002% Trypan blue solution (Sigma-Aldrich) for 10 min to quench extracellular fluorescence and analyzed with a Becton Dickinson FACScan flow cytometer. The same procedure was performed on A20 cells treated with pA2036-9R•siRNA*(DNPs•pA2036-9R•siRNA*) or on A20 cells and PBMCs treated with pA2036- and polyArg•siRNA*-functionalized DNPs (DNPs•pA2036•polyArg•siRNA*) with the respective negative controls. All measurements were carried out in triplicate, in three independent experiments.
Confocal microscopy
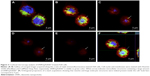
A20 cells (2.0×106) were treated with DNPs•pA2036* (50 μg/mL) for 30, 60, and 120 min. After washing with PBS, the cells were incubated with 0.5% CellMask Deep Red (Molecular Probes, Thermo Fisher Scientific) in Live Cell Imaging Solution according to the manufacturer’s instructions. Successively, cells were fixed with 2% paraformaldehyde at RT for 10 min, washed twice with PBS and, 5 min before confocal analysis, NucBlue (Molecular Probes) was added following manufacture’s instructions. Cells were then spotted on microscope slides and analyzed. Experiments were carried out on an inverted and motorized microscope (Axio Observer Z.1) equipped with a 63×/1.4 Plan-Apochromat objective. The attached laser-scanning unit (LSM 700 4× pigtailed laser 405-488-555-639; Zeiss, Jena, Germany) enabled confocal imaging. For excitation, 405, 488, and 639 nm lasers were used. Fluorescence emission was revealed by Main Dichroic Beam Splitter and Variable Secondary Dichroic Beam Splitter. Triple staining fluorescence images were acquired separately using ZEN 2012 software in the blue, green, and infrared channels at a resolution of 1,024×1,024 pixels, with the confocal pinhole set to one Airy unit and then saved in TIFF format.
Cell viability assays
For MTT and Cell-Titer GLO assays, cells were seeded into 96-well microtiter plates (BD Falcon, USA) at the density of 10×103 cells/well, whereas for the PI assay, cells were seeded into 48-well microtiter plates at the density of 50×103 cells/well. After 24 h, the cells were incubated with DNPs at increasing concentrations (50, 100, 150, and 200 μg/mL) in triplicate. The assays were performed after 24 h and 48 h of incubation, according to the manufacture’s instructions. Briefly, for MTT assay, 10 μL of a 5 mg/mL MTT solution in RPMI 1640 medium, no phenol red (Sigma-Aldrich), was added to each well. The plates were then incubated under cell culture conditions for 3 h. Subsequently, 100 μL of MTT solvent (HCl 0.1 N in isopropanol) was added to each well and incubated for 1 h at RT with stirring to dissolve the formazan crystals. Absorbance of each sample was detected by Microplate Reader 680 (Bio-Rad) at 570 nm. For Cell-Titer GLO, 100 μL of Cell-Titer GLO reagent was added to 100 μL of medium containing cells and incubated at RT for 10 min to stabilize luminescent signal. Luminescence was recorded for 0.25 s per well by Multilabel Reader (PerkinElmer, MA, USA). For PI analysis, cells were washed in PBS and resuspended in 200 μL of a solution containing 0.1% sodium citrate, 0.1% Triton X-100, and 50 μg/mL PI (Sigma-Aldrich). Following incubation at 4°C for 30 min in the dark, the cell DNA content was analyzed with a Becton Dickinson FACScan flow cytometer using the Lysis I program and the percentage of cells in the hypodiploid region was calculated. All measurements were carried out in triplicate, in three independent experiments.
Quantitative real-time polymerase chain reaction (qRT-PCR)
A20 cells (300×103) were incubated with DNPs•pA2036•polyArg•siRNABcl2 and DNPs•pRND•polyArg•siRNA (100 pmol siRNA/200 μg diatomite) for 24 h, collected and washed twice in PBS. Total RNA extraction and qRT-PCR were performed as previously described.35 Briefly, RNA aliquots (1 μg) were reverse transcribed using Random Examers and Superscript IV Reverse Transcriptase (Thermo Fisher Scientific), according to the manufacturer’s protocol. qRT-PCR was carried out with the Quantstudio 7 Flex Real-Time PCR system (Thermo Fisher Scientific) and PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) under the following conditions: 50°C, 2 min; 95°C, 2 min; (95°C, 15 s; 60°C, 60 s) ×40. Primers were as follows: GAPDH, forward 5′-GTCAAGGCCGAGAATGGGAAGC-3′ and reverse 5′-AGAAGGGGCGGAGATGATGACC-3′ and Bcl2, forward 5′-TCGCAGAGATGTCCAGTCAGC and reverse 5′-CCGGTTCAGGTACTCAGTCATC. Bcl2 expression levels were calculated relative to GAPDH messenger RNA (mRNA) levels as endogenous control and expressed as LogΔCt[2−(Ct sample − Ct β-GAPDH)]. Standard deviation (SD) was expressed as % of Ct values of three independent experiments.
Western blot analysis
Cells were treated for 48 h as described earlier, collected, washed twice in PBS, and resuspended in 20–40 μL of radio immunoprecipitation assay (RIPA) buffer (50 mM Tris–HCl pH 7.4, 1% NP40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 μg/mL aprotinin, leupeptin, pepstatin, 1 mM Na3VO4, 1 mM NaF) for 30 min on ice and centrifuged at 14,000× g for 30 min at 4°C. Protein concentration was determined by a modified Bradford method, using the Bio-Rad protein assay and compared with bovine serum albumin standard curve.36 Cytosolic proteins (10 μg) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, electrotransferred to nitrocellulose and reacted with Bcl2 monoclonal antibody (Santa Cruz, TX, USA) and GAPDH monoclonal antibody (Cell Signaling, MA, USA). Blots were then developed using enhanced chemiluminescence detection reagents (SuperSignal West Pico, Pierce) and exposed to X-ray film. All films were scanned for densitometric analysis using ImageJ 1.41o software.
Statistical analysis
Results of the assays are expressed as mean ± SD of three independent experiments. Data are reported as average and SD. The statistical significance of differences among groups was evaluated using analysis of variance, using the software KaleidaGraph v 4.1. The significance was accepted at the confidence level of 95% (P<0.05).
Results
Bare nanoparticle characterization
In this study, DNPs were used to develop a drug delivery system for active targeting of B-cell lymphoma. The morphology of DNPs deposited on crystalline silicon was investigated by AFM. A representative AFM image and the corresponding height profile are presented in Figure 1A. The analysis highlights the presence of nanoparticles of irregular shape and a mean size of ~200 nm. Size values were confirmed by DLS measurements performed on DNPs dispersed in deionized water (pH 7). The results showed an intensity peak corresponding to a size population of 280±80 nm (poly dispersion index =0.1) (Figure 1B). The ζ-potential of the DNPs was −19±3 mV; the negative charge was most likely due to the presence of silanol groups (Si-OH) on the surface of nanoparticles after treatment with hot acids.37
BCR-mediated uptake of DNPs
After silanization, DNPs were conjugated, using EDC/NHS chemistry, to an Id-peptide (pA2036) specifically recognized by the Ig-BCR of A20 cells in mice engrafted with A20 lymphoma.38 pA2036 was isolated from random peptide libraries as the most effective at targeting the A20 Ig-BCR; random peptide (pRND)-conjugated DNPs were used as negative control.29
Particle size distributions determined by DLS after diatomite functionalization with pA2036 and pRND were 390±20 nm and 350±30 nm, respectively, indicating that no particle aggregation had occurred during functionalization. The pA2036- and pRND-conjugated DNPs exhibited ζ-potentials of −10±1 mV and −12±1 mV, respectively.
The uptake of the nanoparticles functionalized with fluorescent (green) pA2036 (DNPs•pA2036*) by A20 cells in vitro was investigated by flow cytometry (Figure 2). On the basis of previous investigations, A20 lymphoma cells were incubated with nanoparticles at 50 μg/mL for 2 h at 37°C.30,39 To exclude nonspecific binding of the nanoparticles to the outer surface of the plasma membrane, extracellular fluorescence was quenched using Trypan blue.40 The results of flow cytometric analysis indicated a fluorescent cell population of 95.5%±3% in non-permeabilized cells (Figure 2B) and 96%±4% in mildly permeabilized cells (Figure 2E). To verify that the uptake of nanoparticles was mediated via the Ig-BCR, the same experiment was performed using nanoparticles functionalized with a fluorescent nonspecific peptide (DNPs•pRND*); the resulting fluorescence was 36%±3% in non-permeabilized and 39%±4% in mildly permeabilized cells (Figure 2C and F). In this case, the limited uptake was not due to receptor-mediated endocytosis, but more likely to caveolae-dependent endocytosis, which occurs when anionic nanoparticles interact with positive sites on membrane proteins.41,42 To further confirm the specificity of pA2036-modified DNPs in targeting A20 cells, the nanoparticle uptake of A20 cells was compared with that of 5T33MM cells, a surface IgG-positive B-cell line unable to bind the pA2036 peptide (Figure 2H).29 The fluorescent cell population of mildly permeabilized 5T33MM cells treated with DNPs•pA2036* was approximately threefold lower than that of A20 cells (34%±3% vs 96%±4%). These data, together with that obtained using DNPs•pRND*, indicate that a portion of DNPs•pA2036* nanoparticles were nonspecifically internalized by the cells.
Cellular uptake of DNPs•pA2036* was also investigated by confocal fluorescence microscopy. Plasma membranes and nuclei of A20 cells were labeled with CellMask Deep Red and NucBlue, respectively, and analysis of cellular internalization was carried out using three different incubation times (30, 60, and 120 min). Figure 3 shows representative confocal microscopy images of cells treated with DNPs•pA2036* after 60 min of incubation, recorded at different focal depths along the z-axis. The endocytosed nanoparticles appear as punctate small vesicles distributed in the cytoplasm. This spot-like intracellular distribution confirms an endocytic mechanism for the uptake of DNPs within vesicular structures. An intracellular structure, containing a large amount of DNPs, was also visible.43 Figure 3F shows orthogonal projections of z-stack acquisition, illustrating the large DNP-containing area and the small vesicles detected inside the cells.
Targeted siRNA delivery
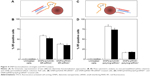
To further explore the potential of DNPs for application in targeted drug delivery, their ability to vehiculate siRNA into lymphoma cells was investigated. To this end, DNPs were conjugated with pA2036 engineered with a C-terminal stretch of basic amino acids (arginine residues) to promote the formation of siRNA–peptide complexes based on electrostatic interactions; addition of these amino acids also protects the nucleic acid from enzymatic degradation.17 This approach, illustrated in Figure 4A, was based on the concept that a single molecule could simultaneously load the siRNA and facilitate active targeting by specific Ig-BCR recognition. The mean particle size of DNPs modified in this way, determined by DLS, was 370±20 nm with a negative ζ-potential of −35±1 mV. The internalization of the complex formed by DNPs, pA2036-9R, and red fluorescent siRNA (DNPs•pA2036-9R•siRNA*) was evaluated by flow cytometry. The result indicated that 58.7%±2% of non-permeabilized and 52.2%±3% of mildly permeabilized cells internalized fluorescent DNPs (Figure 4B). The comparatively lower value of internalization relative to DNPs•pA2036* was probably due to the presence of the poly-arginine-siRNA complex at the C-terminus of pA2036 masking the specific recognition sequence of the A2036 peptide, thus reducing the efficiency of its interaction with Ig-BCR. Use of the pRND control (DNPs•pRND-9R•siRNA*; 350±40 nm, ζ-potential of −39±1 mV) resulted in fluorescence values of 37.8%±2.5% and 35%±3.5% in non-permeabilized and mildly permeabilized cells, respectively (Figure 4B). These data indicate that a limited amount of nonspecific cellular uptake occurred.
To enhance the internalization efficiency and attempt to minimize the influence of nonspecific DNPs uptake, optimization of the surface functionalization was performed. Nanoparticles were conjugated with both the original pA2036 sequence and a poly-Arg peptide complexed to the siRNA molecule (DNPs•pA2036•polyArg•siRNA*). This strategy, depicted in Figure 4C, allowed 1) immobilization of an unmodified pA2036 on the diatomite surface, which should be more efficiently recognized by the Ig-BCR compared to the molecule containing the poly-Arg stretch, and 2) immobilization of the pA2036 and siRNA on different sites of the particle, increasing the accessibility in targeting the Ig-BCR. Following these functionalization steps, the nanoparticles exhibited an increase in size to 400±40 nm and in ζ-potential to −11±1 mV. Figure 4D shows that, when A20 cells were treated with DNPs•pA2036•polyArg•siRNA*, the fluorescent cell populations were 82.4%±4% (non-permeabilized) and 73.8%±3.8% (mildly permeabilized). In contrast, when A20 cells were incubated with DNPs•pRND•polyArg•siRNA* (390±40 nm, ζ-potential =−12±1 mV), the fluorescent cell populations comprised 16.7%±1.5% and 18%±1% of non-permeabilized and mildly permeabilized cells, respectively.
The internalization efficiency of DNPs•pA2036•polyArg•siRNA* was also evaluated in a normal cell population (PBMCs). Compared to untreated cells, the fluorescent lymphocytic population was of 1.9%±0.7%, whereas monocytic population resulted in 9.3%±0.8% (Figure 5).
The size and surface charge of nanoparticles are represented in Figure 6. This result highlights the efficiency of the functionalization strategy, since a significant increase in ligand-mediated targeting was observed along with reduced non-specific internalization of DNPs•pRND•polyArg•siRNA*.
The observed differences in internalization efficiency were likely due to the variation in the size and ζ-potential of nanoparticles (Table 1). These data confirm that caveolae-dependent endocytosis, which occurs during passive targeting, is influenced by the size and surface charge of nanovectors.42
Cell viability
To assess A20 cell viability following DNP exposure, MTT, Cell-Titer GLO, and PI assays were performed.44 A20 cells were incubated in the presence of DNPs functionalized with pA2036 and with a poly-Arg complexed to a nontargeting siRNA (DNPs•pA2036•polyArg•siRNASCR) at four different concentrations (50, 100, 150, and 200 μg/mL) for 24, 48, and 72 h (Figure 7A–C, respectively). The choice of the incubation period reflects the fact that the uptake was relatively rapid and had already occurred after 2 h; therefore, toxicity would be expected to be observed within 24 h.45 In addition, the doses of tested DNPs were chosen as similar to those resulting in good tolerance when injected intravenously in nude mice.46,47 The results showed no differences in cell death in treated cells compared with untreated control cells. Cell viability was also analyzed by light microscopy after 48 h of incubation of A20 cells in the presence of DNPs, and no effects on cell morphology were observed (Insets 1 and 2, Figure 7B). The cytotoxic effect of modified DNPs was also measured by MTT assay using human PBMCs (Figure 7D). Together, these results demonstrate that no significant signs of cellular toxicity were observed as a result of nanoparticle treatment, even after an extended period of time.
Targeted gene silencing of Bcl2 using DNPs•pA2036•polyArg•siRNABcl2
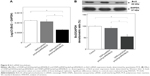
The downregulation of Bcl2 represents a promising potential therapeutic approach in cancer therapy, particularly in hematological malignancies, since its application can enhance the effects of chemotherapeutic agents.48–50 Previously reported pattern of siRNA release from diatomite demonstrated an initial phase with a burst trend within 12 h; therefore, Bcl2 gene silencing was determined by qRT-PCR 24 h after treatment of cells in the presence of 200 μg/mL DNPs functionalized with pA2036 and a poly-Arg peptide complexed to siRNA directed against Bcl2 mRNA (DNPs•pA2036•polyArg•siRNABcl2).17 As depicted in Figure 8A, DNPs•pA2036•polyArg•siRNABcl2 caused a 40.90%±4.5% decrease in the levels of Bcl2 mRNA, whereas only a slight reduction in Bcl2 mRNA expression levels was observed after treatment of cells with the control peptide, DNPs•pRND•polyArg•siRNABcl2 (9.09%±9.70%), relative to untreated cells (negative control). These results were compared to those obtained by transfecting A20 cells using a conventional method (lipofectamine); with respect to basal expression, siRNABcl2 (100 pmol) was able to down-regulate the mRNA level by ~10% after 24 h after transfection (data not shown), demonstrating that treatment of A20 cells with DNPs•pA2036•polyArg•siRNABcl2 resulted in more efficient silencing relative to the results achieved using lipofectamine. Thus, DNPs•pA2036•polyArg•siRNABcl2 could be efficiently and specifically delivered into A20 cells, resulting in significant gene silencing.
The knockdown results were confirmed by Western blot analysis (Figure 8B, upper panel). Compared with untreated cells, 48 h of treatment with DNPs•pA2036•polyArg•siRNABcl2 reduced Bcl2 expression by 45.10%±8.10%, whereas treatment with DNPs•pRND•polyArg•siRNABcl2 resulted in a decrease in Bcl2 protein expression of 7.10%±5.10%, as determined by densitometric analysis of the intensity of the bands (Figure 8B, lower panel). These results suggest that the reduction in Bcl2 expression was due to the function of siRNABcl2, successfully delivered into the cytoplasm by DNPs.
Discussion
A characteristic failure in the treatment of lymphoma is the inability of traditional therapies to reach the anatomic homing of tumorigenic B cells. Effective intracellular drug delivery to target cells is undoubtedly the most difficult hurdle to overcome in the development of a therapy.
The hypervariable regions of the surface Ig-BCR expressed by lymphoma cells are products of the rearrangement of the immunoglobulin genes and are unique to each clonal B-cell population. Hence, the idiotypic determinants of the Ig-BCR expressed by lymphoma cells represent a specific tumor antigen. Ig-BCR was explored in this study because of its potential for use in the development of active personalized targeting therapy.
The A2036 Id-peptide, previously selected by phage display libraries, was chosen since it does not bind to normal mouse PBMCs and bone marrow cells, other B-cell tumor cell lines (MC3, Raji, 5T33MM) or TT cell lines (HPBALL, Jurkat) but is recognized only by A20 lymphoma cells via their Ig-BCR.29 As a ligand of the idiotypic determinants of the Ig-BCR, the specificity of each Id-peptide is strongly restricted to a specific B-cell clone (normal or neoplastic) that harbors the cognate Ig-BCR.
In the context of this novel ligand-mediated targeting system, DNPs were conjugated with pA2036, and their ability to undergo cell-specific internalization was evaluated by cytofluorimetric analysis and confocal microscopy. The results demonstrated efficient internalization of nanoparticles. In addition, comparison with data obtained using nanoparticles conjugated with a random peptide or by treatment of a myeloma cell line (5T33MM) highlighted that a small fraction of nanoparticles underwent nonspecific cell uptake. This result indicates that, in addition to receptor-mediated endocytosis, nanoparticles were taken up by a limited, alternative, nonspecific mechanism, which was probably caveolae-dependent.41
Cytofluorimetric analysis was also performed using siRNA-loaded pA2036-nanoparticles. Two different types of nanoparticles were employed: 1) nanoparticles conjugated with the A2036 sequence engineered with nine arginine residues at the C-terminus, which, besides containing the sequence for selective recognition of B cells, were also able to form a stable electrostatic complex with negatively charged siRNA; and 2) nanoparticles conjugated with both an unmodified pA2036 and a poly-Arg complexed with siRNA. The results demonstrate that the former functionalization strategy was less efficient, probably due to the presence of the conjugated arginine residues masking the specific recognition sequence of the A2036 peptide and thus reducing the efficiency of its interaction with the Ig-BCR. Furthermore, the latter functionalization strategy resulted in both an increase in specific uptake and a decrease in non-specific internalization, probably due to the altered surface charge of the nanovector.42 In addition, the internalization of the so modified nanoparticles was also analyzed on PBMCs resulting in a very low nonspecific uptake.
The toxicity of the nanoparticles modified with the Id-peptide and siRNA to both A20 cells and human PBMCs from healthy donors was tested using cell viability assays. The resulting data demonstrated no significant effects on cell viability after nanoparticle treatment.
Given their high specific internalization efficiency and lack of cytotoxicity, pA2036-modified diatomite particles were used to deliver siRNA direct against the mRNA of the antiapoptotic factor Bcl2 in A20 cells. qRT-PCR and Western blot analyses indicated effective downregulation of Bcl2 protein expression when diatomite served as siRNA carrier. These results demonstrated that, once endocytosed by the cell, diatomite successfully released biologically active siRNA into the cytoplasm; however, further studies are required to evaluate the efficiency of gene silencing in vivo, since previous studies have reported discordant results.51,52
Delivery of siRNABcl2 complexed with A2036 peptide has previously been reported to result in a similar silencing efficiency; therefore, the use of diatomite as a carrier could allow specific co-delivery of nucleic acids and conventional drugs, a promising combination reported to be capable of increasing the sensitivity of malignant cells to chemotherapeutic agents in cancer treatment.30,50,53,54
The strategy presented here represents a highly promising approach for RNAi therapeutic applications in lymphoma treatment. Although the Id-peptide method requires a prior selection procedure for each individual B-cell clone (patient), this does not negate the significance of this work, which provides a proof-of-concept for specific delivery of siRNA cargo into target cells. In addition, this approach could also be applied in the follow-up of cancer patients, using nanoparticles modified with novel imaging agents.
Conclusion
A novel drug delivery strategy for personalized lymphoma therapy, based on DNPs conjugated with an Id-peptide specifically targeting malignant B cells, was developed in this study. Specific internalization was demonstrated in vitro by flow cytometry and confocal microscopy. Cytotoxicity tests demonstrated that diatomite concentrations ranging from 50 to 200 μg/mL have a negligible influence on cell viability. Furthermore, when the siRNA targeting Bcl2 was loaded on to modified DNPs, a significant in vitro downregulation of the target protein was observed. Further research to develop nanoparticles loaded with different anticancer molecules with the potential to work synergistically, and on their applications in vivo, is required.
Acknowledgments
The authors thank Ivo Rendina for helpful suggestions and critical reading and Luca De Stefano for support. The authors also thank Camillo Palmieri and Deref S.p.A for kindly providing Id-peptide and diatomite earth sample, respectively. This work was supported by funds from Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale (2012CK5RPF_004) and POR Campania FSE 2007–2013, Project CRÈME.
Disclosure
The authors report no conflicts of interest in this work.
References
Bangham AD, Standish MM, Watkinsal JC. Diffusion of univalent ions across the lamellae of swollen phospholipids. J Mol Biol. 1965;13(1):238–252. | ||
LaVan DA, McGuire T, Langer R. Small-scale systems for in vivo drug delivery. Nat Biotechnol. 2003;21(10):1184–1191. | ||
Langer R. New methods of drug delivery. Science. 1990;249(4976):1527–1533. | ||
Shi JJ, Xiao Z, Kamaly N, Farokhzad OC. Self-assembled targeted nanoparticles: evolution of technologies and bench to bedside translation. Acc Chem Res. 2011;44(10):1123–1134. | ||
Yang Y, Yu C. Advances in silica based nanoparticles for targeted cancer therapy. Nanomedicine. 2016;12(2):317–332. | ||
Schleich N, Po C, Jacobs D, et al. Comparison of active, passive and magnetic targeting to tumors of multifunctional paclitaxel/SPIO-loaded nanoparticles for tumor imaging and therapy. J Control Release. 2014;194:82–91. | ||
Mitra S, Gaur U, Ghosh PC, Maitra AN. Tumour targeted delivery of encapsulated dextran-doxorubicin conjugate using chitosan nanoparticles as carrier. J Control Release. 2001;74(1–3):317–323. | ||
Steichen SD, Caldorera-Moore M, Peppas NA. A review of current nanoparticle and targeting moieties for the delivery of cancer therapeutics. Eur J Pharm Sci. 2013;48(3):416–427. | ||
Wang X, Li S, Shi Y, et al. The development of site-specific drug delivery nanocarriers based on receptor mediation. J Control Release. 2014;193:139–153. | ||
Thomsen LB, Thomsen MS, Moos T. Targeted drug delivery to the brain using magnetic nanoparticles. Ther Deliv. 2015;6(10):1145–1155. | ||
Xiong L, Bi J, Tang Y, Qiao SZ. Magnetic core-shell silica nanoparticles with large radial mesopores for siRNA delivery. Small. 2016;12(34):4735–4742. | ||
Davis ME, Zuckerman JE, Choi CH, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. | ||
Varshosaz J, Farzan M. Nanoparticles for targeted delivery of therapeutics and small interfering RNAs in hepatocellular carcinoma. World J Gastroenterol. 2015;21(42):12022–12041. | ||
Barreto JA, O’ Malley W, Kubeil M, Graham B, Stephan H, Spiccia L. Nanomaterials: applications in cancer imaging and therapy. Adv Mater. 2011;23(12):H18–H40. | ||
Argyo C, Weiss V, Bräuchle C, Bein T. Multifunctional mesoporous silica nanoparticles as a universal platform for drug delivery. Chem Mater. 2014;26(1):435–451. | ||
Vivero-Escoto JL, Slowing II, Trewyn BG, Lin VS. Mesoporous silica nanoparticles for intracellular controlled drug delivery. Small. 2010;6(18):1952–1967. | ||
Rea I, Martucci NM, De Stefano L, et al. Diatomite biosilica nanocarriers for siRNA transport inside cancer cells. Biochim Biophys Acta. 2014;1840(12):3393–3403. | ||
Ruggiero I, Terracciano M, Martucci NM, et al. Diatomite silica nanoparticles for drug delivery. Nanoscale Res Lett. 2014;9(1):329–335. | ||
Terracciano M, Shahbazi MA, Correia A, et al. Surface bioengineering of diatomite based nanovectors for efficient intracellular uptake and drug delivery. Nanoscale. 2015;7(47):20063–20074. | ||
Zhang H, Shahbazi MA, Mäkilä EM, et al. Diatom silica microparticles for sustained release and permeation enhancement following oral delivery of prednisone and mesalamine. Biomaterials. 2013;34(36):9210–9219. | ||
Migkou M, Dimopoulos MA, Gavriatopoulou M, Terpos E. Applications of monoclonal antibodies for the treatment of hematological malignancies. Expert Opin Biol Ther. 2009;9(2):207–220. | ||
Reed JC. Bcl-2-family proteins and hematologic malignancies: history and future prospects. Blood. 2008;111(7):3322–3330. | ||
Chanan-Khan A. Bcl-2 antisense therapy in B-cell malignancies. Blood Rev. 2005;19(4):213–221. | ||
Abdullah LN, Chow EK-H. Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med. 2013;2(1):3–11. | ||
Banerjee A, Qian P, Wu Z-S, et al. Artemin stimulates radio- and chemo-resistance by promoting TWIST1-BCL-2-dependent cancer stem cell like behavior in mammary carcinoma cells. J Biol Chem. 2012;287(51):42502–42515. | ||
Ebrahim AS, Sabbagh H, Liddane A, Raufi A, Kandouz M, Al-Katib A. Hematologic malignancies: newer strategies to counter the BCL-2 protein. J Cancer Res Clin Oncol. 2016;142(9):2013–2022. | ||
O’ Brien S, Moore JO, Boyd TE, et al. Randomized phase III trial of fludarabine plus cyclophosphamide with or without oblimersen sodium (Bcl-2 antisense) in patients with relapsed or refractory chronic lymphocytic leukemia. J Clin Oncol. 2007;25(9):1114–1120. | ||
Cotter FE, Waters J, Cunningham D. Human Bcl-2 antisense therapy for lymphomas. Biochim Biophys Acta. 1999;1489(1):97–106. | ||
Palmieri C, Falcone C, Iaccino E, et al. In vivo targeting and growth inhibition of the A20 murine B-cell lymphoma by an idiotype-specific peptide binder. Blood. 2010;116(2):226–238. | ||
Migliaccio N, Palmieri C, Ruggiero I, et al. B-cell receptor-guided delivery of peptide-siRNA complex for B-cell lymphoma therapy. Cancer Cell Int. 2015;15:50–58. | ||
Wang Y, Huang HY, Yang L, Zhang Z, Ji H. Cetuximab-modified mesoporous silica nano-medicine specifically targets EGFR-mutant lung cancer and overcomes drug resistance. Sci Rep. 2016;6:25468–25477. | ||
De Stefano L, Oliviero G, Amato J, et al. Aminosilane functionalizations of mesoporous oxidized silicon for oligonucleotide synthesis and detection. J R Soc Interface. 2013;10(83):20130160. | ||
Terracciano M, Rea I, Politi J, De Stefano L. Optical characterization of aminosilane-modified silicon dioxide surface for biosensing. J Eur Opt Soc Rapid Publ. 2013;8:1303. | ||
Kim KJ, Kanellopoulos-Langevin C, Merwin RM, Sachs DH, Asofsky R. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J Immunol. 1979;122(2):549–554. | ||
Spatuzza C, Schiavone M, Di Salle E, et al. Physical and functional characterization of the genetic locus of IBtk, an inhibitor of Bruton’s tyrosine kinase: evidence for three protein isoforms of IBtk. Nucleic Acids Res. 2008;36(13):4402–4416. | ||
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1):248–254. | ||
San O, Gören R, Özgür C. Purification of diatomite powder by acid leaching for use in fabrication of porous ceramics. Int J Mine Process. 2009;93(1):6–10. | ||
Martucci NM, Rea I, Ruggiero I, et al. A new strategy for label-free detection of lymphoma cancer cells. Biomed Opt Express. 2015;6(4):1353–1362. | ||
De Angelis F, Pujia A, Falcone C, et al. Water soluble nanoporous nanoparticle for in vivo targeted drug delivery and controlled release in B cells tumor context. Nanoscale. 2010;2(10):2230–2236. | ||
Loike JD, Silverstein SC. A fluorescence quenching technique using trypan blue to differentiate between attached and ingested glutaraldehyde-fixed red blood cells in phagocytosing murine macrophages. J Immunol Methods. 1983;57(1–3):373–379. | ||
Kou L, Sun J, Zhai Y, He Z. The endocytosis and intracellular fate of nanomedicines: implication for rational design. Asian J Pharmaceut Sci. 2013;8(1):1–10. | ||
Bannunah AM, Vllasaliu D, Lord J, Stolnik S. Mechanisms of nanoparticle internalization and transport across an intestinal epithelial cell model: effect of size and surface charge. Mol Pharm. 2014;11(12):4363–4373. | ||
Vecchione R, Luciani G, Calcagno V, et al. Multilayered silica-biopolymer nanocapsules with a hydrophobic core and a hydrophilic tunable shell thickness. Nanoscale. 2016;8(16):8798–8809. | ||
Love SA, Maurer-Jones MA, Thompson JW, Lin Y, Haynes CL. Assessing nanoparticle toxicity. Annu Rev Anal Chem. 2012;5:181–205. | ||
Rosenholm JM, Meinander A, Peuhu E, et al. Targeting of porous hybrid silica nanoparticles to cancer cells. ACS Nano. 2009;3(1):197–206. | ||
Lu J, Liong M, Li Z, Zink JI, Tamanoi F. Biocompatibility, biodistribution, and drug-delivery efficiency of mesoporous silica nanoparticles for cancer therapy in animals. Small. 2010;6(16):1794–1805. | ||
Varna M, Ratajczak P, Ferreira I, Leboeuf C, Bousquet G, Janin A. In vivo distribution of inorganic nanoparticles in preclinical models. J Biomater Nanobiotechnol. 2012;3(2A):269–279. | ||
Scarfò L, Ghia P. Reprogramming cell death: BCL2 family inhibition in hematological malignancies. Immunol Lett. 2013;155(1–2):36–39. | ||
Garcia-Saez AJ. The secrets of the Bcl-2 family. Cell Death Differ. 2012;19:1733–1740. | ||
Ding L, Hu XM, Wu H, et al. Combined transfection of Bcl-2 siRNA and miR-15a oligonucleotides enhanced methotrexate-induced apoptosis in Raji cells. Cancer Biol Med. 2013;10(1):16–21. | ||
Hatakeyama H, Ito E, Akita H, et al. A pH-sensitive fusogenic peptide facilitates endosomal escape and greatly enhances the gene silencing of siRNA-containing nanoparticles in vitro and in vivo. J Control Release. 2009;139(2):127–132. | ||
Minakuchi Y, Takeshita F, Kosaka N, et al. Atelocollagen-mediated synthetic small interfering RNA delivery for effective gene silencing in vitro and in vivo. Nucleic Acids Res. 2004;32(13):e109. | ||
He D, Fang B. Small hairpin RNA targeting at Bcl-2 increases cytarabine induced apoptosis in Raji cells. Acta Biochim Biophys Sin (Shanghai). 2009;41(2):131–136. | ||
Nagamatsu K, Tsuchiya F, Oguma K, Maruyama H, Kano R, Hasegawa A. The effect of small interfering RNA (siRNA) against the Bcl-2 gene on apoptosis and chemosensitivity in a canine mammary gland tumor cell line. Res Vet Sci. 2008;84(1):49–55. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.