Back to Journals » International Journal of Nanomedicine » Volume 12
Green synthesis of selenium nanoparticles using Acinetobacter sp. SW30: optimization, characterization and its anticancer activity in breast cancer cells
Authors Wadhwani SA, Gorain M , Banerjee P, Shedbalkar UU, Singh R, Kundu GC, Chopade BA
Received 8 April 2017
Accepted for publication 19 July 2017
Published 13 September 2017 Volume 2017:12 Pages 6841—6855
DOI https://doi.org/10.2147/IJN.S139212
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Thomas Webster
Sweety A Wadhwani,1 Mahadeo Gorain,2 Pinaki Banerjee,2 Utkarsha U Shedbalkar,3 Richa Singh,1 Gopal C Kundu,2 Balu A Chopade1,4
1Department of Microbiology, Savitribai Phule Pune University, 2Laboratory of Tumor Biology, Angiogenesis and Nanomedicine Research, National Center for Cell Science, Savitribai Phule Pune University Campus, Pune, 3Department of Biochemistry, The Institute of Science, Mumbai, 4Dr Babasaheb Ambedkar Marathwada University, Aurangabad, Maharashtra, India
Abstract: The aim of this study was to synthesize selenium nanoparticles (SeNPs) using cell suspension and total cell protein of Acinetobacter sp. SW30 and optimize its synthesis by studying the influence of physiological and physicochemical parameters. Also, we aimed to compare its anticancer activity with that of chemically synthesized SeNPs in breast cancer cells. Cell suspension of Acinetobacter sp. SW30 was exposed to various physiological and physicochemical conditions in the presence of sodium selenite to study their effects on the synthesis and morphology of SeNPs. Breast cancer cells (4T1, MCF-7) and noncancer cells (NIH/3T3, HEK293) were exposed to different concentrations of SeNPs. The 18 h grown culture with 2.7×109 cfu/mL could synthesize amorphous nanospheres of size 78 nm at 1.5 mM and crystalline nanorods at above 2.0 mM Na2SeO3 concentration. Polygonal-shaped SeNPs of average size 79 nm were obtained in the supernatant of 4 mg/mL of total cell protein of Acinetobacter sp. SW30. Chemical SeNPs showed more anticancer activity than SeNPs synthesized by Acinetobacter sp. SW30 (BSeNPs), but they were found to be toxic to noncancer cells also. However, BSeNPs were selective against breast cancer cells than chemical ones. Results suggest that BSeNPs are a good choice of selection as anticancer agents.
Keywords: comparison, selective, 4T1, MCF7
Introduction
Selenium (Se) is an essential trace element required by many organisms. It is a crucial cofactor of antioxidant enzymes such as glutathione peroxidases and thioredoxin reductases.1,2 As the selenium nanoparticles (SeNPs) possess antimicrobial and anticancer properties, they can be used as nanomedicines.3,4 Also, they exhibit less toxicity as compared to their inorganic and organic counterparts.5 Thus, their methods of synthesis need to be studied in detail. SeNPs can be synthesized by physical, chemical and biological routes.4,6,7 Methods of biological synthesis of SeNPs are less toxic, ecofriendly and cost-effective, and yield stable SeNPs which do not aggregate because of a protein coating over them.8,9 However, the activity of these SeNPs has not been compared with chemically synthesized SeNPs.
Nanoparticles (NPs) possess size- and shape-dependent properties. Such properties need to be explored and hence, it is the need of the hour to have procedures enabling a higher degree of control over the sizes of SeNPs.10,11 In biological systems, various parameters are known to modulate the synthesis and morphology of NPs.8,12–14 There are many studies on optimization of parameters to control the size and shape of gold and silver NPs.12,13 However, such studies are missing on SeNPs, necessitating a need for a thorough study to enhance the overall properties of SeNPs.
Many bacteria, fungi and plants are reported to synthesize SeNPs.4 However, there is no report on the synthesis of SeNPs by Acinetobacter sp. Acinetobacter is a diverse group of organisms and present ubiquitously in nature.15–21 They have excellent biofilm-forming capability.22–25 They have high degree of antibiotic and metal resistance.16,26,27 Acinetobacter is known to synthesize gold, silver and platinum NPs.12,13,28 Acinetobacter sp. SW30 isolated from activated sewage sludge is reported to produce polyhedral-shaped gold NPs.12 Hence, it can be used to synthesize SeNPs. Use of total cell protein (TCP) for the synthesis would avoid extraction of particles from the cell.
The aim of this study was to synthesize SeNPs using cell suspension and TCP of Acinetobacter sp. SW30 (BSeNPs) and to study the influence of physiological and physicochemical parameters on their synthesis and morphology. This study also compares their anticancer activity with that of chemically synthesized SeNPs (CSeNPs) in breast cancer cells.
Materials and methods
Synthesis of SeNPs using cell suspension of Acinetobacter sp. SW30
A loop-full of culture of Acinetobacter sp. SW30 isolated from activated sewage sludge was inoculated in 200 mL Luria–Bertani (HiMedia, Mumbai, India) broth and incubated at 30°C, 200 rpm for 24 h.12 Cells were harvested by centrifugation (10,000 rpm for 10 min at 10°C) and washed thrice with sterile distilled water (D/W). Cell pellet was suspended in sterile D/W and challenged with sodium selenite (Na2SeO3; SD Fine Chemicals, Mumbai, India) so as to get a final concentration of 1 mM and incubated at 30°C, 180 rpm. Synthesis of SeNPs was observed preliminarily by change in color of the suspension. In all the experiments, after every 24 h, 200 μL aliquots were withdrawn; UV–Visible (UV–Vis) spectra were recorded from 200 to 800 nm on Spectra Max M2 Multimode Microplate Reader (Molecular Devices LLC, Sunnyvale, CA, USA) and those showing maximum synthesis of SeNPs were observed under transmission electron microscopy (TEM).
Optimization of parameters for obtaining SeNPs in nanometer size
TEM observation showed that the biosynthesized SeNPs were more than 100 nm in size. However, in our previous studies, it was observed that various physicochemical parameters such as culture age, cell density, metal salt concentration, temperature and pH have profound effect on the rate of synthesis and morphology of NPs.12 Hence, the effect of such parameters on SeNPs was studied in order to get particles of size <100 nm. The effect of culture age was studied by incubating the culture for 6, 12, 18, 24, 30, 36 and 48 h in Luria–Bertani broth. The culture was harvested and challenged with 1 mM Na2SeO3. Synthesis of SeNPs was monitored up to 120 h using UV–Vis spectral analysis with an interval of 24 h. The outcome of cell density was studied by adjusting the density of 18 h grown culture to <0.3, 0.3, 0.6, 0.9, 1.2, 1.5, 1.8, 2.1, 2.4 and 2.7×109 cfu/mL as per McFarland’s standards.29 The 18 h grown culture with a cell density 2.7×109 cfu/mL was challenged with different concentrations of Na2SeO3, namely, 0.1, 0.3, 0.5, 0.7, 0.9, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5 and 4.0 mM, and incubated at 30°C, 180 rpm. Hereafter, all the experiments were performed using 18 h grown culture with 2.7×109 cfu/mL cell density at 180 rpm, unless otherwise specified.
Temperature of incubation is a very important parameter. The cells were challenged with 1.5 and 3.0 mM Na2SeO3 and were incubated at different temperatures, namely, 20°C, 30°C, 37°C, 50°C and 60°C. Optimized temperature (37°C) and Na2SeO3 (1.5 and 3.0 mM) concentration was used to study the effect of pH. The pH of cell suspension was adjusted at 2, 4, 6, 7, 8, 9 and 10 using 0.1 N HCl and NaOH (Himedia). NPs were observed under TEM to study the effects of respective parameter.
Synthesis of SeNPs using TCP of Acinetobacter sp. SW30
It was found that extraction of SeNPs from bacterial cells was difficult, and therefore, cells suspended in 50 mM phosphate buffer (pH 7.4) were disrupted by Qsonica sonicator at 4°C by keeping the sonifier output at 50 amplitude and giving 10 strokes, each of 60 s, with 60 s interval. The homogenate was centrifuged under cold conditions at 18,000 rpm for 20 min. The clear supernatant obtained was collected and called as TCP. Protein content of TCP was measured by Folin Lowry method30 using bovine serum albumin as the standard. The three concentrations of TCP (1, 2 and 3 mg/mL) were challenged with various concentrations of Na2SeO3, namely, 0.1, 0.3, 0.5, 0.7, 0.9, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5 and 4.0 mM, and incubated at 30°C, 180 rpm.
Characterization of biosynthesized SeNPs
The nature of NPs was analyzed by X-ray diffraction (XRD). Thoroughly dried, thin film of SeNPs solution was taken on a glass slide and observed under D8 Advance Brucker X-ray diffractometer with Cu Kα (1.54 °A) source. The exact morphology of SeNPs was seen under TEM (Technai G2, 20 ultrawin; FEI, Eindhoven, the Netherlands). Selected area electron diffraction pattern of SeNPs was done by selecting the specific area from TEM image. Surface morphology of SeNPs synthesized by cells and TCP was observed by scanning electron microscopy (SEM; JSM-6360A; Jeol, Peabody, MA, USA) and elemental composition was detected by energy dispersive X-ray spectroscopy. Raman spectrum of the SeNPs was recorded on a confocal laser micro-Raman spectrometer (JY Labram-HR) using 24 mW excitation at 514.5 nm.
Fourier transform infrared spectroscopy (FTIR) analysis
To study the functional groups present on the cell surface or in TCP that were involved in NP synthesis, FTIR analysis of culture of Acinetobacter sp. SW30 and TCP challenged with (test) and without (control) Na2SeO3 was performed. These cultures were air dried and FTIR spectrum was recorded from 380 to 4,000 cm−1 at a resolution of 2 cm−1 using Bruker tensor 37 FTIR spectrophotometer.
Cell viability assay
The effect of BSeNPs and CSeNPs on mouse (4T1; obtained from American Type Culture Collection [ATCC], Manassas, VA, USA) and human (MCF-7; obtained from NCCS, Pune, India) breast cancer cells was assessed by MTT assay. Cells (2×104) were seeded in a 96-well, flat-bottomed culture plate and incubated for 24 h, followed by the treatment with NPs (0–100 μg/mL). After incubation, MTT solution (2.75 mg/mL) was added to each well, followed by incubation in dark for 4 h at 37°C. The formazan crystals were dissolved in isopropanol. Absorbance was measured at 570 nm on a microplate reader (Spectra Max M5; Molecular Devices LLC).14 Appropriate controls were used, and percent viability of treated cells was calculated. The anticancer activity of BSeNPs was compared with CSeNPs. To study the cytotoxic effects of NPs on noncancer cells, the same assay was repeated with mouse fibroblast (NIH/3T3; obtained from NCCS) and human embryonic kidney (HEK293; obtained from NCCS) cell lines. Experiments were performed in triplicates. The details of CSeNPs are given in Supplementary material.
Wound migration assay
Wound migration assay was performed using 4T1 breast cancer cells (ATCC [CRL-2539™]). 4T1 (3×105) cells were grown in a monolayer per well in 12-well culture plate; wound with uniform size was made using sterile micro tip after seeding and cells were treated with BSeNPs (0–50 μg/mL) in separate experiments and photographed at t=0, 12 and 24 h by phase contrast microscope (Nikon, Melville, NY, USA). Wound closure was measured by Image-Pro Plus software, analyzed statistically and represented in the form of bar graph.14 The wound migration was measured by Image-Pro Plus software and estimated by the following equation:
|
where Tt is the time after wounding (12 and 24 h) and T0 is the time immediately after wounding (0 h).
Results and discussion
Synthesis of SeNPs using cell suspension of Acinetobacter sp. SW30
Cell suspension of Acinetobacter sp. SW30 could efficiently reduce Se+4 ions in Na2SeO3 to SeNPs intracellularly after 24 h of incubation, which was evident from change in color of the cell suspension to bright orange with absorption peak at 300 and 540 nm. NanoSe showed color change from yellow to red as per the size of synthesized SeNPs.10 Lin and Wang stated that the red shift in the absorption spectra of nanoSe indicates increase in particle size. Also, the absorption spectra in UV range indicated that the particles were below 100 nm in size. Presence of two peaks in our study might indicate that two sizes of NPs were present, with one being ≥100 nm (due to peak at 300 nm) and the other being ≤100 nm (due to peak at 540 nm). Synthesis of SeNPs by bacteria is widespread, and they show large variation in the absorption peak due to differences in sizes of NPs.2,31–33
Optimization of parameters for obtaining SeNPs in nanometer size and their characterization
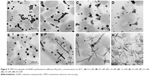
SeNP synthesis was observed in all culture ages, with the maximum observed in 18 h grown culture (Figure 1A). Particles synthesized by 18 h grown culture were uniform and spherical when observed under TEM (Figure 1A inset). Very less synthesis was observed in 6 and 12 h grown culture, which may be due to the presence of less concentration of biomolecules responsible for reduction of sodium selenite. It was found that the synthesis of SeNPs reduced after 18 h of culture age. Hence, 18 h was considered as the optimum culture age.
Concentration of biomolecules in the reaction mixture has a considerable effect on NP synthesis.11 Here, the concentration of biomolecules is in terms of cell densities. At lower cell densities, that is, >0.3–0.6×109 cfu/mL, synthesis of SeNPs was not observed (Figure 1B inset). Synthesis of SeNPs increased with increase in cell densities, with the maximum synthesis observed at 2.7×109 cfu/mL. The concentration of reactants, that is, biomolecules, was most favorable at a cell density of 2.7×109 cfu/mL; thus, for further studies, cell density was adjusted to 2.7×109 cfu/mL (Figure 1B). Similarly, Synechococcus leopoliensis produced significantly more SeNPs at higher cell densities as compared to lower and medium cell densities34 because at higher cell densities, more reducing molecules are present in the solution.
Synthesis of SeNPs was observed with Na2SeO3 at a concentration from 0.3 to 4.0 mM, except at 0.1 mM. Intensity of color increased with increasing Na2SeO3 concentration (Figure 1C). When these particles were observed under TEM, it was found that from 0.3 to 2.0 mM Na2SeO3 concentration, the SeNPs were spherical in shape. The average diameter of SeNPs at 0.3, 0.5, 1.0 and 1.5 mM was 126, 96, 113 and 78 nm, respectively (Figure 2A–D). Thus, at 1.5 mM, smallest particles were synthesized. However, at higher concentrations from 2.5 to 4.0 mM, selenium rods were observed (Figure 2F–H). At 3.0 mM, comparatively shorter rods were observed; therefore, for spherical particles, 1.5 mM was considered as the optimum, and for rods, 3.0 mM Na2SeO3 concentration was considered the optimum. On the contrary, there was no significant effect of different Se concentrations on the morphology of SeNPs synthesized by Shewanella sp. HN-41 and Lactobacillus sp.35,36 Streptomyces bikiniensis can synthesize nanorods of size 17 nm.37
It was observed that during the period of incubation, shape of elemental Se changed from spherical to rod-like structure. After 6 h, spheres were observed, while after 48 h of incubation, length of particles increased in one dimension and they were converted into rods. It may be owing to the Ostwald ripening process evoked by the high free energy of SeNPs.37 In our studies, synthesis of Se rods could be due to the Ostwald ripening process at higher concentration of Na2SeO3. Lortie et al found that increase in selenite concentration increased the reduction rate up to 19 mM of selenite and above 19 mM, selenite reduction rate was decreased due to toxicity of selenite.39
Temperature and pH are known as size and shape modulating agents.39 Synthesis of SeNPs was observed at 30°C and 37°C, with maximum synthesis observed at 37°C (Figure 1D). Our results are similar to Lortie et al; they also could not get synthesis of SeNPs at higher temperature.39 The reason can be that the biomolecules/proteins responsible for the reduction of sodium selenite are active only at 30°C and 37°C. So, 37°C was considered as the optimum temperature. Synthesis of SeNPs by Citrus reticulata peel extract was found to be efficient at 40°C.40 An increase in temperature from 80°C to 100°C causes aggregation of NPs into rods.39
Surprisingly, at 1.5 mM Na2SeO3 concentration, synthesis of SeNPs was noticed at all tested pH except at pH 2, while at 3.0 mM Na2SeO3 concentration, synthesis of SeNPs was noticed at all tested pH values. At pH 2 and 1.5 mM, possibly fewer functional groups are available, which are not sufficient for the reduction process.11 At 1.5 mM Na2SeO3 concentration, synthesis of SeNPs increased with increase in pH up to 9. However, at 3.0 mM Na2SeO3 concentration, synthesis increased up to pH 6, and then it reduced. Also, the spectra changed because of the different morphology of SeNPs (Figure 1E and F). TEM analysis was conducted at pH 6, 7 and 9 for both the Na2SeO3 concentrations. Selenium spheres and rods were observed at 3.0 mM Na2SeO3 concentration, while at 1.5 mM Na2SeO3 concentration, only spheres were observed (Figure 3). From TEM images, it was concluded that there is no significant effect of pH on the size of SeNPs. On the contrary, others have found significant influence of pH on SeNP size. In case of enzyme purified from Geobacillus wiegeii, the size of all NPs was <100 nm at pH 4. Over 50% of NPs were <100 nm in size at pH 5 and 90% were of size <100 nm at pH 6 and 8. At pH 7, NPs were of size 120 nm and only 20% were <100 nm. So, acidic and basic pH favored the formation of NPs with size below 100 nm.39 In Bacillus megaterium and Pseudomonas stutzeri, reduction of sodium selenite was increased with an increase in pH up to 7 and 8.38,41
Synthesis of SeNPs using TCP
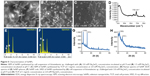
Synthesis of SeNPs was observed at all the tested concentrations of TCP against all the Na2SeO3 concentrations from 0.1 to 4.0 mM. However, the spectra of three of TCP were different from each other due to different shapes and sizes of biosynthesized SeNPs as discussed above. Maximum synthesis of SeNPs was noticed at 2 mM Na2SeO3 concentration for 1 and 2 mg/mL TCP concentrations (Figure 4A and B). However, for 3 mg/mL TCP concentration, 3.5 mM Na2SeO3 concentration gave maximum synthesis (Figure 4C). Interestingly, when SeNPs of 4 mM Na2SeO3 synthesized using 1 mg/mL of TCP were centrifuged, very nice green color was observed in the supernatant; remaining particles were orange colored, which settled at the bottom. When this green supernatant was observed under TEM, polygonal shaped SeNPs were found with average size of 79 nm (Figure 4E). At 1 mg/mL, two peaks, that is, 350 and 530 nm, were found. At 2 mg/mL, there were prominent peaks at 390 and 570 nm. At 3 mg/mL, a single peak at 350 nm was observed. When these particles were observed under TEM and their particle size was measured using ImageJ software, it was found that at 1, 2 and 3 mg/mL TCP concentration, the average particle size was 158, 184 and 211 nm, respectively. This indicates that average particle size increases with increasing concentration of TCP. That is why, 1 mg/mL can be used for further studies. There is no report on synthesis of SeNPs by TCP.
Characterization of BSeNPs
The presence of rods and sphere was confirmed by SEM (Figure 5A and B). In selected area electron diffraction pattern, it was noticed that selenium spheres were amorphous, while the rods were crystalline in nature (Figure 3B and F inset). This was further confirmed by the XRD pattern of SeNP sample containing rods that gave two sharp peaks at 2θ values of 23.3° and 31.7° corresponding to crystal planes of 100 and 101, respectively, which is in agreement with Joint Committee on Powder Diffraction Standards (file no-06-0362), as shown in Figure 5G. However, SeNPs synthesized by 1 mg/mL TCP did not give any sharp peak, as only nanospheres were present in the sample which was amorphous in nature (Figure 5H). Such crystalline SeNPs are also synthesized by many bacteria as well as plants.33,41 Figure 5D shows the Raman spectra of SeNPs. Elemental content of selenium spheres and rods was confirmed by the presence of peak at 1.37 keV in Energy Dispersive X-ray Spectroscopy (Figure 5E and F), which is the characteristic peak for SeLα.34,41
The presence of peak at 1,655 cm−1 revealed amide II, while the peak at 3,289 cm−1 indicated the presence of amino acids/amines in Acinetobacter cell suspension (Figure 6A(i)). After addition of Na2SeO3, few new peaks were observed at 2,919, 1,574, 1,378 and 1,049 cm−1 (Figure 6A(ii)). Peaks at 1,574 and 1,378 cm−1 are due to amide group and C–N stretching of proteins, respectively. Those at 2,919 and 1,049 cm−1 represent C–H and C–O stretch. This shows that SeNPs are coated by proteins. In TCP, 3,301 and 1,635 cm−1 represent primary amines and amide I, respectively (Figure 6B(i)). After addition of Na2SeO3, the peak at 3,301 cm−1 reduced drastically, indicating the participation of amines and amides in the reduction process (Figure 6B(ii)). Comparable peaks were obtained in SeNPs synthesized by Lactobacillus sp. and Bacillus licheniformis JS2.35,42 The carbonyl groups from amino acids can bind strongly to metals, and hence, they can form a coat around NPs to prevent agglomeration.35
Cell viability assay
This study gives a comparison of the anticancer activity of SeNPs synthesized by Acinetobacter sp. SW30 with that of chemical ones. Both types of SeNPs showed antiproliferative activity against 4T1 cells in a dose-dependent manner (Figure 7A). Figure 7B represents the antiproliferative activity of SeNPs against NIH/3T3 cells. However, CSeNPs (20.7%) showed more inhibition of cell viability than BSeNPs (53.5%) at 100 μg/mL. Similar results were observed in MCF-7 cells; CSeNPs (15.8%) exhibited more inhibition of cell viability than BSeNPs (26.6%), as shown in Figure 8A. CSeNPs (97 nm size) had a zeta potential of −13 mV, whereas BSeNPs (94 nm size) had +10 mV (Figure 9). Secretion of large amount of lactate anions from cancer cells and exposure of more phospholipids on the surface of breast cancer cells lead to negatively charged surface of cancer cells as compared to fibroblasts; thus, positively charged BSeNPs may have strong affinity for breast cancer cells, causing enhanced anticancer efficacy of BSeNPs.43–45 Thus, CSeNPs (31%) showed comparatively more inhibition in NIH/3T3 cells than BSeNPs (77%) at 100 μg/mL (Figure 7B). In case of HEK293, CSeNPs (17%) caused more inhibition than BSeNPs (30%), as shown in Figure 8B. CSeNPs showed more cytotoxicity than BSeNPs even in NIH/3T3 and HEK293 cells. Hence, CSeNPs cannot be considered for further studies. MTT assay of Na2SeO3 was also performed, which showed 93% cell viability in MCF-7 cell line (data given in Supplementary material).
SeNPs have anticancer activity against many cancerous cells.46,47 However, there are very few reports on the anticancer activity of SeNPs synthesized by bacteria.37,48 SeNPs synthesized by Bacillus strain ZYK have profound anticancer activity against H157 lung cancer cell line.49 It has been reported that elemental selenium exists as endogenous SeNPs in H157 cell line and induces cytotoxicity by many mechanisms such as inhibition of glycolysis, glycolysis-dependent mitochondrial dysfunction, and so on.50
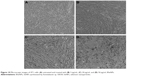
Microscopic observation of 4T1 cells treated with SeNPs showed a dose-dependent reduction in cell number, loss of cell-to-cell contact and cell shrinkage (Figure 10A–D). The same cellular morphologic changes were observed when MCF-7 cells were treated with selenium nanorods synthesized by S. bikiniensis strain Ess_amA-1.37 Ahmad et al proposed the possible mechanism of anticancer activity of SeNPs, which could be due to their ability to bind to Cu (II), leading to its reduction to Cu (I), and regeneration of Cu (II) causes the production of reactive oxygen species, which selectively kill cancer cells as there is high level of copper in cancerous cells.38 BSeNPs are also reported to exhibit significant anticancer activity against H157 lung cancer cell line, fibrosarcoma cell line and A375 human melanoma.5,48,50,51 Oral administration of SeNPs can enhance immune response in mice bearing 4T1 breast cancer tumor.52
Wound migration assay
The assessment of effect of BSeNPs on cell migration of 4T1 cells was performed by wound migration assay (Figure 11). There was significant inhibition of cell migration after 12 h of incubation. SeNPs with 5, 20 and 50 μg/mL concentrations showed percent inhibition (64%, 54% and 27%, respectively) in cell migration as compared to the control cells (Figure 12). Similar results were observed when SeNPs conjugated with anisomycin suppressed cell migration in HePG2 cells.53
  | Figure 12 Percentage migration of 4T1 cells after 24 h treatment with 0, 5, 20 and 50 μg/mL SeNPs. |
Conclusion
Acinetobacter sp. SW30 was found to synthesize intracellular SeNPs which were characterized by different physicochemical techniques such as UV–Vis spectrophotometry, XRD, SEM, energy dispersive X-ray spectroscopy and TEM. The 18 h grown culture with 2.7×109 cfu/mL could synthesize amorphous nanospheres of size 78 nm at 1.5 mM and crystalline nanorods at 2.0 mM Na2SeO3 concentration onward. The pH had no significant effect on the morphology of SeNPs. Polygonal-shaped SeNPs of average size 79 nm were obtained in the supernatant of 4 mg/mL of TCP. FTIR studies have confirmed that proteins are the essential molecules for the reduction and coating of SeNPs. Though CSeNPs have shown more anticancer activity than BSeNPs, they are toxic to noncancer cells also; hence, BSeNPs are a good alternative to anticancer agents.
Acknowledgment
SAW and RS acknowledge the University Grants Commission (UGC), New Delhi, India, for awarding research fellowship.
Disclosure
The authors report no conflicts of interest in this work.
References
Husen A, Siddiqi KS. Plants and microbes assisted selenium nanoparticles: characterization and application. J Nanobiotechnol. 2014;12:28. | ||
Srivastava N, Mukhopadhyay M. Biosynthesis and structural characterization of selenium nanoparticles mediated by Zooglea ramigera. Powder Technol. 2013;244:26–29. | ||
Forootanfar H, Adeli-Sardou M, Nikkhoo M, et al. Antioxidant and cytotoxic effect of biologically synthesized selenium nanoparticles in comparison to selenium dioxide. J Trace Elem Med Biol. 2013;28(1):75–79. | ||
Wadhwani SA, Shedbalkar UU, Singh R, Chopade BA. Biogenic selenium nanoparticles: current status and future prospects. Appl Microbiol Biotechnol. 2016;100(6):2555–2566. | ||
Shakibaie M, Khorramizadeh M, Faramarzi M, Sabzevari O, Shahverdi A. Biosynthesis and recovery of selenium nanoparticles and the effects on matrix metalloproteinase-2 expression. Biotechnol Appl Biochem. 2010;56(1):7–15. | ||
Iranifam M, Fathinia M, Sadeghi Rad T, Hanifehpour Y, Khataee AR, Joo SW. A novel selenium nanoparticles-enhanced chemiluminescence system for determination of dinitrobutylphenol. Talanta. 2013;107:263–269. | ||
Zhang Y, Wang J, Zhang L. Creation of highly stable selenium nanoparticles capped with hyperbranched polysaccharide in water. Langmuir. 2010;26(22):17617–17623. | ||
Shedbalkar U, Singh R, Wadhwani S, Gaidhani S, Chopade BA. Microbial synthesis of gold nanoparticles: current status and future prospects. Adv Colloid Interf Sci. 2014;209:40–48. | ||
Singh R, Shedbalkar U, Wadhwani S, Chopade BA. Bacteriagenic silver nanoparticles: synthesis, mechanism, and applications. Appl Microbiol Biotechnol. 2015;99(11):4579–4593. | ||
Zong-Hong Lin, Chris Wang CR. Evidence on the size-dependent absorption spectral evolution of selenium nanoparticles. Mater Chem Phy.2005;92(2–3):591–594. | ||
Shah M, Fawcett D, Sharma S, Tripathy S, Poinern G. Green synthesis of metallic nanoparticles via biological entities. Materials. 2015;8(11):7278–7308. | ||
Wadhwani SA, Shedbalkar UU, Singh R, Karve MS, Chopade BA. Novel polyhedral gold nanoparticles: green synthesis, optimization and characterization by environmental isolate of Acinetobacter sp. SW30. World J Microbiol Biotechnol.2014;30(10):2723–2731. | ||
Singh R, Wagh P, Wadhwani S, et al. Synthesis, optimization and characterization of silver nanoparticles from Acinetobacter calcoaceticus and their enhanced antibacterial activity with antibiotics. Int J Nanomedicine. 2013;8:4277–4290. | ||
Ghosh S, More P, Derle A, et al. Diosgenin functionalized iron oxide nanoparticles as novel nanomaterial against breast cancer. J Nanosci Nanotechnol. 2015;15(12):1533–4880. | ||
Maindad DV, Kasture VM, Chaudhari H, Dhavale DD, Chopade BA, Sachdev DP. Characterization and fungal inhibition activity of siderophore from wheat rhizosphere associated Acinetobacter calcoaceticus strain HIRFA32. Indian J Microbiol.2014;54(3):315–322. | ||
Towner KJ, Chopade BA. Biotyping of Acinetobacter calcoaceticus using the API 20NE system. J Hosp Infect. 1987;10(2):145–151. | ||
Huddedar SB, Shete AM, Tilekar JN, Gore SD, Dhavale DD, Chopade BA. Isolation, characterization, and plasmid pUPI126-mediated indole-3-acetic acid production in Acinetobacter strains from rhizosphere of wheat. Appl Biochem Biotechnol. 2002;102–103(1–6):21–39. | ||
Jagtap SC, Yavankar SP, Pardesi KR, Chopade BA. Isolation, characterization and antibiotic sensitivity of Acinetobacter genospecies from animals. Indian Vat J. 2009;86(10):996–999. | ||
Mujumdar SS, Bashetti SP, Chopade BA. Plasmid pUPI126-encoded pyrrolnitrin production by Acinetobacter haemolyticus A19 isolated from the rhizosphere of wheat. World J Microbiol Biotechnol. 2014;30(2):495–505. | ||
Rokhbakhsh-Zamin F, Sachdev D, Kazemi-Pour N, et al. Characterization of plant-growth-promoting traits of Acinetobacter species isolated from rhizosphere of Pennisetum glaucum. J Microbiol Biotechnol. 2011;21(6):556–566. | ||
Patil JR, Chopade BA. Distribution and in vitro antimicrobial susceptibility of Acinetobacter species on the skin of healthy humans. Natl Med J India. 2000;14(4):204–208. | ||
Sahu PK, Iyer PS, Gaikwad MB, Talreja SC, Pardesi KR, Chopade BA. An MFS transporter-like ORF from MDR Acinetobacter baumannii AIIMS 7 is associated with adherence and biofilm formation on biotic/abiotic surface. Int J Microbiol. 2012;2012: Article ID 490647. | ||
Yavankar SP, Pardesi KR, Chopade BA. Species distribution and physiological characterization of Acinetobacter genospecies from healthy human skin of tribal population in India. Ind J Microbiol. 2007;24(4):336–345. | ||
Fulsundar S, Kulkarni HM, Jagannadham MV, et al. Molecular characterization of outer membrane vesicles released from Acinetobacter radioresistens and their potential roles in pathogenesis. Microb Pathog. 2015;83:12–22. | ||
Singh R, Nadhe S, Wadhwani S, Shedbalkar U, Chopade BA. Nanoparticles for control of biofilms of Acinetobacter species. Materials. 2016;9:383. | ||
Shakibaie MR, Dhakephalker PK, Kapadnis BP, Salajaghe GA, Chopade BA. Plasmid mediated silver and antibiotic resistance in Acinetobacter baumannii BL54. Iranian J Med Sci. 1998;23(1):30–36. | ||
Shakibaie MR, Dhakephalker BA, Kapadnis BP, Chopade BA. Silver resistance in Acinetobacter baumannii BL54 occurs through binding to a Ag-binding protein. Iranian J Biotechnol. 2003;1(1):41–46. | ||
Gaidhani S, Yeshvekar RV, Shedbalkar US, Bellare JH, Chopade BA. Bio-reduction of hexachloroplatinic acid to platinum nanoparticles employing Acinetobacter calcoaceticus. Process Biochem.2014;49:2313–2319. | ||
Shakibaie MR, Kapadnis BP, Dakephalkar P, Chopade BA. Removal of silver from photographic waste water effluent using Acinetobacter baumannii BL54. Can J Microbiol. 1999;45(12):995–1000. | ||
Scott S. Measurement of microbial cells by optical density (Winter). J Valid Technol.2011;17:46–49. | ||
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with folin phenol reagent. J Biol Chem. 1951;193(1):265–275. | ||
Biswas KC, Barton LL, Tsui WL, Shuman K, Gillespie J, Eze CS. A novel method for the measurement of elemental selenium produced by bacterial reduction of selenite. J Microbiol Methods. 2011;86(2):140–144. | ||
Fesharaki PJ, Nazari P, Shakibaie M, Rezaie S, Banoee M, Shahverdi AR. Biosynthesis of selenium nanoparticles using Klebsiella pneumoniae and their recovery by a simple sterilization process. Braz J Microbiol. 2010;41(2):461–466. | ||
Prasad S, Vyas P, Prajapati V, Patel P, Selvaraj K. Biomimetic synthesis of selenium nanoparticles using cell-free extract of Microbacterium sp. ARB05. Micro Nano Lett. 2012;8:11. | ||
Hnain A, Brooks J, Lefebvre DD. The synthesis of elemental selenium particles by Synechococcus leopoliensis. Appl Microbiol Biotechnol. 2013;97(24):10511–10519. | ||
Radhika R, Gayathri S. Extracellular biosynthesis of selenium nanoparticles using some species of Lactobacillus. Indian J Mar Sci. 2015;43(5). | ||
Filipe V, Hawe A, Jiskoot W. Critical evaluation of nanoparticle tracking analysis (NTA) by nanosight for the measurement of nanoparticles and protein aggregates. Pharm Res.2010;27(5):796–810. | ||
Ahmad MS, Yasser MM, Sholkamy EN, Ali AM, Mehanni MM. Anticancer activity of biostabilized selenium nanorods synthesized by Streptomyces bikiniensis strain Ess_amA-1. Int J Nanomedicine. 2015;10:3389–3401. | ||
Lortie L, Gould WD, Rajan S, McCready RG, Cheng KJ. Reduction of selenate and selenite to elemental selenium by a Pseudomonas stutzeri isolate. Appl Environ Microbiol. 1992;58(12):4042–4044. | ||
Correa-Llanten DN, Munoz-Ibacache SA, Maire M, Blamey JM. Enzyme involvement in the biosynthesis of selenium nanoparticles by Geobacillus wiegelii strain GWE1 isolated from a drying oven. Int J Biol Biomol Agri Food Biotechnol Eng. 2014;8(6):637–641. | ||
Sasidharan S, Sowmiya R, Balakrishnaraja R. Biosynthesis of selenium nanoparticles using Citrus reticulata peel extract. World J Pharma Res. 2014;4(1):1322–1330. | ||
Mishra RR, Prajapati S, Das J, Dangar TK, Das N, Thatoi H. Reduction of selenite to red elemental selenium by moderately halotolerant Bacillus megaterium strains isolated from Bhitarkanika mangrove soil and characterization of reduced product. Chemosphere. 2011;84(9):1231–1237. | ||
Sonkusre P, Ravikanth Nanduri R, Pawan Gupta P, Singh CS. Improved extraction of intracellular biogenic selenium nanoparticles and their specificity for cancer chemoprevention. J Nanomed Nanotechnol. 2014;5:2. | ||
Dobrzyńska I, Skrzydlewska E, Zbigniew A, Figaszewski ZA. Changes in electric properties of human breast cancer cells. J Membr Biol. 2013;246(2):161–166. | ||
Chen B, Le W, Wang Y, et al. Targeting negative surface charges of cancer cells by multifunctional nanoprobes. Theranostics. 2016;6(11):1887–1898. | ||
Yu B, Zhang Y, Zheng W, Fan C, Chen T. Positive surface charge enhances selective cellular uptake and anticancer efficacy of selenium nanoparticles. Inorg Chem. 2012;51(16):8956–8963. | ||
Zhang J, Wang H, Bao Y, Zhang L. Nano red elemental selenium has no size effect in the induction of seleno-enzymes in both cultured cells and mice. Life Sci. 2014;75(2):237–244. | ||
Gao X, Kong L. Treatment of cancer with selenium nanoparticles. United States patent US20110262564 A1. 2010 Jun 18. | ||
Bao P, Xiao KQ, Wang HJ, et al. Characterization and potential applications of a selenium nanoparticle producing and nitrate reducing bacterium Bacillus oryziterrae sp. nov. Sci Rep.2016;6:34054. | ||
Bao P, Chen SC, Xiao KQ. Dynamic equilibrium of endogenous selenium nanoparticles in selenite-exposed cancer cells: a deep insight into the interaction between endogenous SeNPs and proteins. Mol Bio syst. 2015;11(12):3355–3361. | ||
Yang F, Tang Q, Zhong X, et al.Surface decoration by Spirulina polysaccharide enhances the cellular uptake and anticancer efficacy of selenium nanoparticles. Int J Nanomedicine. 2012;7:835–844. | ||
Yazdi MZ, Mahdavi M, Varastehmoradi B, Faramarzi MA, Shahverdi AR. The immunostimulatory effect of biogenic selenium nanoparticles on the 4T1 breast cancer model: an in vivo study. Biol Trace Elem Res. 2012;149(1):22–28. | ||
Xia Y, You P, Xu F, Liu J, Xing F. Novel functionalized selenium nanoparticles for enhanced anti-hepatocarcinoma activity in vitro. Nanoscale Res Lett. 2015;10(1):1051. |
Supplementary materials
Methodology
Chemical synthesis of selenium nanoparticles
Selenium nanoparticles (SeNPs) were synthesized by adding 2 mL of 100 mM Na2SeO3 to 100 mL of 10 mM heated sodium borohydride kept on a magnetic stirrer. Then, 100 mM triton X-100 was added as a stabilizer.1 Chemically synthesized SeNPs were characterized by ultraviolet–visible spectrophotometer and transmission electron microscopy.
MTT assay for Na2SeO3
MTT assay of Na2SeO3 (2.5 mM) on MCF-7 cell line was performed.
Results
Chemical synthesis of SeNPs
Synthesis of SeNP was observed immediately after adding Na2SeO3 but after few minutes color disappears. The stable particles were obtained after addition of triton X-100. The specific peak was observed at 500 nm when observed under UV–Vis spectrophotometer (Figure S1A). Under transmission electron microscopy, crystalline rods and amorphous spheres were found (Figure S1B). These nanoparticles were concentrated by ultracentrifugation and their concentration was determined by dry weight analysis and used for comparison on anticancer activity with biological ones after sterilizing in autoclave.
  | Figure S1 (A) UV-Vis spectra and (B) TEM of chemically synthesized SeNPs. |
MTT assay for sodium selenite
Na2SeO3 showed 93% cell viability in MCF-7 cell line (Figure S2).
Reference
Nath S, Ghosh SK, Panigahi S, Thundat T, Pal T. Synthesis of selenium nanoparticle and its photocatalytic application for decolorization of methylene blue under UV irradiation. Langmuire. 2004;20(18):7880–7883. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.