Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 12
g-Nomic: a new pharmacogenetics interpretation software
Authors Sabater A , Ciudad CJ , Cendros M, Dobrokhotov D, Sabater-Tobella J
Received 30 January 2019
Accepted for publication 27 March 2019
Published 29 May 2019 Volume 2019:12 Pages 75—85
DOI https://doi.org/10.2147/PGPM.S203585
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Ana Sabater,1 Carlos J Ciudad,2 Marc Cendros,1 Denis Dobrokhotov,1 Juan Sabater-Tobella1
1Department of Information Technology, EUGENOMIC, Barcelona 08012, Spain; 2Department of Biochemistry and Physiology, University of Barcelona, Barcelona 08028, Spain
Abstract: We present g-Nomic, a pharmacogenetics interpretation software that analyzes globally a prescribed medication taking into account the personal background genetics, drug–drug interactions, lifestyle, nutritional supplements, inhibitors, inducers, and other risks to analyze primary or secondary metabolism pathways. G-Nomic provides a set of recommendations describing the suitability of a given combination of drugs for each patient according to their genes and polymedication. G-Nomic is updated monthly including data from the new drugs to be included, their known interactions, and the relevant pharmacokinetic biomarkers. For the interactions, the list is curated manually, only keeping those with clinical relevance. For each drug, their FDA and EMA drug labels are accessed, to check for relevant enzymes and transport proteins that influence its pharmacokinetics, and for their ability to induce or inhibit other enzymes, particularly the CYP-450 system. When this information is not available, a PubMed search is made to look for these characteristics. In addition, a distinction is made between drugs and prodrugs. A query on the g-Nomic software begins with entering the medication by either their common or commercial name. Non-pharmacological substances can be also added or selected under “lifestyle habits”. The lifestyle list is dynamic, showing only the substances known to interact with the drugs that are currently selected, and includes herb compounds, such as St. John’s wort, as well as proper lifestyle substances such as grapefruit or cigarette smoking. The software provides a list of the genes classified as primary biomarkers as candidates for genetic testing, and a list of the interactions that have been detected. If genetic information is available then, or is made available at a later point, these results can also be entered and the software returns pharmacogenetics recommendations regarding specific genotypes. g-Nomic takes all the above-mentioned parameters in an easy and user-friendly tool making prescription safer.
Keywords: pharmacogenetics, SNP, drug-drug interaction, drug-lifestyle, drug-herb, software
Introduction
Adverse drug reactions (ADRs) are one of the leading causes of death in developed countries. In the United States of America, it is reported that more than 100.000 people died each year in the USA only, due to ADR even if they take medicines correctly prescribed according to the protocol.1 In Spain, more than 5% of ADR-related hospitalizations had a fatal outcome.2 According to statistics, senior Americans take, on average, more than five drugs.3 and 40% of the ADR could be avoided.4 if pharmacogenetics criteria were taken into account.
Nevertheless, many practitioners confirm that the problem with medication persists even after doing a pharmacogenetics test such as Roche’s Amplichip.5 the first FDA-approved pharmacogenetics test, designed to detect variants in genes involved in drug metabolism. The main reason for this issue is that pharmacogenetics tests alone are significant only in the case of monotherapy, but in combination therapy – most of the cases – the final effect of the drug must be assessed by taking into account the interactions with the other medications taken by the patient as well as the lifestyle. Consequently, approximately 30% of the variability in response to prescriptions is due to genetics, but 60% of the problems are due to drug–drug interactions, drug–lifestyle interactions, and inhibitions and inductions produced by polymedication. The remaining 10% corresponds to problems linked to age, absorption, and drug side effects among others.
Pharmacogenetics studies the actions of the pharmacological response to drugs depending on the genetic background of a patient. Single nucleotide polymorphisms (SNPs) are at the basis of human variability originating the classical drug-metabolizing categories of normal or extensive metabolizers, intermediate, poor and rapid or ultra-rapid metabolizers. The changes produced by these non-pathological mutations may be silent and they only will be manifested when a drug for its therapeutic action, depends on the normal functioning of the enzymes, transporters or therapeutic targets emitted by these genes with these SNPs.
This can affect receptors like VKORC1, transporters like the ABC and SLC families, or enzymes involved in the metabolism of drugs belonging to Phase I such as the P450 family or Phase II like UGT, NAT, and GST. Therefore, the correct application of pharmacogenetics reduces the costs of medical assistance, minimizes adverse reactions to medications, avoids therapeutic failures, and allows prescribing in a safe and more efficient way.
There are already organizations and consortiums worldwide with the aim of achieving safe and effective pharmacological treatments: PGRN (Pharmacogenomics research network), PharmGKB, CPIC, and Dutch International Working Group among others.6–8 Drug agencies worldwide already recognize SNPs involved in the metabolism and transport of certain drugs. In addition, new drug labels already include pharmacogenetics markers and drug–drug or drug–lifestyle interactions based on pharmacogenetics markers. This has contributed significantly to make pharmacogenetics a viable milestone by listing the genetic markers that are relevant and their clinical significance.
However, as stated above, aside from the data gathered from pharmacogenetics studies, there is an important issue of exploring and taking into account the effects of drug–drug interactions. While the presence of a loss-of-function variation in the gene encoding for an enzyme will abolish its activity, it is known that substances with inhibitory capacity can also impair the enzymatic activity even in the absence of loss-of-function variants. Therefore, a person who would be categorized as a normal metabolizer based solely on a genetic test would experience phenoconversion to a poor metabolizer when exposed to a strong inhibitor.
Likewise, enzyme inducers can increase enzymatic activity over what a genetic test might indicate.9 This rationale can be extended to drug–herb and drug–lifestyle interactions as potential modulators of drug metabolism and therefore of drug response. Taken the above in consideration, it is hard to achieve personalized prescription without considering the medication of the patient as a whole including lifestyle, food, nutritional supplements, tobacco, abuse drugs, and herbal plants.
Therefore, with the aim of making prescription more secure to both, doctor and patient, we have developed g-Nomic, a novel pharmacogenetics interpretation software.
The g-Nomic pharmacogenetics interpretation software can be accessed via de website
Methods
The pharmacogenetics interpretation software, g-Nomic, is an initiative in the area of biotechnology, that already integrates information of more than 2,000 active ingredients (equivalent to more than 10,000 drug brand names) with genetic data in order to grant practitioners safety in the medical prescription.
Active ingredients
We have defined as active ingredient either medicine, food, food supplement, vitamins, natural herbs, or abuse drug. In g-Nomic, there is information related to more than 2,000 active ingredients. Of these 1,206 active ingredients do not have any pharmacogenetics information and 934 have pharmacogenetics information either as a substrate, inducer, or inhibitor.
The active ingredients present in g-Nomic without pharmacogenetics information are relevant as the software gives evidence regarding relevant drug–drug interactions, drug with lifestyle, inhibitors, inducers, risks associated with medication as well as genes related to the medication.
All data entered are for in vivo. No data are entered to g-Nomic if is in vitro or in animals.
Input
g-Nomic pharmacogenetics interpretation software inputs for each medication possible substrates (drug or prodrug), primary and secondary pathways, inhibitions (competitive or allosteric, normal or strong) and inductions (normal or strong), the interaction of these drugs with other drugs, interactions with lifestyle, the possible risks associated with medication and alerts about published guidelines regarding medication, and finally the genetic information in terms of SNPs (if known) of the individual to be categorized as EM, IM, UM, or PM (Figure 1).
Development
The development of g-Nomic took place in two phases. Phase I had the objective of compiling the initial information of the database, that is the drugs to be included, their known interactions, and the relevant pharmacokinetic biomarkers. Once the database was populated, and the software released, Phase II deals with updating it regularly with new information.
For the interactions database, a preliminary list was elaborated by reviewing focused literature.10,11 This list was curated manually, only keeping the relevant interactions to avoid the phenomenon known as “alert fatigue”,12 observed when there is an overload of drug interaction alerts with limited relevance that leads professionals to ignore notifications. Relevant interactions were defined as those that warrant taking some sort of measure, from monitorization to a strict contraindication of concurrent use, which roughly matches categories 1–3 of the Hansten and Horn significance scale.
Data on metabolic pathways were compiled by selecting all the drugs present in the Spanish vademecum as a starting point later it has been updated will all new released drugs and updated with the FDA and EMA information. For each drug, we accessed their FDA and EMA drug labels, when available, to check for relevant enzymes and transport proteins and for their ability to induce or inhibit other enzymes, particularly the CYP-450 system. When this information was not available, a PubMed search was made to look for these characteristics. In addition, a distinction was made between drugs and prodrugs (ie, whether the parent drug is active, or it requires an activating transformation first).
Prodrugs that in vivo are transformed into active drugs without enzymes that can have altered activity due to SNPs in their codifying genes, are considered as drugs and not as prodrugs. Therefore, only substances that are activated by enzymatic reactions are considered prodrugs in our database.
After the initial population of the database, the information is kept up-to-date by a monthly review of reported changes in drug labels, both from FDA and EMA, as well as automated PubMed searches, and constant implementation of the results once they are reviewed. In-depth review of bibliography, targeted to particular drugs of interest, can also be performed outside this schedule. Only data from human studies (both patients and healthy subjects) are eligible to be included in the software. Results from in vitro studies, as well as experimentation in animals, are considered too preliminary to be included.
g-Nomic takes all the above-mentioned parameters in an easy and user-friendly tool making prescription safer. The first release was available in 2014. Since then more than 3,000 users have accessed this tool. Currently, it holds at least 8,000 drug–drug interactions from clinical publications, 1,900 drug–lifestyle interactions, 1,500 risks associated with medication, and 600 polymorphisms in 50 genes. What makes g-Nomic pharmacogenetics interpretation software so powerful is the interacting algorithm that is capable of mixing all the inputs in just a few seconds giving presently more than 2,300,000 possible combinations.
Algorithm
gNomic is a Web-based Application developed in Java. For each gene, information is provided in terms of which drugs are substrate, inhibitor, and inducers. For each gene, there is also definition of the genotypes, phenotypes and polymorphisms that define the activity. Each gene also has in the definition if it belongs to Phase I, Phase II, or if is a transport protein and the type, as well as other type of activity. As this will be defining a set of rules in the algorithm, there is a specific way of determining the exceptions to the so-called standard messages, following the directions of the guidelines provided by medicine agencies.
Workflow
A query on the gNomic software begins with entering the medication. Each drug can be entered by either their common name (eg, topiramate) or a commercial name (eg, Topamax).
It contains the commercial names of the Spanish Vademecum and many of the USA and some of France so, outside of Spain, it is recommendable to enter the drugs by their generic or technique name and not by the commercial name.
Non-pharmacological substances can be selected in a “lifestyle habits” tab. The lifestyle list is dynamic, showing only the substances known to interact with the drugs that are currently selected, and includes herb compounds, such as St. John’s wort, as well as proper lifestyle substances such as grapefruit or cigarette smoking. These substances can influence the outcome of a therapy, yet they are likely to be overlooked as they are not part of the actual pharmacologic therapy. In 2018, there are a total of 250 herbs in the software.
The software provides a list of the genes classified as primary biomarkers as candidates for genetic testing, as well as a list of the interactions (drug–drug and drug–lifestyle) that have been detected. If genetic information is available then or is made available at a later point, these results can also be entered and the software may return pharmacogenetics recommendations regarding specific genotypes. Effects on lower relevance biomarkers, or secondary pathways, are hidden by default but can be toggled on by the user. The report messages are generated on the fly; the user can navigate back and forth between the different tabs, changing medication or adding genetic results, and the messages will update in real time without the need to start over.
The interactions can come from clinical reports, but can also be a consequence of the known pharmacokinetic properties of the drugs. Specifically, a substance can be a known inhibitor or inducer of an enzyme that is a primary biomarker of a drug. These kinds of interactions are usually mentioned in drug labels, but normally the precipitating substances are not referred by name but by their effect on pharmacokinetic pathways (eg, avoid CYP3A4 inhibitors). The software works on a semiquantitative model for these kinds of interactions, so the presence of a single substance whose inhibitory potency has been labeled as “strong” would trigger an alert, whereas for weaker strength categories, an increasing number of co-administered inhibitors are required to reach the alert threshold. The notifications are as concise as possible, as we have tried to convey the most crucial information in short messages that can be read and understood quickly. This is because we have conceived the software to be used effectively in a doctor’s office, where time is usually limited.
Disclaimer
The g-Nomic pharmacogenetics interpretation software is a useful tool to be used on the point of care with the sole aim to help the practitioner to take a decision regarding patient’s medication. The last word is for the doctor as the only one who has the complete medical history together with the physical exploration and other relevant data that cannot be interpreted at this time by a machine.
Results
Below it will be described different outputs of the results that can be obtained by the use of g-Nomic interpretation software applied to daily basis medication examples taking into account drug–drug interaction, drug–lifestyle, inhibitions, inductions, pharmacogenetics, and risks associated with medication:
Input data to g-Nomic
Patient’s data can be entered to g-Nomic software using three alternative ways:
- Manual entry
The user enters manually patient’s data. In this case, name and surname or reference number and date of birth are required. After this can be entered for the patient as well as studied genes.
This is functionality is currently only available for internal use. It allows to import pharmacogenetics data from an excel file containing data from several patients, with the following format: patient name, surname, clinic, reference number, gene, and genotype.
This is functionality is currently only available for internal use. It allows to import pharmacogenetics data from an excel file with the following information: rsnumber, gene, position, genotype via upload of this file the system generates a patient record with all the genes found in the file and the corresponding predictive phenotype. A file with 925,000 records takes less than 2 mins to upload and generates the predictive phenotypes for the known genes in g-Nomic.
Case of a drug and a patient with PM phenotype
Aripiprazole is a drug used for schizophrenia metabolized primarily by CYP2D6 and CYP3A4. According to the dosage and administration given in the Drug label from FDA,13 it is recommended administering half of the usual dose in case of CYP2D6 PM patients. When checking g-Nomic pharmacogenetics interpretation software for this eventuality, the relevant information obtained is shown in Figure 2
 | Figure 2 g-Nomic pharmacogenetics report for aripiprazole of a patient showing a PM CYP2D6 phenotype. |
Case of a prodrug
Case of a patient taking Tamoxifen (used for the treatment of breast cancer type ER+). Tamoxifen is considered a prodrug and is converted to its active drug endoxifen via the CYP2D6 and CYP3A4 genes. If the patient is poor metabolizer for CYP2D6 (*5*5 for example), this activation will happen very slowly and there will be a much lower dose of the active ingredient, endoxifen, thus leading to an inefficient treatment. This information can be obtained in the report obtained by using g-Nomic (Figure 3)
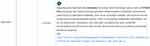 | Figure 3 g-Nomic pharmacogenetics report for a CYP2D6 PM individual prescribed with tamoxifen. |
Case of drug interactions
This occurrence is represented by a medication combining Ticagrelor + Simvastatin. Ticagrelor, with the brand name of Brilinta, it is used as a platelet inhibitory drug, and in common with simvastatin, is metabolized by CYP3A4. Therefore, when the two drugs are administered together, there is a decrease in the metabolism of simvastatin reducing its clearance.14 The AUC of simvastatin is expected an increase to two to threefold, increasing the risk of muscle damage.15 In Figure 4, it can be observed the information given by g-Nomic when consulting about this drug combination.
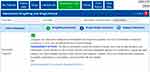 | Figure 4 Drug interaction between simvastatin and ticagrelor: The two drugs are metabolized by CYP3A4. |
Case of a drug in combination with lifestyle
Tamoxifen + calcium
Calcium increases the side effects of hypercalcemia produced by tamoxifen. Therefore, in the case of breast cancer patients treated with tamoxifen, it is advisable not to give calcium, as a lifestyle supplement+, in combination with tamoxifen because life-threatening metabolic complication of flare hypercalcemia develops (Figure 5). In the case of hypercalcemia, gallium nitrate allows restarting tamoxifen safely.16
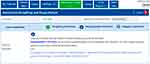 | Figure 5 Interpretion of g-Nomic in the case of drug–lifestyle interaction: Tamoxifen plus calcium. |
Case of a prodrug inhibition
As a representative of this situation, we can take as an example the case of tamoxifen when taken together with paroxetine (Figure 6). As stated in the FDA drug label of paroxetine: Tamoxifen is a prodrug requiring metabolic activation by CYP2D6.17 Inhibition of CYP2D6 by paroxetine may lead to reduced plasma concentrations of an active metabolite (endoxifen) and hence reduced the efficacy of tamoxifen.
 | Figure 6 g-Nomic report in the case of the prodrug inhibition caused by paroxetine on the formation of endoxifen from tamoxifen. |
Case of an inducer drug
This case can be illustrated through the example of the pharmacological response produced by the combination of Paroxetine + Rifampicin. Basically, Rifampicin, also known as rifampin, is an antibiotic used to treat several types of bacterial infections. However, this drug is a potent inducer of certain cytochrome P-450 enzymes including CYP2D6,18 which will accelerate the metabolism of paroxetine, and in consequence, it could lead to therapeutic failure (Figure 7). To prevent this effect, drugs metabolized by P-450 enzymes may require adjustments when rifampin is administered concomitantly.
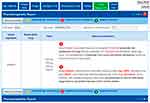 | Figure 7 g-Nomic report in case of the drug induction of several cytochrome P-450 enzymes produced by Rifampin which may low plasma levels of drugs such as Paroxetine. |
Case of risks associated with medication
There can be many risks associated with medication which are illustrated here by the case of administrating drugs such as: Metformin for treating type 2 diabetes, Quetiapine to treat schizophrenia, Fluticasone used to relieve allergic and non-allergic nasal symptoms, Salmeterol as a Bronchodilator Agent, Valsartan widely used to treat high blood pressure or Haloperidol as an antipsychotic. In the case of Haloperidol, Quetiapine and Salmeterol: These drugs may prolong the QT interval and/or induce Torsades de Pointes (Figure 8). If one of the prescribed drugs is an antiarrhythmic, it might be problematic to discern whether the individual developed the arrhythmia due to his heart condition or due to the medication.
 | Figure 8 g-Nomic report showing risks associated with specific medications. |
Discussion
The vast information available nowadays regarding pharmacogenetics needs to be implemented in the clinical practice to accomplish the objective of providing a personalized therapy which is efficient and secure.19 In this respect, several organizations and consortiums have been created worldwide with the aim of achieving safe and effective pharmacological treatments: PGRN, PharmGKB, CPIC, and D.I.W.G among others. Accordingly, and with the aim of making prescription more secure to both, doctor and patient, we have developed a pharmacogenetics software, g-Nomic, which is described and presented in this article.
Performing a drug prescription at first attempt is a complex situation and both, practitioners and pharmacists take a lot of responsibility especially considering the very short time due to lack of time in medical consultations. In this regard, knowing the genetic polymorphisms of a patient helps to adjust the dose for the intended pharmacological effect and whether it would be needed to seek a therapeutic alternative. In general, patients are polymedicated and the effect of various drugs administered together may be different (produce ineffectiveness or toxicity) compared to when they are administered alone. Frequently, even if a person could take specific drugs individually according to their genetic background, the combination of drugs themselves could inhibit or induce the other producing a drug adverse reaction. Almost, all drug treatments have the risk of not being suitable for that particular person, and traditional medicine practice uses the try-and-see method until the patient has a successful treatment. Unfortunately, the try-and-see method sometimes does not work because the patient already had a fatal effect. In g-Nomic, we have added customized lists of possible interacting lifestyle per each drug so that physician should only click to those confirmed by the patient.
To successfully apply pharmacogenetics and make a prescription safely and accurately, many parameters should be taken into consideration, such as drug–drug interactions, drug–lifestyle, inhibitions and inductions, and dose variation according to patient studied genes. All this information can only be interpreted together, using a pharmacogenetics interpretation software like g-Nomic.
There are other pharmacogenomic software tools such as PharmCAT, that extracts all CPIC guideline gene variants from a genetic database, interpret the variant alleles, and generates a report,20 GDA Pharmacogenetics, which allows integrative study of drug response data, mutations, and gene expression profiles focusing on a panel of more than 70 cancer cell lines treated with 50,800 compounds,21 or Virtual Pharmacist, a web tool that interpret personal genome for the impact of genetic variation on drug response.22 However, even if these are very valuable tools, they basically focus of genetic variation or are very specific oriented, whereas gNomic pharmacogenetic interpretation crosses information concerning prescribed drugs, patient’s genetic variations to give a personalized report with all necessary information: drug interactions, drug with lifestyle, inhibitions, inductions, and dose variation according to the patient’s genes. G-Nomic is designed as a practical tool to be used in clinical practice.
Cardiology is one of the specialties where pharmacogenetics has a more direct impact. Taking anticoagulants as an example, a patient starting on warfarin (acenocoumarol), can have fatal side effects when prescribed on a standard initial dose. On the other hand, the new oral anticoagulants, said to be safer and more reliable than warfarin, do not have a lab test to measure if the patient has correct drug levels, and certain patients could have an impaired extrusion protein function (P-gp, coded by the gene ABC1B1) that in long term could lead to internal bleeding. In both cases, the usage of pharmacogenetics could have prevented these adverse events. When prescribing a patient with an antiplatelet drug, such as clopidogrel, because of insufficient clopidogrel-induced platelet inhibition, since clopidogrel requires hepatic bioactivation via several cytochromes, the patient could be at risk. This effect could also be produced by polymedication. Thus, prediction is essential to prescribe safer and avoid predictable side effects. Furthermore, in the case of hypertension, the number of drugs that could be prescribed is large. In this particular therapy, it is crucial to know how if poly-pharmacy will inhibit or induce the medication, leading to inefficiency or toxicity risk.
Applied pharmacogenetics can indeed avoid many emergency cases, save lives and money. But to correctly apply pharmacogenetics, interpretation software is needed since there are too many variables to consider, including the information provided in modern drug labels regarding possible inhibitors or inducers.
The future of medical practice cannot be envisioned without taking into account pharmacogenetics. However, in reality, it is also very difficult to imagine all medical doctors applying pharmacogenetics. In addition, patient can buy drugs over the counter or take food and medical plants that could cause an adverse reaction. Therefore, the puzzle gets complex when it comes to making a full interpretation. Pharmacogenetics is still very new, and it will take time to be fully implemented. A possible near future would be to create a new service of “drug surveillance” to be offered in hospitals. Trained pharmacists, pharmacologist, or physicians, who will act as an interface between the patient and all his general practitioners and specialists, will lead this new service. Doctors would suggest their patients who experienced an ADR visiting the “drug surveillance unit”. At this new “drug surveillance” service, medication will be checked from a personalized point of view, and they will have the capacity to consult with different doctors to seek the best suitable treatment. At this point, the aid of a pharmacogenetics interpretation software like g-Nomic would be of great help. More in the long term, we cannot forget that we are in the digital century and for sure in years to come artificial intelligence will be resolving most of all problems related to drug adverse reactions. However, until then, actions should be taken to prevent patients from experiencing side effects with their prescribed medication.
It is entirely in the hands of hospitals, elderly houses, and medical practitioners on how they would like to treat their patients: like in the last century using the try-and-see method or with the aid of pharmacogenetics and a pharmacogenetics interpretation softwares like g-Nomic. The software presented in this article addresses this issue directly by integrating information from different sources and providing the corresponding alerts in a process that is quick, simple, and concise.
Conclusions
Pharmacogenetics is still a young science, but it should be applied in benefit of both doctors and patients. Although important organizations perform research and publish recommendations in the pharmacogenetics field, this guidance has later no impact, because prescribers state that there are not protocols or that it is still too early to make any decisions in this direction. In addition, it is clear that many factors occur in combination when drugs are prescribed. In this regard, a tool like g-Nomic could be a software providing a necessary aid. Nevertheless, it has to be taken into consideration that g-Nomic cannot replace medical doctors, as they are the only ones handling all the medical and clinical data from the patient.
Disclosure
The name given to the pharmacogenetics interpretation software, g-Nomic, has been registered and has a copyright registered in the EUIPO with file number 012876868. Since g-Nomic is a software, it cannot be patented, but the screens do have safe stamp copyright licensed under the company brand of EUGENOMIC®. Marc Cendros, Juan Sabater-Tobella and Ana Sabater work for the company EUGENOMIC, but g-Nomic pharmacogenetics software has been built according to guidelines and publications without any possible conflict of interest. The authors report no other conflicts of interest in this work.
References
1. Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta- analysis of prospective studies. J Am Med Assoc. 1998. doi:10.1001/jama.279.15.1200
2. Carrasco-Garrido P, De Andrés LA, Barrera VH, De Miguel GÁ, Jiménez-García R. Trends of adverse drug reactions related-hospitalizations in Spain (2001–2006). BMC Health Serv Res. 2010. doi:10.1186/1472-6963-10-287
3. Charlesworth CJ, Smit E, Lee DSH, Alramadhan F, Odden MC. Polypharmacy among adults aged 65 years and older in the United States: 1988–2010. J Gerontol A Biol Sci Med Sci. 2015;70:989–995. doi:10.1093/gerona/glv013
4. Jain S, Jain P, Sharma K, Saraswat P. A prospective analysis of drug interactions in patients of intensive cardiac care unit. J Clin Diagn Res. 2017. doi:10.7860/JCDR/2017/23638.9403
5. Jain KK. Applications of AmpliChip CYP450. Mol Diagn. 2005;9:119–127.
6. Swen, J. J., et al. Pharmacogenetics: from bench to byte – an update of guidelines. Clin Pharmacol Ther. 2011;89:662–673. doi:10.1038/clpt.2011.34
7. Caudle, K. E., et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab. 2014;15:209–217. doi:10.2174/1389200215666140130124910
8. Whirl-Carrillo, M., et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–417. doi:10.1038/clpt.2012.96
9. Lynch T, Price A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am Fam Physician. 2007. doi:10.1046/j.1365-2125.1999.00073.x
10. Tatro DS. Drug Interaction Facts 2014 : The Authority on Drug Interactions. Philadelphia: Lippincott Williams & Wilkins; 2013.
11. Hansten PD. TOP 100 Drug Interactions : A Guide to Patient Management. Clearwater, FL: H & H PUBLISHING CO; 2014.
12. Phansalkar, S., et al. Drug–drug interactions that should be non-interruptive in order to reduce alert fatigue in electronic health records. J Am Med Informatics Assoc. 2013;20:489–493. doi:10.1136/amiajnl-2012-001089
13.
14.
15. Mrotzek SM, Rassaf T, Totzeck M. Ticagrelor leads to statin-induced rhabdomyolysis: a case report. Am J Case Rep. 2017;18:1238–1241. doi:10.12659/AJCR.905974
16. Arumugam GP, Sundravel S, Shanthi P, Sachdanandam P. Tamoxifen flare hypercalcemia: an additional support for gallium nitrate usage. J Bone Miner Metab. 2006;24:243–247. doi:10.1007/s00774-005-0678-4
17.
18.
19. Sales GS, Tobella JS. Nutrigenética y nutrigenómica. in Medicina Personalizada Posgenómica. Conceptos Prácticos Para Clínicos. 2010. doi:10.1016/B978-84-458-2025-4.00008-5
20. Klein TE, Ritchie MD. PharmCAT: a pharmacogenomics clinical annotation tool. Clin Pharmacol Ther. 2018;104:19–22. doi:10.1002/cpt.v104.1
21. Caroli J, Sorrentino G, Forcato M, Del Sal G, Bicciato S. GDA, a web-based tool for genomics and drugs integrated analysis. Nucleic Acids Res. 2018;46:W148–W156. doi:10.1093/nar/gky434
22. Cheng, R., et al. Virtual pharmacist: a platform for pharmacogenomics. PLoS One. 2015;10:e0141105. doi:10.1371/journal.pone.0120491
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

