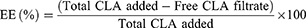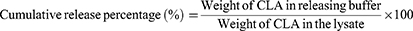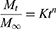Back to Journals » International Journal of Nanomedicine » Volume 19
Kaempferia parviflora Extracellular Vesicle Loaded with Clarithromycin for the Treatment of Helicobacter pylori Infection
Authors Nemidkanam V , Banlunara W, Chaichanawongsaroj N
Received 8 November 2023
Accepted for publication 21 February 2024
Published 27 February 2024 Volume 2024:19 Pages 1967—1983
DOI https://doi.org/10.2147/IJN.S444686
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor R.D.K. Misra
Variya Nemidkanam,1 Wijit Banlunara,2 Nuntaree Chaichanawongsaroj3
1Department of Clinical Chemistry, Graduate Program in Clinical Biochemistry and Molecular Medicine, Faculty of Allied Health Sciences, Chulalongkorn University, Bangkok, 10330, Thailand; 2Department of Pathology, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, 10330, Thailand; 3Department of Transfusion Medicine and Clinical Microbiology, Research Unit of Innovative Diagnosis of Antimicrobial Resistance, Faculty of Allied Health Sciences, Chulalongkorn University, Bangkok, 10330, Thailand
Correspondence: Nuntaree Chaichanawongsaroj, Department of Transfusion Medicine and Clinical Microbiology, Research Unit of Innovative Diagnosis of Antimicrobial Resistance, Faculty of Allied Health Sciences, Chulalongkorn University, Bangkok, 10330, Thailand, Tel +66 838199988, Email [email protected]
Purpose: Kaempferia parviflora extracellular vesicles (KPEVs) have been reported as promising nanovesicles for drug delivery. This study aimed to load clarithromycin (CLA) into KPEVs (KPEVS-CLA) and determine the physical properties, drug-releasing efficiency, gastric cell uptake, anti-H. pylori activities, and anti-inflammatory responses in comparison with free CLA and KPEVs.
Methods: The size and surface charge of KPEVs-CLA were evaluated using dynamic light scattering and visualized using a transmission electron microscope. The encapsulation efficiency (EE%), loading capacity (LC%), and drug release of KPEVs-CLA were examined using HPLC. Anti-H. pylori growth and anti-adhesion were evaluated. IL-8 gene expression, NF-κB signaling proteins, and anti-inflammatory profiles were examined using qRT-PCR, Western blotting, and Bio-Plex immunoassay, respectively. Anti-chemotaxis was then examined using a Transwell assay.
Results: KPEVs-CLA were intact and showed a negative surface charge similar to that of KPEVs. However, slightly enlarged KPEVs were observed. CLA was successfully loaded into KPEVs with EE of 93.45% ± 2.43%, LC of 9.3% ± 3.02%. CLA release in the PBS and gastric mimic buffer with Fickian diffusion (n ≤ 0.43) according to Korsmeyer-Peppas kinetic model (R2=0.98). KPEVs-CLA was localized in the gastric cells’ cytoplasm and perinuclear region. Anti-H. pylori growth and anti-H. pylori adhesion of KPEVs-CLA were compared with those of free CLA with no cytotoxicity to adenocarcinoma gastric cells. KPEVs-CLA significantly reduced IL-8, G-CSF, MIP-1α, and MIP-1β levels. Moreover, KPEVs-CLA showed a superior effect over CLA in reducing G-CSF, MIP-1α, and NF-κB phosphorylation and monocyte chemotactic activities.
Conclusion: KPEVs serve as potential carriers of CLA. They exhibited a higher efficiency in inhibiting gastric cell inflammation mediated by H. pylori infection than free CLA. The establishment of KPEVs-CLA as a nanodrug delivery model for H. pylori treatment could be applied to other plant extracellular vesicles or loaded with other cancer drugs for gastric cancer treatment.
Keywords: extracellular vesicle, clarithromycin, Kaempferia parviflora, Helicobacter pylori, inflammation
A Letter to the Editor has been published for this article.
Introduction
Kaempferia parviflora (KP) or Krachaidam, a Thai medicinal plant, has been used locally for treating gastrointestinal tract diseases since ancient times. KP rhizome, also known as black ginger, contains anti-microorganism, anti-inflammatory, anticancer, antiallergic, anti-osteoarthritis, antioxidant, cardioprotective, neuroprotective, sexual-enhancing, and physical-enhancing activities.1–6 The major active compounds are methoxyflavones and their derivatives.6 Our previous study demonstrated the inhibitory activity of crude KP extract against Helicobacter pylori, an etiological agent of gastric cancer.7,8 Additionally, KP significantly reduced H. pylori internalization into gastric cells, inflammatory response, and leukocyte chemotaxis.9
Plant-derived extracellular vesicles (PDEVs) are natural nanoparticles (approximately 50–1000 nm) that are biologically released from plant cells and accumulate within the apoplastic space.10 These PDEVs deliver nutrients, growth factors, and bioactive chemical compounds to different plant parts. PDEVs have recently become a novel alternative nanoparticle in the study of nanomedicine because they are biologically friendly and cost-effective. PDEVs can be loaded with drugs and miRNA or can modify the extracellular vesicles (EVs) surface like an engineered synthetic nanoparticle. The attractive advantages of PDEVs over synthetic nanoparticles are their high biocompatibility and low toxicity due to their natural origin.11,12 The active compounds from medicinal plants were spontaneously packed into PDEVs, and their therapeutic properties have been extensively revealed.13–15 Based on our previous findings, Kaempferia parviflora extracellular vesicles (KPEVs) were successfully isolated from fresh KP rhizomes using differential and sucrose density gradient centrifugation, which is the most common method for isolating PDEVs.16,17 The characteristics of KPEVs demonstrate promising drug delivery features that are compatible with treating gastrointestinal diseases, such as its nanosize (200–300 nm) with negative surface charge (14.7 ± 3.61 mV), internalization into gastric cells with acid tolerance, dimethoxyflavone packaging, and low cytotoxicity.18
H. pylori is an initiation factor of gastric diseases (eg, gastritis, gastric ulcers, and gastric cancer).19,20 The infection modulates the immune response by NF-κB pathway-related receptors, thereby releasing proinflammatory cytokines and chemokines.19,21 Subsequently, the released chemokines recruit more inflammatory cells to the infected area, enhancing tissue inflammation. Chronic gastric inflammation further develops into gastric cancer. In prolonged infection, reactive oxygen species induced by oxidative stress also promote host DNA damage. Currently, antimicrobial drugs are the most common and effective therapy for treating H. pylori infection. An up-to-date treatment guideline recommends clarithromycin (CLA)-based triple therapy as the first-line treatment.22 Unfortunately, CLA has extreme hydrophobicity and low solubility in aqueous solution. Hence, to improve drug systemic drug biodistribution, polymers have been combined with CLA to improve their solubility in body fluids. The polymers have the side effect of elevating the viscosity of the gastric mucus, thereby impairing the efficiency of drug penetration into the H. pylori niche in the mucus layer.23 On the other hand, CLA-loaded amine-functionalized mesoporous silica nanoparticles (MSNs-NH2) were found to distribute and accumulate in the liver, kidney, and lung tissues of NMRI mouse models, enhancing drug delivery to these tissues efficiently.24 Additionally, CLA-loaded nanostructured lipid carriers exhibited mucoadhesive properties that could potentially increase the ocular bioavailability of CLA.25
Thus, this study aimed to use KPEVs as a vehicle for CLA delivery. The physical properties of KPEVs-CLA and drug release profiles were compared with those of free CLA. The feasibility of passive loading of CLA into KPEVs (KPEVs-CLA) for H. pylori treatment was examined, including cytotoxicity, anti-H. pylori activities, and anti-inflammatory properties. These findings could provide a theoretical basis for an alternative strategy for treating H. pylori infection, and KPEVs could be applied as a promising therapeutic approach to other infectious and inflammatory diseases.
Materials and Methods
Materials
Clarithromycin and sucrose were purchased from Glentham Life Sciences (Corsham, UK). Phosphate buffer saline (PBS) was purchased from GE Healthcare (Chicago, IL, USA). Acetonitrile HPLC grade and modified Giemsa stains were purchased from Merck Millipore (Burlington, MA, USA). Recombinant MCP-1 was purchased from ImmunoTools, Germany.
KPEVs Isolation and Purification
KPEVs were isolated according to our previous method,18 which was modified from Sung et al.26 Fresh KP rhizomes were ordered from a local farm at Khao Kho, Phetchabun, Thailand. The plant material was identified by Mrs. Parinyanoot Klinratana of the Department of Botany, Faculty of Science, Chulalongkorn University, Bangkok, Thailand, and a specimen voucher (number A17644) was deposited at the department’s herbarium. No additional approvals from institutional/local regulations were required to conduct research with plant material. The KP rhizomes were washed and pressed to obtain the KP juice. The supernatant was then collected and centrifuged using Sorvall Legend XT/XF centrifuge under the following conditions: 1000 × g, 2000 × g, 4000 × g, and 10,000 × g for 10, 20, 30, and 60 min, respectively, followed by ultracentrifugation at 150,000 × g for 2 h (Sorvall WX100 + ultracentrifuge, T647.5 rotor, Kadj = 201). The 8%/30%/45% sucrose density gradient was performed at 150,000 × g (Hitachi Koki himac CS150NX, S55A rotor, Kadj = 73.4) for 1.5 h. The protein concentration of KPEVs was measured using the Bradford protein assay (Bio-Rad Laboratories, CA, USA), which was used to represent the KPEVs concentration.
Preparation and Quantification of KPEVs Loaded with CLA (KPEVs-CLA)
CLA stock (20 mg/mL) was prepared in acetone and then diluted in 1X PBS to final concentrations of 10 µg/mL, 100 µg/mL, and 1000 µg/mL. The amount of KPEVs and CLA was prepared in the ratio between the total protein of KPEVs (µg) and the total weight of CLA (µg). Passive loading of CLA into KPEVs was performed by mixing each CLA concentration with the KPEVs at ratios (KPEVs: CLA) 1:1, 1:10, and 1:100 (w/w). The mixtures were incubated at room temperature for one hour on a rotator. This incubation method was modified from that described by Yang et al.27 Free CLA was separated from the KPEVs-CLA fraction by centrifugation using a Nanosep 10 kDa MWCO filter (Pall, NY, USA) at 10,000 × g for 20 min. KPEVs-CLA was then washed twice with 1X PBS and resuspended in 1X PBS. KPEVs-CLA was filtered sterile through a 0.45-µm filter and stored at −80°C until use.
Encapsulation Efficiency and Loading Capacity
The amount of CLA loaded in KPEVs was determined using HPLC and calculated based on CLA standard curves. The isocratic run was performed using an acetonitrile mixture and 0.1-M potassium dihydrogen phosphate buffer (55:45 v/v), pH 4.4, as the mobile phase with a flow rate of 0.6 mL/min and temperature of 40°C. A Purospher reverse phase column C18 with a 5-µm particle size of 4.0 × 250 mm (Merck Millipore, USA) was used as the solid phase. KPEVs-CLA lysate was prepared by dissolving in acetonitrile at 4°C overnight, then centrifuged at 7000 × g for 10 min. The KPEVs-CLA lysate and free CLA fractions were first concentrated using SpeedVac vacuum concentrators (Thermo Scientific, MA, USA) and further injected into the HPLC system. Each sample injection volume was set at 20 µL, and CLA was detected using UV-VIS at 210 nm. CLA standards (1 µg/mL–1 mg/mL) were prepared in the mobile phase to generate the standard curve. The peak area represents the CLA concentration in each sample and standard. Encapsulation efficiency (EE%) and loading capacity (LC%) were calculated using the following formulas:
Characterization of KPEVs-CLA by DLS and Transmission Electron Microscopy (TEM)
The particle size distribution and surface charge of KPEVs-CLA, prepared at a ratio of 1:100, were evaluated using a Zetasizer Nano ZS dynamic light scattering (Malvern Instruments, Malvern, UK) with a 4-mW He–Ne laser light source of 633 nm, 25°C, and 173° angle. The size distribution, polydispersity index (PDI), and zeta potential were calculated using Zetasizer software. The parameters of PBS (refractive index at 1.330, viscosity at 0.8882 cP, and dielectric constant at 79.0) were set in a sample dispersant setting. TEM imaging was performed using a JEM-1400 electron microscope (JEOL, Japan) to observe KPEVs-CLA morphology.
KPEVs-CLA Lipophilic Staining
KPEVs (500 µg/mL) were stained with Dil (1,1′-Dioctadecyl-3,3,3′,3′-Tetramethylindocarbocyanine Perchlorate) at a final concentration of 1 µg/mL and then incubated at 37°C in the dark for 20 min. The labeled KPEVs were washed with 1X PBS and spun through a Nanosep 10 kDa MWCO filter (Pall, NY, USA) at 10,000 × g for 15 min to remove unbounded fluorescence dye. Labeled KPEVs were prepared fresh or maintained at 4°C for cellular localization assay.
KPEVs-CLA Cellular Localization
Adenocarcinoma gastric cells (AGS cells, brought from ATCC, VA, USA) were cultured in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (Hyclone, UT, USA) at 37°C in 5% CO2 and 80% humidity. Cells (8 × 104 cells/well) were seeded on sterile coverslips, placed in a 24-well plate, and cultured in complete RPMI for 24 h. Each coverslip was washed with 1X PBS before treatment. Subsequently, 50-µg/mL Dil-labeled KPEVs-CLA was added to RPMI and incubated at 37°C for 12 h. Cells were washed with 1X PBS, fixed with 4% paraformaldehyde, and the nuclei were stained with DAPI. Labeled KPEVs-CLA in gastric cells were imaged in z-stack mode using a confocal laser scanning microscope (Zeiss, Germany) at 549/565 nm (Dil) and 358/461 nm (DAPI).
In vitro Release Efficiency of Loaded Clarithromycin
The CLA release study was conducted using dialysis under sink condition. Simulated gastric fluid (SGF) buffer (pH 1.2) and 1X PBS (pH 7.4) were used as releasing buffers to study the efficiency of CLA release under gastric-like and neutral conditions, respectively. The drug-releasing method was modified from that described by Zhang et al.12 KPEVs-CLA (100-µg CLA) was loaded into a D-Tube Dialyzer tube MWCO 10 kDa (Sigma-Aldrich, MO, USA), placed into 10-mL each releasing buffer, and incubated in a shaking incubator at 100 rpm, 37°C. Then, 100-µL each releasing buffer was collected periodically at 0, 1, 2, 4, 6, 8, 24, 48, and 72 h and replaced with the same volume of fresh buffer at each time point. To ensure the analytical sensitivity, the remaining KPEVs-CLA in the dialysis tube and releasing buffer were further concentrated using a SpeedVac vacuum concentrator (Thermo Scientific, MA, USA). The samples were then resuspended in acetonitrile and injected into HPLC. The cumulative release percentage (%) was calculated using the following formula:
The release mechanism of CLA from KPEVs was evaluated using the Korsmeyer-Peppas kinetic mathematical model as this model is applicable for the unknown release mechanism.
where, Mt is the amount of drug released at time t, M∞ is the amount of drug released after time ∞, K is a kinetic constant, n is a release exponent that indicates the mechanism of drug release from the KPEVs. The value of n signifies the mechanism of drug release; a value of 0.43 or below 0.43 for a sphere suggests diffusion-controlled release (Fickian diffusion). A value between 0.43 and 0.85 suggests anomalous diffusion (non-Fickian diffusion), while a value of 0.85 suggests relaxation-controlled release. A value higher than 0.85 indicates super transport.28
H. pylori Growth Inhibition Assay and Minimum Bactericidal Concentrations (MBC) of KPEVs-CLA
The effect of KPEVs-CLA on H. pylori growth was determined using a broth microdilution assay and a resazurin indicator. Three-day-old cultures of H. pylori were suspended in a Brain Heart Infusion (BHI) broth supplemented with 10% FBS and adjusted to 1 × 108 cfu/mL using McFarland densitometers (Biosan, Riga, Latvia). CLA (20 mg/mL) and KPEVs-CLA stock (100-µg CLA) were diluted in BHI broth to final concentrations at 0.015–0.12 µg/mL. The diluted solutions were then added to a 96-well plate, followed by inoculation with 10 µL (1 × 106 cfu) of H. pylori. The plate was then incubated for 24 h under microaerophilic conditions (Anaero Pack-MicroAero, Mitsubishi gas chemical, Japan). CLA at 0.25 µg/mL was used as a control, according to CLSI guidelines. Resazurin fluorescent dye (6 mg/mL) was then added to each well and incubated for 3–4 h. The fluorescence intensity was measured at 550/600 nm. Percentage growth inhibition and 50% inhibitory concentration (IC50) were calculated.
To determine the minimum bactericidal concentrations (MBC) of KPEVs-CLA, H. pylori was treated with CLA or KPEVs-CLA for 24 h using a broth microdilution assay. Then, 10-µL culture in each well was grown on BHI agar supplemented with 7% (v/v) sheep blood for three days. The lowest concentration that exhibited no H. pylori growth was considered an MBC.
H. pylori Adhesion Assay
Urease assay was used to detect H. pylori adherence as described by Tharmalingam et al.29 AGS cells were seeded onto a 96-well plate at 1 × 104 cells/well in antibiotic-free RPMI medium with 10% FBS for 24 h. Three-day-old H. pylori were prepared in an RPMI medium and co-incubated with AGS cells (multiplicity of infection (MOI) = 1:100). CLA or KPEVs-CLA stocks were diluted in RPMI to a final concentration of 0.01–0.48 µg/mL and further co-incubated for two hours. RPMI and KPEVs alone were used as untreated and vehicle controls. The cells were then washed thrice with 1X PBS, and adherent H. pylori was quantified by adding 100-μL BBL Urease broth (Becton Dickinson, MD, USA) to each well then incubated at 37°C for two hours. The absorbance was measured at 560 nm using a microplate reader (BioTek, USA). The percentage of H. pylori adhesion was calculated and compared with the media control.
Cytotoxicity Assay
Thiazolyl Blue Tetrazolium Blue (MTT) assay was performed to assess cell viability. AGS and THP-1 (human leukemia monocytes) cell lines were cultured in an RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (Hyclone, UT, USA) at 37°C in 5% CO2 and 80% humidity. THP-1 cells were generously provided by Asst. Prof. Viroj Boonyaratanakornkit, Department of Clinical Chemistry, Faculty of Allied Health Sciences, Chulalongkorn University. Both cells were seeded onto a 96-well plate at a seeding density of 1 × 104 cells/well and 1 × 105 cells/well, respectively. The CLA or KPEVs-CLA were diluted to 0.01–0.12 µg/mL with the culture medium and added to each well. PBS and KPEVs (0.12 µg/mL) were added to the untreated and vehicle control wells. AGS cells were co-incubated with each treatment for 6, 12, and 24 h. THP-1 cells were treated only at 24 h to further evaluate chemotaxis activity. MTT solution (5 mg/mL) was added and incubated for four hours in the dark. The cells were permeabilized in 100-µL 10% (w/v) SDS to solubilize the formazan crystals. The absorbance was measured at 570 nm using a microplate reader (BioTek, USA). Viable cell percentage was calculated and compared with the untreated control cells.
Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
IL-8 as a proinflammatory cytokine marker gene was quantified using qRT-PCR. AGS cells (1 × 105 cells/well) in a 6-well plate were co-cultured with 3-day-old H. pylori (MOI = 1:100). CLA or KPEVs-CLA at 0.01–0.12 µg/mL prepared in RPMI were added and incubated for 12 h. RPMI and KPEVs alone were used as untreated and vehicle controls. Cells were lysed and extracted using GENEzol (Geneaid Biotech, Taiwan) according to the manufacturer’s instructions. mRNA was converted into cDNA using Maxime RT PreMix Oligo(dT)15 Primer (iNtRON Biotechnology, South Korea). cDNA was amplified using SYBR iTaq DNA Polymerase reagent (Bio-Rad Laboratories, CA, USA) with a CFX96 real-time quantitative PCR system (Bio-Rad Laboratories, CA, USA). All primers were purchased from U2Bio, South Korea, and their sequences were as follows: IL-8-F, 5′-TCC AAA CCT TTC CAC CCC AA-3′; IL-8-R, 5′- ACT TCT CCA CAA CCC TCT GC-3′; GAPDH-F, 5′-CTG ACT TCA ACA GCG ACA CC-3′; GAPDH-F, 5′-GTG GTC CAG GGG TCT TAC TC-3′. The thermal cycling program was set as follows: pre-denature at 95°C for 5 min, denature at 95°C for 30s, annealing/extension at 58°C for 30s, and repeated for 40 cycles. The relative mRNA expression level was determined through the cycle threshold (Ct) normalized against GAPDH using the 2−ΔΔCt formula.
Western Blotting of the NF-κB Signaling Pathway
AGS cells (1 × 106 cells/well) were seeded in a 6-well plate and incubated for 24 h in complete RPMI without antibiotics. Cells were induced with 3-day-old H. pylori (MOI = 1:100), then CLA or KPEVs-CLA at 0.06 µg/mL prepared in RPMI were added and further incubated for 1 h. RPMI and KPEVs alone were used as untreated and vehicle controls. AGS cells were lysed, and total protein was determined using the Bradford assay. Each sample was subjected to 5%/10% SDS-PAGE and transferred to a PVDF membrane at 100 V for 70 min. The membrane was blocked with 5% BSA/TBS-T for one hour and incubated overnight at 4°C with the following primary antibodies: p-NF-κB p65, NF-κB p65, and GAPDH (Cell signaling technology, USA). The membrane was washed thrice with 1x TBS-T, for ten minutes. HRP-conjugated anti-mouse secondary antibody was added, incubated for 60 min, and washed with 1x TBS-T. Protein signals were developed using the Amersham ECL prime Western blotting detection kit (GE Healthcare, USA). Protein bands were captured using Amersham ImageQuant 800 (Cytiva, MA, USA) and quantified using ImageJ software.
Bio-Plex Multiplex Immunoassay
To study the anti-inflammatory profile of KPEVs-CLA, cytokine levels were determined using Bio-Plex Pro Human Cytokine 27-plex (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s instructions. The AGS cells were co-incubated with each treatment at 0.06 µg/mL for 6, 12, and 24 h as described above. The cultured media was collected and centrifuged at 700 x g for five minutes in a refrigerated centrifuge. The quantity of all 27 cytokines (FGF basic, Eotaxin, G-CSF, GM-CSF, IFN-γ, IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12, IL-13, IL-15, IL-17A, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF-BB, RANTES, TNF-α, and VEGF) were analyzed using Bio-Plex Data Pro Software (Bio-Rad, USA).
Monocyte Chemotaxis Assay
Antimonocyte chemotaxis activity was demonstrated using Transwell assay.9 A transwell with a 5-µm polycarbonate filter (Corning, NY, USA) was inserted into a 24-well cell culture plate. THP-1 cells were washed and resuspended in an RPMI medium without FBS. The cells (105 cells) were co-cultured with CLA or KPEVs-CLA at 0.06 µg/mL in the upper chamber of the Transwell. RPMI and KPEVs were used as untreated and vehicle controls, respectively. In the lower chamber, RPMI was added for non-chemotaxis control, and RPMI was supplemented with 10-ng/mL MCP-1 as a chemoattractant for every test well. Subsequently, the transwells were incubated at 37°C for 1.5 h and washed twice with PBS. Then, ice-cold methanol was added to fix the cell for 10 min, stained with modified Giemsa for 30 min, and imaged for ten fields using a light microscope with a 20X objective lens. Chemotaxis cells were counted using ImageJ software.
Statistical Analysis
The replicated data are presented as a mean ± SD. Graph plotting, heatmap generation, and statistical testing were performed using GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA). The difference between the control and test groups was calculated using one-way ANOVA followed by Bonferroni post test. Statistical significance was considered for p ≤ 0.05, p ≤ 0.01, or p ≤ 0.001. For heatmap generation, average cytokine concentrations were initially normalized using z-score normalization to scale cytokine levels within each cytokine and incubation time.
Results
Preparation of KPEVs-CLA, Physical Characterization, and Drug Release
At an equal amount of KPEVs and CLA (KPEVs: CLA = 1:1), the highest EE% of 92.10% ± 3.75% was found (Figure 1A). However, it gave the lowest LC% at only 1.60% ± 0.46%. CLA at 1:10 and 1:100 reduced EE% to 57% ± 11.70% and 28.81 ± 1.36%, whereas LC% increased to 9.3% ± 3.02% and 93.45% ± 2.43%, respectively. Therefore, the KPEVs: CLA ratio of 1:100 with highest LC% was chosen for KPEVs-CLA for further DLS, TEM, and biological experiments.
The integrity, size distribution, and surface charge of KPEVs-CLA, at a ratio of 1:100, were analyzed using DLS. The size range of KPEVs-CLA was 352.2 ± 22.83 nm with a polydispersity index (PDI) of 0.3 ± 0.069. The surface charge of KPEVs-CLA was negative, represented by a zeta potential of −14.8 mV ± 0.56 mV (Figure 1C). The TEM image showed the corresponding size and intact KPEVs after CLA loading (Figure 1B). The CLA release kinetics were time-dependently sustained in neutral pH (PBS, pH 7.4) and acidic buffer (stimulated gastric buffer (SGF), pH 1.2). Rapid drug release in the first hour was observed at an exponential rate (37.67% ± 8.36% and 24.38% ± 10.72% in PBS and SGF, respectively) (Figure 1D). The releasing rate was slower and constant during 8–48 h and then elevated again during 48–72 h. CLA release under gastric-like conditions (25.96% ± 9.97%) was 1.67-fold slower than that in 1X PBS (43.36% ± 10.65%) at 2 h. Additionally, the drug release profiles fit the Korsmeyer-Peppas kinetic model, indicating Fickian diffusion in both PBS and SGF media, as the n values were less than 0.43 (Table S1).
Localization of KPEVs-CLA in Gastric Cells
Dil-labeled KPEVs and KPEVs-CLA exhibited the same localization (Figure 2A). Accordingly, z-stack 3D imaging demonstrated that KPEVs-CLA (Figure 2B) could be localized in the cytoplasm compartment of AGS cells close to the DAPI-stained nuclear membrane (blue) as KPEVs.
Anti-H. pylori Growth of KPEVs-CLA
KPEVs-CLA exhibited a similar dose-dependent inhibition effect as free CLA (Figure 3A). Anti-H. pylori growth was significantly observed at the lowest CLA concentration of 0.01 µg/mL, and more notably at 0.03–0.12 µg/mL. The inhibition effect of KPEVs-CLA was significantly detectable at 0.03–0.12 µg/mL. The half-inhibitory concentrations (IC50) of CLA (0.029 ± 0.002 µg/mL) and KPEVs-CLA (0.032 ± 0.003 µg/mL) were similar. Moreover, the MBC of CLA and KPEVs-CLA were 0.06 µg/mL.
Anti-H. pylori Adhesion Activities of KPEVs-CLA on Gastric Cells
After two hours of co-incubation, CLA and KPEVs-CLA showed a significant concentration-independent reduction effect on H. pylori adhesion (Figure 3B). At the lowest concentration (0.01 µg/mL), KPEVs-CLA and CLA reduced H. pylori adhesion to 64.80% ± 11.72% (1.52-fold decrease) and 82.12% ± 11.40% (1.20-fold decrease), respectively. The highest anti-adhesion effect was observed with KPEVs-CLA at 0.48 µg/mL (2.46-fold decrease) and CLA at 0.24 µg/mL (1.79-fold decrease). Additionally, KPEVs alone showed a slight inhibition effect, approximately a 1.16-fold decrease.
Cytotoxic Effects of KPEVs-CLA on Gastric Cells
CLA and KPEVs-CLA at 0.01–0.12 µg/mL, which are in the MBC range, were tested against AGS cells. The cell viability percentage of CLA and KPEVs-CLA treatments was between 105.92% and 91.57% at 6–24 h (Figure 3C). Therefore, all treatments insignificantly reduced cell viability.
Anti-Inflammatory Effect of KPEVs-CLA Against H. pylori-Induced IL-8 Gene Expression and the NF-κB Signaling Pathway
To investigate the anti-inflammatory effect of KPEVs-CLA on H. pylori infection at the transcriptional level, IL-8 mRNA expression was determined using qRT-PCR at 12 h. As shown in Figure 4A, the IL-8 gene was highly upregulated in AGS cells induced by H. pylori. CLA and KPEVs-CLA significantly concentration-independently reduced IL-8 expression. Treatment with CLA at 0.06 µg/mL showed the highest inhibition with a 3.44-fold reduction, whereas the most efficient concentration of KPEVs-CLA was at 0.03 µg/mL with a 4.02-fold reduction. However, the reduction in IL-8 expression by CLA and KPEVs-CLA at 0.03–0.12 µg/mL was insignificant. Furthermore, KPEVs alone at 0.12 µg/mL significantly reduced IL-8 expression with a 1.47-fold reduction. Therefore, CLA and KPEVs-CLA were chosen for the subsequent experiments at a concentration of 0.06 µg/mL.
NF-κB is an upstream signaling protein that regulates the expression of most proinflammatory cytokine genes. CLA did not have a significant inhibitory effect on pNF-κB expression. However, KPEVs and KPEVs-CLA significantly downregulated pNF-κB, of which KPEVs-CLA exhibited the most significant downregulation, with a 1.79-fold decrease (Figure 4B). These results indicated that KPEVs-CLA effectively inhibits anti-inflammatory activity through the NF-κB signaling pathway.
Effect of KPEVs-CLA on Cytokine Production in H. pylori-Infected Gastric Cells
Multiplex cytokine analysis was performed to study the effect of KPEVs-CLA on H. pylori-induced cytokine release. A heatmap was generated to provide a comprehensive overview of the effect of H. pylori and treatments on cytokine release at different incubation times (Figure 5A). Secreted cytokines with levels below the limit of detection of the assay were excluded (ie, FGF basic, IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-7, IL-13, IL-15, IL-17A, MCP-1, PDGF-BB, and TNF-α). H. pylori significantly induced the secretion of various proinflammatory cytokines (ie, IL-6, IL-8, IL-12, and IFN-γ), chemokines (ie, MIP-1α and MIP-1β), and growth factors (ie, VEGF, G-CSF, and GM-CSF). Additionally, IL-10 was the only anti-inflammatory cytokine induced by H. pylori. Cytokine levels of IL-9, eotaxin, RANTES, and IP-10 were slightly induced by H. pylori; however, they were statistically insignificant compared with uninfected AGS cells (Figure S1).
IL-8 was the most prominently responsive proinflammatory cytokine against H. pylori. CLA and KPEVs-CLA showed similar reduction effects on IL-8 levels starting from 6 h, with the most inhibitory effect observed at 24 h compared with the H. pylori-infected group (Figure 5B). KPEVs-CLA exhibited a slightly stronger inhibitory effect at 24 h with a 9.5-fold decrease compared with CLA, which demonstrated a 5.8-fold decrease. Furthermore, KPEVs at 0.06 µg/mL exhibited no inhibitory effect.
Anti-chemokine release of KPEVs-CLA was also observed in MIP-1α and MIP-1β (Figure 5B). KPEVs-CLA exhibited a notable inhibition effect on MIP-1α at 6–24 h and MIP-1β at 12–24 h. The highest inhibition effect on each chemokine was a 4.4-fold and 2.9-fold decrease, respectively. Conversely, CLA exhibited a less significant inhibition effect at a later incubation time. It exhibited a 1.6-fold decrease in both MIP-1α and MIP-1β at 24 h. Additionally, KPEVs at 0.06 µg/mL significantly reduced MIP-1α release at 24 h.
Interestingly, KPEVs-CLA demonstrated superiority in reducing growth factor release compared with free CLA. KPEVs-CLA exhibited an inhibitory effect on G-CSF at 12 and 24 h with a 2.17-fold and 4.54-fold decrease, respectively, whereas CLA had no effect (Figure 5B). Additionally, KPEVs could decrease G-CSF secretion at 24 h.
In summary, KPEVs-CLA significantly reduced IL-8, MIP-1α, MIP-1β, and G-CSF levels. KPEVs-CLA exhibited more inhibitory effect than free CLA on releasing MIP-1α, MIP-1β, and G-CSF.
Reduction of Monocyte Chemotaxis by KPEVs-CLA in Response to MCP-1 Attraction
The cytotoxicity of CLA and KPEVs-CLA did not affect THP-1 viability at 24 h (Figure 6A). To study the anti-chemotaxis ability, MCP-1 was used as a chemoattractant. MCP-1 induced massive monocyte chemotaxis activity with a 10.28-fold increase (Figure 6B). KPEVs exhibited an anti-chemotaxis effect with a 2.21-fold decrease. CLA also significantly reduced the number of chemotactic cells (2.32-fold decrease), whereas KPEVs-CLA showed the highest anti-chemotaxis activity (3.84-fold decrease). Thus, KPEVs-CLA has an enchantment effect on anti-chemotaxis activity.
Discussion
The current treatment for H. pylori infection still relies on antibiotics, particularly clarithromycin (CLA). CLA is typically part of a first-line therapy known as triple therapy, quadruple therapy, or concomitant therapy. However, the use of CLA in treatments has its limitations. Because of its low bioavailability and short half-life, CLA often requires high doses and prolonged treatment. Consequently, a high prevalence of adverse effects is observed in 30–70% of patients with symptoms such as nausea, headache, diarrhea, and gastric discomfort.19 There is also an increasing prevalence of H. pylori resistance to CLA in the ASEAN region, with rates ranging from 17% to 43%.30 The development of CLA in nanodrug platforms has shown promising effects on the treatment of infectious diseases. Encapsulating CLA into a nanovector can improve the effectiveness of CLA against resistant bacteria while reducing its adverse effects.31,32
Our previous findings showed that KPEVs exhibit promising properties as a new drug delivery vector. In this study, CLA was spontaneously loaded into KPEVs (KPEVs-CLA) using the co-incubation method. This loading method favors the encapsulation of a hydrophobic/lipophilic drug or various small molecules into the exosome lipid bilayer. Moreover, membrane integrity is maintained by co-incubation loading in contrast to physical forces or chemical treatment (eg, ultrasound, electroporation, freeze-thaw, extrusion, and saponin).33,34 To achieve the highest LC% for maximized drug delivery per unit of KPEVs, a high drug concentration typically required in the co-incubation method. Thus, the highest KPEVs: CLA ratio of 1:100 was needed for the highest LC%, consistent with a previous report on doxorubicin co-incubation loading into exosomes.35 However, when CLA saturated the hydrophobic region of KPEVs, excess unloaded CLA remained, consequently reducing the percentage of successfully loaded CLA into KPEVs (EE%). The loading capacity of doxorubicin into lemon PDEVs was 1.35 µg per 1 µg of PDEVs, which is comparable to the loading weight observed in our KPEVs-CLA. Additionally, the slight increase in size after drug loading was similarly observed.36 It is worth noting that applying mechanical force on EVs may improve drug loading and size distribution but could result in a loss of cell internalization ability, their unique biocomposition, and bioavailability.33,37,38
An expansion in the size of KPEVs-CLA was observed compared with the original KPEVs,18 and PDI also slightly increased. Nevertheless, the PDI of KPEVs-CLA is still considered homogeneous, which is suitable for drug delivery applications.39 A similar size increase was also observed when doxorubicin was loaded into grapefruit PDEVs.40 EVs generally exhibit a negative surface charge, contributed by their structural components (ie, phospholipids and glycans) and surface-associated biomolecules such as nucleic acids.41 Although, CLA having a positive charge at pH 7, loading CLA into KPEVs did not change their negative surface charge.42 Similarly, minimal changes in zeta potential were observed in ginger and lemon EVs loaded with doxorubicin.12,36 The negative surface charge could offer an advantage of using KPEVs-CLA for targeting H. pylori infection. Negatively charged nanoparticles demonstrate enhanced mucus penetration activity, exhibiting higher mobility within the extracellular matrix and intestinal mucus compared to positive charge.43,44 Accordingly, KPEVs-CLA could be anticipated to efficiently penetrate through mucus layer, reaching the site of H. pylori infection on the gastric epithelium surface. However, the properties of drug-loaded PDEVs may vary depending on the specific PDEVs, drug type, and loading method. The KPEVs-CLA stability was mainly related to KPEVs features which are the outermost shell of nanocarriers. In our previous study, it was demonstrated that KPEVs stably maintained at −20°C and −80°C for 8 weeks with no freeze–thaw cycle.18 A stability study of milk EVs loaded with paclitaxel showed that storing loaded EVs at −80°C preserved particle size, PDI, and drug content for up to 4 weeks.45 Furthermore, doxorubicin-loaded lemon EVs revealed that both loaded and unloaded EVs remained comparably stable in simulated blood conditions.36
The drug solubility in PBS and SGF buffer is 0.12 mg/mL and 0.5 mg/mL, respectively.46,47 Therefore, we used a releasing media volume at least 6 times greater than the drug solubility for ensuring sink conditions. The kinetic releases of CLA from KPEVs were similar under neutral and gastric mimic environments. These results suggest that KPEVs-CLA exhibits no burst effect, and the release is controlled by concentration gradient (Fickian diffusion). The results showed an exponential release during the initial 1–8 h, followed by constant release until 72 h, with only a slightly slower rate observed in the gastric mimic environment at 2 h. A similar release pattern was found in CLA encapsulated in liposomes and doxorubicin-loaded lemon PDEVs in PBS buffer. They have a high release rate in the first 10 h followed by a slow-release rate until 72 h.32,36,48 This prolonged release suggests that CLA remained inside KPEVs for the duration required for KPEVs cell internalization, starting at 6 h and reaching its peak at 12 h.18 This enabling the direct delivery of CLA into human gastric cells or H. pylori, further modulate host cell-bacterial response. Additionally, as the gastrointestinal tract (GIT) serves as the site of drug absorption through oral administration, encapsulating drugs within nanocarriers enhances their transport and absorption across the GIT barriers, including the gastric barrier in the stomach, mucus, and epithelial barriers in the intestine. This facilitates their entry into the systemic circulation via both the blood circulation and the lymphatic pathway.49 This phenomenon has been demonstrated by a novel curcumin-clarithromycin nano-emulsion (Cuur-CLR-NE), which showed superior effects in abolishing gastric epithelium necrosis, reducing symptoms of gastritis, and diminishing inflammatory cell infiltration in the lamina propria.50 Furthermore, CLA solubility increased and became more stable with a decreasing pH; the dissociation constant was 8.76.51 Taken together, the enhanced solubility, stability, and sustained release of CLA from KPEVs in acidic environments contribute to the compatibility of KPEVs-CLA for oral administration.
Antimicrobial activity is a pivotal property of CLA in combating H. pylori. The drug efficacy in KPEV nanovesicles was maintained, and the growth inhibition and MBC of KPEVs-CLA against H. pylori corresponded to free CLA. Adhesion of H. pylori to the gastric cell membrane is crucial for colonization and pathogenesis. A slight inhibitory effect was observed in KPEVs, whereas CLA and KPEVs-CLA exhibited a stronger anti-H. pylori adhesion effect. Similar findings have been reported for nanoparticles loaded with Garcinia mangostana extract or CLA, where the encapsulated form showed anti-H. pylori adhesion to HEp-2 cells.52,53 The major H. pylori adhesins in the outer membrane proteins family, including blood-group antigen-binding adhesin and sialic acid-binding adhesin (SabA),54 may be the target of KPEVs-CLA. However, the anti-adhesion mechanism requires further elucidation.
KPEVs and KPEVs-CLA could be taken up by gastric cells, and localization in the cytoplasm was observed. Dil staining is well-known as an effective stain for studying EVs trafficking due to its stability and low background. Labeling EVs with Dil staining did not affect EVs size distribution.55 Importantly, Dil can be used for studying biological event without being cytotoxic and interfering with their bioactivity.56,57 The internalization of PDEVs by recipient cells involves three mechanisms: endocytosis, receptor-mediated cell signaling, and phagocytosis. Doxorubicin-loaded lemon PDEVs were shown to be internalized into the cytoplasm of ovarian adenocarcinomas through caveolin-mediated endocytosis.36 The surface properties of nanocarriers, such as hydrophilicity and electroneutrality, also enhance mucus penetration and mucoadhesion in the gastrointestinal tract. This leads to prolonged drug retention time and improved drug delivery to mucosal surfaces and underlying tissues.49
H. pylori pathogenesis is closely associated with the induction of inflammation in gastric cells. The multiple virulence factors of H. pylori play a pivotal role in inducing the NF-κB signaling pathway, the central upstream signal transduction pathway in inflammation. The NF-κB transcription factor regulates the expression of genes encoding inflammatory cytokines and subsequently releases proinflammatory cytokines from gastric cells. Consequently, these cytokines induce the recruitment of leukocytes to the infection site, and they are activated to execute their immune functions, resulting in an elevated inflammatory level. Chronic inflammation related to H. pylori infection can damage DNA, upregulate growth factor release, and alter cellular phenotypes.19 Notably, CLA and KP extracts have been shown to downregulate several inflammatory-related signaling pathways, including NF-κB, p38, ERK, JNK, and MAPK pathways.4,58–60 Our study indicated that KPEVs-CLA can downregulate the H. pylori-induced IL-8 cytokine gene marker through the NF-κB signaling pathway. In contrast, CLA may have downregulated the IL-8 gene through a different signaling pathway. Tsugawa et al reported that CLA could not suppress pNF-κB expression in LPS-induced monocytic cells but inhibited p38 MAPK and MEK1/2 phosphorylation, thereby reducing MCP-1 production.61 However, the effect of CLA on NF-κB activation remains controversial. In the NF-κB signaling pathway of monocytic cell lines induced by H2O2 and human bronchial epithelial cells induced by IL-13, higher CLA concentrations (8–32 µg/mL) and longer incubation time (2 h) have been shown to downregulate NF-κB activation, thereby attenuating mucin production and hypersecretion.62,63 This suggests that CLA may downregulate the release of cytokines through various inflammatory-related pathways, including NF-κB, MEK/ERK, and STAT6.62,64,65
The chemokines response to H. pylori infection was observed through the induction of MIP-1α and MIP-1β. These chemokines are well known for their chemoattractant properties, particularly toward monocytic cell lines. H. pylori-infected cells showed an upregulation of GM-CSF and G-CSF release, which play a role in activating the granulocytic cells. VEGF, a growth factor that plays a role in angiogenesis, was found to be released by H. pylori infection. In contrast, IL-1β, TNF-α, and MCP-1 were not secreted in H. pylori-induced AGS cells. Other H. pylori-infected gastric cell lines, such as MKN-45 and HGC-27 cells, as well as monocytic cell lines, such as RAW 264.7 cells, have been shown to produce high levels of IL-1β, IL-6, and TNF-α compared with our AGS model.66,67 Thus, some cytokines may be expressed differently against H. pylori infection across different cell lines.
Both CLA or KPEVs-CLA treatment inhibited IL-8 and MIP-1β secretion levels. KPEVs-CLA reduced the MIP-1α and G-CSF levels by approximately 2-fold compared with KPEVs and free CLA. This indicates that KPEVs-CLA have combined effects in downregulating MIP-1α and G-CSF. The MIP-1α is involved in the progression of multiple diseases. The reduction in MIP-1α levels can attenuate tissue inflammation by inhibiting other pro-inflammatory cytokines release and recruitment of monocytes/macrophages, further improving tissue functional recovery.68 Additionally, decreasing G-CSF levels can downregulate granulocyte maturation and tissue infiltration, possibly minimizing gastric carcinoma proliferation, metastasis, and severity.69
The monocyte recruitment response to H. pylori infection depends on the chemotactic interaction between monocyte chemoattractant protein-1 (MCP-1)/C-C chemokine ligand type 2 (CCL2) and C-C chemokine receptor type 2 (CCR2).70 A correlation between mononuclear recruitment and gastric carcinogenesis has been suggested in H. pylori infection71 as well as in other cancers.72 Additionally, it has been discovered that H. pylori causes hyperactive monocytes, which may contribute to gastric cancer progression.73 Information on CLA and PDEVs against monocyte chemotaxis is limited. CLA reportedly inhibits fibroblast migration and neutrophil chemotaxis.74,75 Broccoli PDEVs indirectly inhibit monocyte recruitment by regulating dendritic cell maturation.14 Hence, our study is the first to show that CLA and KPEVs effectively inhibit MCP-1-induced monocyte chemotaxis. Interestingly, KPEVs-CLA have more anti-chemotaxis activity than free CLA and KPEVs treatments. Reducing infiltrated monocytes in gastric tissue provides an advantage in lowering the risk of developing chronic gastric inflammation and cancer.76
Conclusion
In summary, our study demonstrated that KPEVs-CLA have physical and biological properties as nanomedicines. The localization of KPEVs-CLA in gastric cells was demonstrated, and CLA was spontaneously released from KPEVs in acidic environments. The anti-H. pylori and anti-adhesion activities were comparable between KPEVs-CLA and free CLA. Moreover, KPEVs-CLA showed superior anti-inflammatory activity by inhibiting IL-8, MIP-1, and G-CSF secretion and monocyte chemotaxis through the NF-κB signaling pathway. Thus, KPEVs-CLA could serve as an alternative treatment for gastric cancer induced by H. pylori infection. In the future, it is important to further study the effectiveness and toxicity of KPEV-CLA in animal models to explore their therapeutic impact and safety. Additionally, conducting incompatibility studies using various techniques such as Fourier Transform Infrared Spectroscopy (FT-IR), Differential Scanning Calorimetry (DSC), Thermogravimetric Analysis (TGA), or X-ray Diffraction (XRD) would be beneficial for investigating the interaction between CLA and KPEVs.
Abbreviations
CFU, colony-forming unit; CLA, clarithromycin; CLSI, Clinical and Laboratory Standards Institute; EE, encapsulation efficiency; EVs, extracellular vesicles; FBS, fetal bovine serum; HP, Helicobacter pylori; HPLC, high-performance liquid chromatography; kDa, kilodalton; KP, Kaempferia parviflora; KPEVs, Kaempferia parviflora extracellular vesicles; KPEVs-CLA, Kaempferia parviflora extracellular vesicles loaded with CLA; LC, loading capacity; MBC, minimum bactericidal concentration; MOI, multiplicity of infection; MWCO, molecular weight cut off; PBS, phosphate-buffered saline; PDEVs, plant-derived extracellular vesicles; PDI, polydispersity index; qRT-PCR, quantitative reverse transcription polymerase chain reaction.
Acknowledgments
We would like to thank Professor Richard Ferrero for giving us the opportunity to participate in the research exchange program.
Funding
This work was supported by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. program (Grant No. PHD/0217/2559), 90th Anniversary Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund, GCUGR1125633043D) and Thailand Science Research and Innovation (TSRI) (CU_FRB640001_01_37_1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wattanathorn J, Tong-Un T, Thukham-Mee W, Weerapreeyakul N. A functional drink containing Kaempferia parviflora extract increases cardiorespiratory fitness and physical flexibility in adult volunteers. Foods. 2023;12(18):3411. doi:10.3390/foods12183411
2. Hairunisa I, Bakar MFA, Da’i M, Bakar FIA, Syamsul ES. Cytotoxic activity, anti-migration and in silico study of black ginger (Kaempferia parviflora) extract against breast cancer cell. Cancer. 2023;15:10. doi:10.3390/cancers15102785
3. Sitthichai P, Chanpirom S, Maneerat T, et al. Kaempferia parviflora rhizome extract as potential anti-acne ingredient. Molecules. 2022;27(14). doi:10.3390/molecules27144401
4. Takuathung MN, Potikanond S, Sookkhee S, et al. Anti-psoriatic and anti-inflammatory effects of Kaempferia parviflora in keratinocytes and macrophage cells. Biomed Pharmacother. 2021;143:112229. doi:10.1016/j.biopha.2021.112229
5. Paramee S, Sookkhee S, Sakonwasun C, et al. Anti-cancer effects of Kaempferia parviflora on ovarian cancer SKOV3 cells. BMC Complement Altern Med. 2018;18(1):178. doi:10.1186/s12906-018-2241-6
6. Chen D, Li H, Li W, Feng S, Deng D. Kaempferia parviflora and its methoxyflavones: chemistry and biological activities. Evid Based Complement Alternat Med. 2018;2018:4057456. doi:10.1155/2018/4057456
7. Chaichanawongsaroj N, Amonyingcharoen S, Saifah E, Poovorawan Y. The effects of Kaempferia parviflora on anti-internalization activity of Helicobacter pylori to HEp-2 cells. Afr J Biotechnol. 2010;9:30.
8. Chaichanawongsaroj N, Amonyingcharoen S, Pattiyathanee P, R-k V, Poovorawan Y. Anti-Helicobacter pylori and anti-internalization activities of Thai folk remedies used to treat gastric ailments. J Med Plants Res. 2012;6(8):1389–1393. doi:10.5897/jmpr10.552
9. Nemidkanam V, Kato Y, Kubota T, Chaichanawongsaroj N. Ethyl acetate extract of Kaempferia parviflora inhibits Helicobacter pylori-associated mammalian cell inflammation by regulating proinflammatory cytokine expression and leukocyte chemotaxis. BMC Complement Med Ther. 2020;20(1):124. doi:10.1186/s12906-020-02927-2
10. de la Canal L, Pinedo M. Extracellular vesicles: a missing component in plant cell wall remodeling. J Exp Bot. 2018;69(20):4655–4658. doi:10.1093/jxb/ery255
11. Wang B, Zhuang X, Deng ZB, et al. Targeted drug delivery to intestinal macrophages by bioactive nanovesicles released from grapefruit. Mol Ther. 2014;22(3):522–534. doi:10.1038/mt.2013.190
12. Zhang M, Xiao B, Wang H, et al. Edible ginger-derived nano-lipids loaded with doxorubicin as a novel drug-delivery approach for colon cancer therapy. Mol Ther. 2016;24(10):1783–1796. doi:10.1038/mt.2016.159
13. Zhuang X, Deng ZB, Mu J, et al. Ginger-derived nanoparticles protect against alcohol-induced liver damage. J Extracell Vesicles. 2015;4(1):28713. doi:10.3402/jev.v4.28713
14. Deng Z, Rong Y, Teng Y, et al. Broccoli-derived nanoparticle inhibits mouse colitis by activating dendritic cell AMP-activated protein kinase. Mol Ther. 2017;25(7):1641–1654. doi:10.1016/j.ymthe.2017.01.025
15. Woith E, Guerriero G, Hausman JF, et al. Plant extracellular vesicles and nanovesicles: focus on secondary metabolites, proteins and lipids with perspectives on their potential and sources. Int J Mol Sci. 2021;22(7). doi:10.3390/ijms22073719
16. Yang C, Zhang M, Merlin D. Advances in plant-derived edible nanoparticle-based lipid nano-drug delivery systems as therapeutic nanomedicines. J Mater Chem B. 2018;6(9):1312–1321. doi:10.1039/C7TB03207B
17. Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi:10.1080/20013078.2018.1535750
18. Nemidkanam V, Chaichanawongsaroj N. Characterizing Kaempferia parviflora extracellular vesicles, a nanomedicine candidate. PLoS One. 2022;17(1):e0262884. doi:10.1371/journal.pone.0262884
19. Malfertheiner P, Camargo MC, El-Omar E, et al. Helicobacter pylori infection. Nat Rev Dis Primers. 2023;9(1):19. doi:10.1038/s41572-023-00431-8
20. Kumar S, Patel GK, Ghoshal UC. Helicobacter pylori-Induced inflammation: possible factors modulating the risk of gastric cancer. Pathogens. 2021;10(9). doi:10.3390/pathogens10091099
21. Salvatori S, Marafini I, Laudisi F, Monteleone G, Stolfi C. Helicobacter pylori and Gastric Cancer: pathogenetic Mechanisms. Int J Mol Sci. 2023;24(3). doi:10.3390/ijms24032895
22. Chey WD, Leontiadis GI, Howden CW, Moss SF. ACG clinical guideline: treatment of Helicobacter pylori infection. Am J Gastroenterol. 2017;112(2):212–239. doi:10.1038/ajg.2016.563
23. Grübel P, Cave DR. Factors affecting solubility and penetration of clarithromycin through gastric mucus. Aliment. Pharmacol. Ther. 1998;12(6):569–576.
24. Khosravian P, Khoobi M, Ardestani MS, et al. Enhancement antimicrobial activity of clarithromycin by amine functionalized mesoporous silica nanoparticles as drug delivery system. Lett Drug Des Discovery. 2018;15(7):787–795. doi:10.2174/1570180815666180117155818
25. Makoni PA, Khamanga SM, Walker RB. Muco-adhesive clarithromycin-loaded nanostructured lipid carriers for ocular delivery: formulation, characterization, cytotoxicity and stability. J Drug Delivery Sci Technol. 2021;61. doi:10.1016/j.jddst.2020.102171
26. Sung J, Yang C, Viennois E, Zhang M, Merlin D. Isolation, purification, and Characterization of Ginger-derived Nanoparticles (GDNPs) from ginger, rhizome of Zingiber officinale. Bio-Protocol. 2019;9(19):e3390. doi:10.21769/BioProtoc.3390
27. Yang X, Shi G, Guo J, Wang C, He Y. Exosome-encapsulated antibiotic against intracellular infections of methicillin-resistant Staphylococcus aureus. Int J Nanomed. 2018;13:8095–8104. doi:10.2147/IJN.S179380
28. Askarizadeh M, Esfandiari N, Honarvar B, Sajadian SA, Azdarpour A. Kinetic modeling to explain the release of medicine from drug delivery systems. ChemBioEng Rev. 2023;10(6):1006–1049. doi:10.1002/cben.202300027
29. Tharmalingam N, Kim S-H, Park M, et al. Inhibitory effect of piperine on Helicobacter pylori growth and adhesion to gastric adenocarcinoma cells. Infect Agent Cancer. 2014;9:43.
30. Aumpan N, Mahachai V, Vilaichone RK. Management of Helicobacter pylori infection. JGH Open. 2023;7(1):3–15. doi:10.1002/jgh3.12843
31. Jain SK, Haider T, Kumar A, Jain A. Lectin-conjugated clarithromycin and acetohydroxamic acid-loaded PLGA nanoparticles: a novel approach for effective treatment of H. pylori. AAPS Pharm Sci Tech. 2016;17(5):1131–1140. doi:10.1208/s12249-015-0443-5
32. Geng S, Liu X, Xu H, et al. Clarithromycin ion pair in a liposomal membrane to improve its stability and reduce its irritation caused by intravenous administration. Expert Opin Drug Deliv. 2016;13(3):337–348. doi:10.1517/17425247.2016.1123247
33. Zeng H, Guo S, Ren X, Wu Z, Liu S, Yao X. Current strategies for exosome cargo loading and targeting delivery. Cells. 2023;12(10). doi:10.3390/cells12101416
34. Fu S, Wang Y, Xia X, Zheng JC. Exosome engineering: current progress in cargo loading and targeted delivery. NanoImpact. 2020;20. doi:10.1016/j.impact.2020.100261
35. Gong C, Tian J, Wang Z, et al. Functional exosome-mediated co-delivery of doxorubicin and hydrophobically modified microRNA 159 for triple-negative breast cancer therapy. J Nanobiotechnology. 2019;17(1):93. doi:10.1186/s12951-019-0526-7
36. Xiao Q, Zhao W, Wu C, et al. Lemon-Derived extracellular vesicles nanodrugs enable to efficiently overcome cancer multidrug resistance by endocytosis-triggered energy dissipation and energy production reduction. Adv Sci. 2022;9(20):e2105274. doi:10.1002/advs.202105274
37. Sukreet S, Silva BVRE, Adamec J, Cui J, Zempleni J. Sonication and short-term incubation alter the content of bovine milk exosome cargos and exosome bioavailability (OR26-08-19). Curr. Dev. Nutr. 2019;3. doi:10.1093/cdn/nzz033.OR26-08-19
38. Nizamudeen ZA, Xerri R, Parmenter C, et al. Low-power sonication can alter extracellular vesicle size and properties. Cells. 2021;10(9). doi:10.3390/cells10092413
39. Danaei M, Dehghankhold M, Ataei S, et al. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics. 2018;10(2). doi:10.3390/pharmaceutics10020057
40. Moon K, Hur J, Kim KP, Lee K, Kang JY. Surface‐functionalizable plant‐derived extracellular vesicles for targeted drug delivery carrier using grapefruit. Adv. Mater. Interfaces. 2023;10(22). doi:10.1002/admi.202300220
41. Hallal S, Tuzesi A, Grau GE, Buckland ME, Alexander KL. Understanding the extracellular vesicle surface for clinical molecular biology. J Extracell Vesicles. 2022;11(10):e12260. doi:10.1002/jev2.12260
42. Myers AG, Clark RB. Discovery of macrolide antibiotics effective against multi-drug resistant gram-negative pathogens. Acc Chem Res. 2021;54(7):1635–1645. doi:10.1021/acs.accounts.1c00020
43. Akkus ZB, Nazir I, Jalil A, Tribus M, Bernkop-Schnurch A. Zeta potential changing polyphosphate nanoparticles: a promising approach to overcome the mucus and epithelial barrier. Mol Pharm. 2019;16(6):2817–2825. doi:10.1021/acs.molpharmaceut.9b00355
44. Lieleg O, Baumgartel RM, Bausch AR. Selective filtering of particles by the extracellular matrix: an electrostatic bandpass. Biophys J. 2009;97(6):1569–1577. doi:10.1016/j.bpj.2009.07.009
45. Agrawal AK, Aqil F, Jeyabalan J, et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine. 2017;13(5):1627–1636. doi:10.1016/j.nano.2017.03.001
46. Zaid Alkilani A, Musleh B, Hamed R, Swellmeen L, Basheer HA. Preparation and characterization of patch loaded with clarithromycin nanovesicles for transdermal drug delivery. J Funct Biomater. 2023;14(2). doi:10.3390/jfb14020057
47. Kumar PR, Shyale S, Gouda MM, Kumar SS. A sensitive UV spectrophotometric analytical method development, validation and preformulation studies of clarithromycin. Research J Pharm and Tech. 2011;4(2):242–246.
48. Li Y, Su T, Zhang Y, Huang X, Li J, Li C. Liposomal co-delivery of daptomycin and clarithromycin at an optimized ratio for treatment of methicillin-resistant Staphylococcus aureus infection. Drug Deliv. 2015;22(5):627–637. doi:10.3109/10717544.2014.880756
49. Wang D, Jiang Q, Dong Z, et al. Nanocarriers transport across the gastrointestinal barriers: the contribution to oral bioavailability via blood circulation and lymphatic pathway. Adv Drug Deliv Rev. 2023;203:115130. doi:10.1016/j.addr.2023.115130
50. Mosallam FM, Bendary MM, Elshimy R, El-Batal AI. Curcumin clarithromycin nano-form a promising agent to fight Helicobacter pylori infections. World J Microbiol Biotechnol. 2023;39(12):324. doi:10.1007/s11274-023-03745-7
51. Nakagawa Y, Itai S, Yoshida T, Nagai T. Physicochemical properties and stability in the acidic solution of a new macrolide antibiotic, clarithromycin, in comparison with erythromycin. Chem Pharm Bull. 1992;40(3):725–728. doi:10.1248/cpb.40.725
52. Pan-In P, Banlunara W, Chaichanawongsaroj N, Wanichwecharungruang S. Ethyl cellulose nanoparticles: clarithomycin encapsulation and eradication of H. pylori. Carbohydr Polym. 2014;109:22–27. doi:10.1016/j.carbpol.2014.03.025
53. Pan-in P, Tachapruetinun A, Chaichanawongsaroj N, Banlunara W, Suksamrarn S, Wanichwecharungruang S. Combating Helicobacter pylori infections with mucoadhesive nanoparticles loaded with Garcinia mangostana extract. Nanomedicine. 2014;9(3):457–468. doi:10.2217/nnm.13.30
54. Xu C, Soyfoo DM, Wu Y, Xu S. Virulence of Helicobacter pylori outer membrane proteins: an updated review. Eur J Clin Microbiol Infect Dis. 2020;39(10):1821–1830. doi:10.1007/s10096-020-03948-y
55. Pantazopoulou M, Lamprokostopoulou A, Karampela DS, et al. Differential intracellular trafficking of extracellular vesicles in microglia and astrocytes. Cell Mol Life Sci. 2023;80(7):193. doi:10.1007/s00018-023-04841-5
56. Chen C, Cai N, Niu Q, Tian Y, Hu Y, Yan X. Quantitative assessment of lipophilic membrane dye-based labelling of extracellular vesicles by nano-flow cytometry. J Extracell Vesicles. 2023;12(8):e12351. doi:10.1002/jev2.12351
57. Lee SH, Jin KS, Bang OY, et al. Differential migration of mesenchymal stem cells to ischemic regions after middle cerebral artery occlusion in rats. PLoS One. 2015;10(8):e0134920. doi:10.1371/journal.pone.0134920
58. Ichiyama T, Nishikawa M, Yoshitomi T, et al. Clarithromycin inhibits NF-kappaB activation in human peripheral blood mononuclear cells and pulmonary epithelial cells. Antimicrob Agents Chemother. 2001;45(1):44–47. doi:10.1128/AAC.45.1.44-47.2001
59. Reijnders TDY, Saris A, Schultz MJ, van der Poll T. Immunomodulation by macrolides: therapeutic potential for critical care. Lancet Respir Med. 2020;8(6):619–630. doi:10.1016/S2213-2600(20)30080-1
60. Thaklaewphan P, Ruttanapattanakul J, Monkaew S, et al. Kaempferia parviflora extract inhibits TNF-alpha-induced release of MCP-1 in ovarian cancer cells through the suppression of NF-kappaB signaling. Biomed Pharmacother. 2021;141:111911. doi:10.1016/j.biopha.2021.111911
61. Tsugawa K, Imaizumi T, Watanabe S, Tsuruga K, Yoshida H, Tanaka H. Clarithromycin attenuates the expression of monocyte chemoattractant protein-1 by activating toll-like receptor 4 in human mesangial cells. Clin Exp Nephrol. 2017;21(4):573–578. doi:10.1007/s10157-016-1333-1
62. Tanabe T, Kanoh S, Tsushima K, Yamazaki Y, Kubo K, Rubin BK. Clarithromycin inhibits interleukin-13-induced goblet cell hyperplasia in human airway cells. Am J Respir Cell Mol Biol. 2011;45(5):1075–1083. doi:10.1165/rcmb.2010-0327OC
63. Kobayashi Y, Wada H, Rossios C, et al. A novel macrolide solithromycin exerts superior anti-inflammatory effect via NF-kappaB inhibition. J Pharmacol Exp Ther. 2013;345(1):76–84. doi:10.1124/jpet.112.200733
64. Shinkai M, Tamaoki J, Kobayashi H, et al. Clarithromycin delays progression of bronchial epithelial cells from G1 phase to S phase and delays cell growth via extracellular signal-regulated protein kinase suppression. Antimicrob Agents Chemother. 2006;50(5):1738–1744. doi:10.1128/AAC.50.5.1738-1744.2006
65. Shinkai M, Lopez-Boado YS, Rubin BK. Clarithromycin has an immunomodulatory effect on ERK-mediated inflammation induced by Pseudomonas aeruginosa flagellin. J Antimicrob Chemother. 2007;59(6):1096–1101. doi:10.1093/jac/dkm084
66. Wen J, Chen C, Luo M, et al. Notch signaling ligand jagged1 enhances macrophage-mediated response to Helicobacter pylori. Front Microbiol. 2021;12:692832. doi:10.3389/fmicb.2021.692832
67. Zhao C, Lu X, Bu X, Zhang N, Wang W. Involvement of tumor necrosis factor-α in the upregulation of CXCR4 expression in gastric cancer induced by Helicobacter pylori. BMC Cancer. 2010;10(1). doi:10.1186/1471-2407-10-419
68. Pelisch N, Rosas Almanza J, Stehlik KE, Aperi BV, Kroner A. Use of a self-delivering anti-CCL3 FANA oligonucleotide as an innovative approach to target inflammation after spinal cord injury. eNeuro. 2021;8(2). doi:10.1523/ENEURO.0338-20.2021
69. Karagiannidis I, Salataj E, Said Abu Egal E, Beswick EJ. G-CSF in tumors: aggressiveness, tumor microenvironment and immune cell regulation. Cytokine. 2021;142:155479. doi:10.1016/j.cyto.2021.155479
70. Arnold IC, Zhang X, Urban S, et al. NLRP3 Controls the development of gastrointestinal CD11b(+) dendritic cells in the steady state and during chronic bacterial infection. Cell Rep. 2017;21(13):3860–3872. doi:10.1016/j.celrep.2017.12.015
71. Schneider BG, Piazuelo MB, Sicinschi LA, et al. Virulence of infecting Helicobacter pylori strains and intensity of mononuclear cell infiltration are associated with levels of DNA hypermethylation in gastric mucosae. Epigenetics. 2014;8(11):1153–1161. doi:10.4161/epi.26072
72. Cendrowicz E, Sas Z, Bremer E, Rygiel TP. The role of macrophages in cancer development and therapy. Cancer. 2021;13(8). doi:10.3390/cancers13081946
73. Frauenlob T, Neuper T, Mehinagic M, Dang HH, Boraschi D, Horejs-Hoeck J. Helicobacter pylori infection of primary human monocytes boosts subsequent immune responses to LPS. Front Immunol. 2022;13:847958. doi:10.3389/fimmu.2022.847958
74. Gouzos M, Ramezanpour M, Bassiouni A, Psaltis AJ, Wormald PJ, Vreugde S. Antibiotics affect ROS production and fibroblast migration in an in-vitro model of sinonasal wound healing. Front Cell Infect Microbiol. 2020;10:110. doi:10.3389/fcimb.2020.00110
75. Zimmermann P, Ziesenitz VC, Curtis N, Ritz N. The immunomodulatory effects of macrolides—a systematic review of the underlying mechanisms. Front Immunol. 2018;9. doi:10.3389/fimmu.2018.00302
76. Kaparakis M, Walduck AK, Price JD, et al. Macrophages are mediators of gastritis in acute Helicobacter pylori infection in C57BL/6 mice. Infect Immun. 2008;76(5):2235–2239. doi:10.1128/IAI.01481-07
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.