Back to Journals » Infection and Drug Resistance » Volume 14
Antibiotics Combinations and Chitosan Nanoparticles for Combating Multidrug Resistance Acinetobacter baumannii
Authors Banoub NG, Saleh SE, Helal HS , Aboshanab KM
Received 10 July 2021
Accepted for publication 6 August 2021
Published 20 August 2021 Volume 2021:14 Pages 3327—3339
DOI https://doi.org/10.2147/IDR.S328788
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Nancy G Banoub,1 Sarra E Saleh,2 Hala S Helal,1 Khaled M Aboshanab2
1Department of Microbiology and Immunology, Faculty of Pharmacy, Heliopolis University, Cairo, Egypt; 2Department of Microbiology and Immunology, Faculty of Pharmacy, Ain Shams University, Cairo, Egypt
Correspondence: Khaled M Aboshanab
Department of Microbiology and Immunology, Faculty of Pharmacy, Ain Shams University, Cairo, 11566, Egypt
Tel + 20-228429040; +20-1007582620
Fax + 20-224051107
Email [email protected]
Background: Successful treatment of Acinetobacter (A.) baumannii-associated infection is complicated by the emergence of multidrug resistance (MDR), particularly in clinical settings. This urges searching for new alternatives to encounter such health problem.
Aim: This study aimed to evaluate certain antibiotic combinations and CNPs either alone or in combination of some selected antibiotics for the purpose of combating MDR A. baumannii clinical isolates.
Methods: A total of 51 A. baumannii clinical isolates were recovered from discharged clinical specimens of the Clinical Microbiology Central Laboratory of AL Kasr Al Aini hospital, Cairo, Egypt. Conventional standard Lab tests were used for identification followed by recA gene testing for confirmation. Antimicrobial susceptibility tests were conducted out according to CLSI guidelines. Genotypic analysis using Enterobacterial Repetitive Intergenic Consensus-polymerase chain reaction (ERIC-PCR) of the respective isolates showed that they were clustered in nine clones. The prepared CNPs were characterized by dynamic light scattering and HR-transmission electron microscope imaging. Antibiotic combinations and co-effect of CNPs with some selected antibiotics (either each alone or in combination of two) were evaluated using the Checkerboard microdilution and minimum inhibitor concentration decrease factor (MDF) methods, respectively.
Results: The recovered 51 A. baumannii clinical isolates were MDR (100%) of these 92% (47/51) were extensively drug resistance (XDR). Combinations of colistin (CT)+meropenem (MEM) and MEM+tigecycline (TGC) showed synergism in 77.7% and 44.4% and additive effects in 22.3% and 55.6% of the tested MDR A. baumannii isolates (n=51), respectively. However, CT+TGC combination showed antagonism. CNPs exhibited good inhibitory activity (inhibition zones ranged from 24 to 31 mm) against selected nine MDR A. baumannii isolates (one isolate from each clone). The MIC of CNPs at concentrations (ranging from 1 to 5 mg/mL) were from 0.16 to 0.25 mg/mL, indicating good in vitro antimicrobial activities. CNPs (5 mg/mL) when combined with CT, TGC or MEM, CT+MEM and TGC+MEM significantly increased the susceptibilities of the MDR A. baumannii isolates to these antibiotics by 88.8%, 66.6%, 100%, 77.7%, and 44.4%, respectively. No significant effects were observed when CNPs (5 mg/mL) were combined with CT+TGC.
Conclusion: The current study demonstrated the significant in-vitro activities of CNPs either alone or in combination with CT, TGC or MEM, CT+MEM and TGC+MEM and the successful combinations of MEM either with CT or with TGC against the MDR A. baumannii pathogens. However, further in vivo studies should be conducted to verify such activities and their potential use in human.
Keywords: A. baumannii, multidrug resistance, chitosan nanoparticles, antibiotic combinations, meropenem, colistin, tigecycline
Introduction
A. baumannii is a Gram-negative, non-fermentative, coccobacillus and is considered to be one of the major causative agents of nosocomial infections. It is figured in the “critical” category of World Health Organizations (WHO) priority pathogens list for development of new antibiotics.1 A. baumannii is one of the six superbugs‟ identified by the Infectious Diseases Society of America as “ESKAPE”.2 A. baumannii has been implicated in a diverse range of infections including, pneumonia, bacteremia, wound and burn infection, urinary tract infection and meningitis.3,4 It is conspicuously prevalent in intensive care units where frequent epidemics have been tremendously problematic to control.3,4 The rapid emergence and worldwide distribution of drug resistant A. baumannii as a foremost nosocomial pathogen highlights its successful adaptation clinical settings and health-care ecosystem.5 Many studies have shown that the biofilm formation is the reason behind the survival of A. baumannii in harsh environments and high resistance to various antibiotics. Several mechanisms are considered key factors in the high resistance of biofilms such as: (a) impaired drug diffusion, (b) enzyme-caused neutralizations, (c) heterogeneous function, (d) slow rate of growth, (e) persistent cells, and (f) alterations in microbial phenotypic and genotypic features.6–9
Phenotypic identification of A. baumannii clinical isolates should be confirmed using genotypic methods via detection of recA gene by polymerase chain reaction (PCR)10 followed by genotyping confirmation using the Enterobacterial Repetitive Intergenic Consensus-PCR (ERIC-PCR). ERIC-PCR analysis can differentiate MDR A. baumannii strains at the clonal level to confirm their clonal relationship. This helps controlling these resistant strains and tracing their epidemics.11
As known, the main strategy for treating infectious diseases is antimicrobial agents; however, the misuse of antimicrobial agents accelerates the emergence of resistance which in turn leads to serious losses such as financial cost, societal cost, and cost of death.12,13 A. baumannii can evolve antibiotic resistance through several mechanisms, including enzymes inactivating antibiotics, alteration of the target or cellular functions due to mutations, formation of biofilms and reduced entry into the target site of bacteria (Efflux pump).14 To combat the bacterial resistance, many scientists are trying to develop and explore new antimicrobials, however, it is not an easy process to have efficient and approved one.15–18
As known in literature, carbapenems, colistin and tigecycline still retain activities against MDR A. baumannii.19–21 Carbapenems such as meropenem cause bacterial cell death by binding covalently to penicillin-binding proteins (PBPs) involved in bacterial cell wall biosynthesis. Colistin, owing to its unique mechanism of action, that is disruption of bacterial outer membrane lipids as well as tigecycline which inhibits bacterial protein synthesis by acting on the 30S ribosomal subunit and prevents amino acids from incorporating and elongating peptide chains.19–21
Another promising strategy for overcoming the microbial resistance is the use of nanoparticles. Chitosan nanoparticles generally consist of biodegradable polymers or lipids which are biocompatible and are non-toxic. But, the possible toxic effect can never be denied due to their large surface area and smaller size relative to the cellular components, and proteins can lead to adverse tissue reaction and cause toxicity such as toxicity towards a murine melanoma cell line, several tumor cell lines and gastric carcinoma cell line.22–24
The major processes underlying the antibacterial effects of NPs are disruption of the bacterial cell membrane, penetration of the bacterial cell membrane and induction of intracellular antibacterial effects, including interactions with DNA and proteins.25–27 Chitosan nanoparticles (CNPs) acquired extraordinary biological action particularly against MDR pathogens due to its minute size, quantum effect, elevated sorption capacity and good antimicrobial activity by binding positively charged chitosan to negatively charged bacterial cell wall surfaces such as lipopolysaccharides. This binding has led to the alteration of the bacterial membrane permeability, causing leakage of intracellular constituents and cell death, binding to DNA in bacteria causing inhibition of DNA replication and cell death and Chitosan acts as a chelating agent that selectively binds to trace metal elements causing toxin production and inhibiting microbial growth.27–29 Characterization CNPs can be performed appropriately by Dynamic Light Scattering (DLS) and High resolution -Transmission electron microscope imaging (HR-TEM).30–32 Till now, few studies had been conducted to explore the antimicrobial activities of either antibiotic combinations or combination of CNPs with certain antibiotics against MDR or XDR clinical pathogens.19–22,28 Therefore, this study aimed to evaluate certain antibiotic combinations and CNPs either alone or in combination of antibiotics for the purpose of combating MDR A. baumannii clinical isolates, the life threatening pathogens with limited therapeutic options.
Methods
Specimen Collection and Identification of the Recovered Clinical Isolates
A total of 51 A. baumannii isolates were obtained from the Microbiology Central Lab of AL Kasr Al Aini hospital, Cairo, Egypt between January, and June 2020. These isolates were recovered from 730 different discharged clinical specimens including, pus, urine, sputum, bronchial lavage, and cerebrospinal fluid according to the hospital records. Identification of the isolates were carried out using conventional techniques (colony morphology, culture using a specific media ChromAgar, and biochemical tests). The isolates were cultured on chromAgar (when the color changes from yellow to red, it indicates that the isolate is A. baumannii). The identification of isolates was also confirmed using the automated system, Vitek-2 (bioMérieux, Marcy L’Etoile, France) and PCR analysis of the recA gene as previously described.33 Negative control (PCR reaction with chromosomal DNA of A. baumannii ATCC 17978 but without recA primers) and positive control (PCR reaction with chromosomal DNA of A. baumannii ATCC 17978 as a PCR template plus recA primers) were used for quality control.
Antimicrobial Susceptibility Testing and MDR Definition
The obtained bacterial isolates were evaluated for susceptibility to the antimicrobial agents recommended by the Clinical and Laboratory Standards Institute, 2018.34 Susceptibility tests were performed using the Kirby–Bauer disk diffusion method on Mueller–Hinton agar (Hi media, India) using the following antimicrobial disks (Bioanalyse, Turkey): Piperacillin (PIP, 100 μg), piperacillin/tazobactam (TPZ, 10/100 μg), ampicillin-sulbactam (SAM, 10/10 μg), cefepime (FEP, 30 μg), ceftriaxone (CRO, 30 μg), Amikacin (AK, 30), gentamicin (CN, 30 mcg), ciprofloxacin (CIP, 5 mcg), imipenem (IMP, 10 μg), meropenem (MEM, 10 μg), doxycycline (DO, 30 μg), Trimethoprim-sulfamethoxazole (SXT, 1.25/23.7 μg), tigecycline (TGC, 15 μg). Susceptibility to colistin (CT) was examined via minimum inhibitory concentration (MIC) measurement using E-test (Bioanalyse, Turkey) according to manufacturer’s recommendations. The reference A. baumannii ATCC 17978 and E. coli ATCC 25922 strains were used as a quality control. The MIC of TGC, CT, MEM (products of Merck, Darmstadt, Germany) were carried out using microbroth dilution method according to CLSI guidelines, 2018).34 MDR phenotype was inferred as described by Magiorakos et al.35
Molecular Typing of Recovered Isolates
ERIC-PCR was carried out on 51 A. baumannii isolates to investigate the clonal relationship, clonal expansion, and their diversity.36 Genomic DNA was extracted using the Genomic DNA Purification Kit (Thermo Fisher Scientific, UK) according to the manufacturer’s recommendations. ERIC-PCR was carried out using the ERIC-1 primer (5ʹ-ATGTAAGCTCCTGGGGATTCAC-3ʹ) and ERIC-2 primer (5ʹ-AAGTAAGTG ACTGGGGTGAGCG-3ʹ) primers as previously described.36 Analysis of ERIC-PCR dendrogram was constructed using the UPGMA clustering method, Bionumerics program version 7.6 (Applied Maths). The Percentage of similarity among 51 isolates of A. baumannii was calculated using Jaccard’s Coefficient.37
Evaluation of Antibiotic Combinations
The MIC of MEM, TGC and CT was determined using the broth microdilution technique according to the CLSI guidelines 2018.34 In vitro combinations of MEM+TGC, MEM+CT and TGC+CT were performed in 96-well microdilution plates and evaluated using the checkerboard method. Two-fold Serial dilutions of antibiotic aqueous solutions were prepared starting from (1024 µg/mL) to (0.25 µg/mL) using standard laboratory powders of the antibiotics (CT, MEM and TGC). A 0.5 McFarland standards of bacteria used and inoculated into Mueller-Hinton broth medium. The plates were incubated at 37 °C for 24hr. The sum of the fractional inhibitory concentration (ƩFICs) was calculated as described by Hsieh et al.38 The combination is considered synergistic when ƩFIC is ≤ 0.5, additive when ƩFIC is > 0.5 and ≤ 1, indifferent when ƩFIC is >1 and ≤ 4, and antagonistic when ƩFIC is > 4.39
Preparation of Chitosan Nanoparticles (CNPs)
About 5 mg/mL of low molecular weight chitosan (Sigma-Aldrich, Darmstadt, Germany, CAT, 448869) was suspended in 10 mL of 1% v/v acetic acid and the pH was adjusted between 4.6 and 4.8 using 10 N NaOH. A total of 0.1 g of sodium tripolyphosphate was dissolved in 100 mL of distilled water. CNPs were produced suddenly while adding the tripolyphosphate solution dropwise to the chitosan solution under uninterrupted mixing. The manufactured CNPs were purified at 10,000 g for 20 min by centrifugation. Then, the pellet was collected and the CNPs were washed with distilled water then freeze-dried.40
Characterization of CNPs
Dynamic Light Scattering (DLS; Zeta Sizer Characterization)
The prepared CNPs were characterized by DLS where, the particle size distribution and zeta potential were measured through DLS with Zetasizer Nano S (Malvern, UK). The analysis was carried out at a scattering angle of 90° at a temperature of 25°C using nanoparticles dispersed in deionized distilled water (1 g of sample was dissolved in 25 mL of deionized water and then sonication is done in sonics Vibra cell Sonicator, UK for 15 min). Particle size distribution of the nanoparticles is reported by intensity as previously reported.41
Transmission Electron Microscope Imaging (HR-TEM)
Chitosan nano- suspension was prepared in Ultrasonicator (SB-120DTN, Taiwan) for 15 min then particles were deposited from a dilute aqueous suspension onto (200 mesh) Cu grid with the support of a 10 nm thickness carbon film. After solvent evaporation, Cu grid was placed in double title grid holder and tested under Transmission Electron Microscope (HR-TEM Tecnai G20, FEI, Netherlands) as previously described.42
Antimicrobial Activity of CNPs
This was done using two methods:
Well-Cut Diffusion Technique
Purified colonies of MDR A. baumannii isolates from overnight plates were picked and inoculated on Mueller Hinton medium. After solidifying, wells were punched out using 0.7 cm cork borer. Then, 100 μL of chitosan nanoparticles were pipetted into each well. All plates were incubated at 37°C for 12 h. After incubation, the radius of clear inhibition zone around each well was measured in mm as previously determined.43–45
Broth Dilution Technique
Broth dilution assays were used to determine the MIC of the CNPs against MDR A. baumannii isolates. Two-fold serial dilutions of CNPs were prepared using Mueller Hinton broth (starting concentrations were 1, 2.5 and 5 mg/mL). To prepare the inoculum, all the bacterial cell suspensions were adjusted to 0.5 McFarland0.5 (1–2 × 108 cfu/mL), then 100 μL of each MDR A. baumannii was used for inoculating the tubes under aseptic condition. The tubes were then mixed and incubated for 24 hr at 37°C. After 24 hr of incubation, the MIC was calculated.40,46
Evaluation of CNPs-Antibiotic Combinations
In vitro evaluation of CNPs (5 mg/mL) in combination with MEM, TGC and CT (each alone or MEM+TGC, MEM+CT and TGC+CT) against the selected MDR A. baumannii was determined by calculating the MIC decrease factor (MDF) as previously reported.46 In brief, the MIC of each of MEM, TGC or CT (each alone or MEM+TGC, MEM+CT and TGC+CT) was determined using the agar diffusion technique according to the CLSI guidelines 2018.34 Then, the same MICs of the respective antibiotics was determined but in the presence of CNPs (5 mg/mL) in each well. The MDF of each isolate was calculated according to the following formula MDF=MICwithout CNPs/MICwith CNPs. An MDF value equal or greater than 4 was defined as a significant inhibition according to the protocol of Huguet et al.47
Results
Specimen Collection and Identification of the Recovered Clinical Isolates
A total of 51 identified A. baumannii isolates were obtained from the Microbiology Central Lab of AL Kasr Al Aini hospital. The PCR analysis of the recA gene (425 bp) of the respective isolates is shown in Figure S1 (Supplementary File).
Antimicrobial Susceptibility Findings
Antibiogram analysis of the 51 A. baumannii clinical isolates against the 14 tested antimicrobial agents is delineated in Table S1 (Supplementary File). Results revealed that, all the tested A. baumannii clinical isolates were MDR (100%) and 92% (47/51) were XDR. The MDR isolates (n=51) were 92–100% resistance to PIP, TPZ, SAM, FEP, CRO, AK, CN, CIP, IMP, MEM, DO, and SXT. However, lowest resistance was observed to TGC and CT (4% each) (Table S1 Supplementary File).
ERIC-PCR Analysis of Recovered Isolates
ERIC‑PCR analysis of the 51 MDR A. baumannii isolates is shown in Figure S2 (Supplementary File). Dendrogram analysis using BioNumerics fingerprint data software and unweighted pair group method with arithmetic averages at 97% similarity on 51 isolates of A. baumannii; the different clusters at 97% similarity are arbitrarily designated as Clusters 1–9, clusters 1 and 7 are the largest group representing the most prevalent clones of A. baumannii and its variants among the tested isolates (Figure 1).
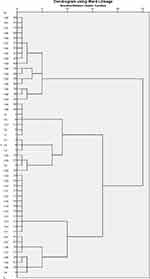 |
Figure 1 Dendrogram of ERIC-PCR analysis of the 51 isolates of A. baumannii; the different clusters at 97% similarity are arbitrarily designated as Clusters 1–9. |
Evaluation of Antibiotic Combinations
Based on dendrogram analysis, nine selected isolates coded, A31, A35, A20, A8, A3, A25, A11, A26, and A42 representing the 9 clusters (1–9) were selected for evaluating the two antibiotic combinations (CT+MEM, TGC+MEM and CT+TGC). The MIC values (µg/mL) of the tested antibiotics either alone or in combinations are shown in Table S2 (Supplementary File). The FICI of each of the tested isolates is delineated in Table 1. The FICI values of two tested antibiotic combinations against the nine MDR A. baumannii are demonstrated in Figure S3 (Supplementary File). Total percentage of synergy, additive, and antagonistic effects of two tested antibiotic combinations (CT+MEM, TGC+MEM and TGC+CT) against MDR A. baumannii (n=9) is shown in Figure S4 (Supplementary File). Results revealed that the CT+MEM and TGC+MEM combinations gave synergy in 77.7% (7/9) and 44.4% (4/9) of the tested isolates (n=9). On the other hand, CT+TGC gave 100% antagonism on the tested isolates (n=9).
 |
Table 1 FICI Values of Two Tested Antibiotic Combinations (CT+MEM, TGC+MEM and TGC+CT) Against Nine Selected MDR A. baumannii |
Characterization of CNPS
DLS (Zeta Sizer Characterization)
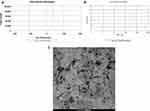
As shown in Figure 2A and B), the zeta potential was positive at 37.7mV and the average size of CNPs at selected concentration was 441.7 ± 58 nm.
Transmission Electron Microscope Imaging (HR-TEM)
As displayed in Figure 2C, the TEM images have displayed the morphological properties and surface appearance of CNPs. The CNPs have virtually spherical shape, smooth surface, and size range of about 80–500 nm.
Antimicrobial Activity of CNPs
Well-Cut Diffusion Technique
Based on dendrogram analysis, nine MDR A. baumannii isolates (coded A31, A35, A20, A8, A3, A25, A11, A26, and A42) representing the 9 clusters (1–9) were used for this test. Results of this test is delineated in Figure S5 (Supplementary File) and in Table 2.
 |
Table 2 Average of Inhibition Diameter (Mm) ±SD of CNPs Against MDR A. baumannii Isolates (n=9) |
Broth Dilution Technique
The result of the MIC by broth dilution assay of the CNPs prepared at three concentrations 1, 2.5 and 5 mg/mL is outlined in Table 3. The MIC of the tested MDR A. baumannii isolates (n=51) was in the range of 0.16 to 0.5 mg/mL as presented in Table 3.
 |
Table 3 MIC Values CNPs (1, 2.5, 5 mg/ml) Against the MDR A. baumannii Isolates (n=51) by Broth Dilution |
Evaluation of CNPs-Antibiotic Combinations
Evaluation of CNPs (5 mg/mL) in combination with MEM, TGC and CT (each antibiotic alone) or with combination of two antibiotics including, MEM+TGC, MEM+CT and TGC+CT against nine MDR A. baumannii isolates (coded A31, A35, A20, A8, A3, A25, A11, A26, and A42) representing the 9 clusters (1–9) are shown in Table 4. Of these, 8 isolates (88.8%) were XDR. Results revealed that, CNPs 5 mg/mL, when combined with CT, TGC or MEM, significantly increased the susceptibilities of the MDR A. baumannii isolates to these antibiotics by 88.8%, 66.6% and 100%, respectively. Moreover, CNPs (5 mg/mL), when combined with CT+MEM, TGC+MEM, significantly increased the susceptibilities of the MDR A. baumannii isolates to these antibiotics by 77.7%, and 44.4%, respectively. No significant effects were observed when CNPs (5 mg/mL) were used in combination with CT+TGC (Table 4).
 |
Table 4 Effects of CNPs (5 mg/ml) on the MIC of MEM, TGC and CT (Each Alone or in Combinations) |
Discussion
A. baumannii is an opportunistic pathogen of relevant medical importance responsible for the various recalcitrant nosocomial infections worldwide, predominantly in the critically ill patients.48 A. baumannii is a MDR “red alert” pathogen with limited therapeutic options and therefore, impose life threatening conditions.49 In this study, we aimed to explore the activity of chitosan nanoparticles (CNPs) and to evaluate certain antibiotic combinations for the purpose of combating resistance mediated by this nightmare pathogen. Accordingly, a total of 51 A. baumannii clinical isolates were recovered from 730 different discharged clinical specimens including, pus, urine, sputum, bronchial lavage, and cerebrospinal fluid according to the hospital records of the Clinical Microbiology Central Laboratory of AL Kasr Al Aini hospital, Cairo, Egypt. Conventional standard Lab tests were used for identification followed by testing for the recA gene for confirmation. Detection of A. baumannii in microbiological laboratories is usually based on both phenotypic and genotypic methods.10,50 The accuracy, speed, identification, and interpretation of genotypic methods are higher than those of phenotypic identification methods.11 Bacterial genomes containing repeated sequences such as the ERIC sequence which can be used for epidemiological purpose to evaluate similarity between the isolates and their diversity as well as origin.51,52 In our study, the ERIC-PCR analysis of the recovered 51 MDR A. baumannii clinical isolates indicated that they bunched in nine clusters based on the obtained fingerprinting. This indicates that, there was cross‑transmission within hospitalized patients. Based on the obtained findings, it can be stated that ERIC‑PCR is a reliable method to demonstrate the clonal relatedness among A. baumannii recovered from different specimens of different patients.36,37 The findings of this study are similar to the studies reported by Ying et al and Hammoudi et al who were able to cluster A. baumannii strains based on their genetic relatedness and confirmed cross contamination in the clinical settings.51,53
Antimicrobial susceptibility tests were carried out on the obtained 51 MDR A. baumannii clinical isolates against a panel of antimicrobial agents composed of 14 different antimicrobial agents according to CLSI guidelines, 2018.34 The rational of antimicrobial selection to be enrolled in this study, was based on the international and empirical guidelines which defined the antimicrobial agents involved in the treatment protocols of A. baumannii-associated infections.1,34 The tested isolates exhibited high percentage of resistance which ranged from 92 to 100% towards cefepime, gentamicin, ceftriaxone, amikacin, imipenem, ciprofloxacin, trimethoprim sulfamethoxazole, ampicillin /sulbactam, doxycycline, piperacillin /tazobactam and meropenem. Results revealed that, all the tested A. baumannii clinical isolates were MDR (100%) of these, 92% were XDR. However, they showed lower resistance towards TGC and CT (4% of isolates were resistant). Accordingly, all A. baumannii isolates included in our study were MDR and 92% of these were XDR according to international standard definitions for the acquired resistance.31 The MDR status reported in the current study agrees with the findings of other two recent studies carried in Iraq.54,55 In this regard, two strategies have been performed to combat the resistance of A. baumannii which are the use of antibiotics combinations as well as CNPs either alone or in combination of the antibiotics that showed activities against the respective pathogens according to the results obtained from antimicrobial susceptibility tests. The antibiotic combinations were evaluated by checkerboard microdilution method as previously determined.38,39 The MICs of the antibiotics in the combinations were significantly reduced as compared to the MICs of each drug alone, and thereby gave synergism. In this study, the CT-MEM combination demonstrated a synergistic effect in 77.7% of the isolates, the findings of this study are like studies reported by other researchers.56,57 The TGC-MEM combination showed a synergistic and additive effects for 44.4% and 55.6% of the tested isolates, respectively. The findings of this study are like that of a recent study conducted by Li et al who evaluated the addition of TGC in combination with MEM against A. baumannii isolates in Heilongjiang Province in China.18 The CT-TGC combination showed antagonist effect for 100% of the isolates. This finding disagrees with a study carried by Li et al who evaluated the addition of TGC in combination with CT against A. baumannii isolates.18 The variation in the results may be due to differences in the time at which the studies were conducted, as well as differences in the geographic areas or it could be due to physicochemical interaction of the two antibiotics. The second strategy for overcoming MDR resistance that has been evaluated in this study was the use of nanoparticles. CNPs have antimicrobial activity against bacteria, fungi, and viruses. Interestingly, nanoparticles containing low molecular weight chitosan were previously verified to have high activity against Gram negative bacteria than Gram positive bacteria.25 In general, nanoparticles offer many distinctive advantages such as, reducing acute toxicity, overcoming resistance, and lowering cost, when compared to conventional antibiotics.58,59 There are different methods used for synthesis and characterization of nanoparticles either alone or in combination with different materials aiming to improve their antimicrobial activity.60–67 In this study, CNPs were prepared and characterized by DLS (to measure hydrodynamic diameter in the nanometer range) and confirmed via imaging the formed CNPs through HR-TEM. The size of CNPs at selected concentration was 441.7 nm and Zeta potential positive 37.7mV which mean that the formed nanoparticles were stable and of smaller particle size with positive charge on the surface of the CNPs indicating high sorption capacity and good antimicrobial properties.29 The HR-TEM images have shown the morphological properties and surface appearance of nanoparticles. Morphologically, the CNPs prepared were found to be spherical in shape and of smooth surface indicating good properties of the synthesized CNPs as previously reported.68,69 The CNPs formed in this study exhibited good antimicrobial activities against nine selected MDR A. baumannii (representing the nine ERIC-PCR clusters). The MIC of CNPs at concentrations in the range of 1–5 mg/mL, were found to be in the range of 0.16–0.25 mg/mL which were very promising results. The difference in the antimicrobial activities against MDR A. baumannii were dependent on the different concentrations of the tested CNPs. Our results are in line with other two previous studies conducted by Cobrado et al70 and Pourhajibagher et al.71 Interestingly, Pourhajibagher et al proved that CNPs produced significant reduction of 93.2% on the viable count of planktonic and of 55.3% on the biofilm formation of A. baumannii strains as compared to the control group.71 Furthermore, the activities of CNPs (5 mg/mL) when combined with CT, TGC or MEM, CT+MEM and TGC+MEM were evaluated via measuring the MDF as previously reported.48 Based on our findings, the CNPs at a concentration 5 mg/mL gave maximum antibacterial activity against the 51 MDR A. baumannii clinical isolates (MIC was 0.16 mg/mL against 44 isolates and 0.31 mg/mL against 7 isolates). This result was in accordance with a previous study conducted in 2020.40 Accordingly, CNPs at a concentration 5 mg/mL was selected to study their co-effects with above-mentioned antibiotics. Our results showed the respective CNPs-antibiotics combinations significantly increased the susceptibilities of the MDR A. baumannii isolates by 88.8, 66.6, 100, 77.7, and 44.4%, respectively. No significant effects were observed when CNPs (5 mg/mL) were combined with CT+TGC. The obtained results were in accordance with many recent studies conducted in 2021 on the antimicrobial activities of different nanoparticles either alone or in combination with other antimicrobials have proved activity against various pathogens. These included, Casein-silver NPs combined with TGC against A. baumannii,72 antibiofilm and anti-virulence potential of silver NPs against MDR A. baumannii,73 Cu:Ag bimetallic NPs for antibiotic-resistant bacteria,74 Lignin-Capped silver NPs,75 Smaller Copper Oxide Nanoparticles against MDR Bacteria,76 colistin-integrated chitosan nanoparticles.77 Our results revealed that CNPs when combined with MEM significantly increased the susceptibilities of the MDR A. baumannii isolates by 100% as compared to MEM alone. Our result is supported by the findings of another study that revealed, meropenem-loaded CNPs exhibited both in vitro and in vivo activities against a wide range of Gram positive and Gram negative MDR pathogens with a great potential for overcoming antimicrobial resistance.78 The encapsulation of CNPs plus antibiotics as well as their in vivo evaluations will be an important our prospective work.
Conclusion
The current study demonstrated the significant in vitro activities of CNPs either alone or in combination with CT, TGC, MEM, CT+MEM and TGC+MEM antibiotics. Combinations of CT+MEM and MEM+TGC showed synergism in 77.7% and 44.4% and additive effects in 22.3 and 55.6% of the tested MDR A. baumannii isolates (n=51), respectively. CNPs (5 mg/mL) exhibited good inhibitory activities (MIC was from 0.16 to 0.31 mg/mL) against nine MDR A. baumannii isolates that were selected according to the results of the ERIC-PCR. CNPs (5 mg/mL) when combined with CT, TGC or MEM, CT+MEM and TGC+MEM significantly increased the susceptibilities of the MDR A. baumannii isolates by 88.8%, 66.6%, 100%, 77.7%, and 44.4%, respectively. However, no significant effects were observed when CNPs (5 mg/mL) were combined with CT+TGC. The obtained finding will guide the physicians for the management of MDR A. baumannii-associated infections. However, further in vivo studies should be conducted to verify such activities and their potential use in human.
Data Sharing Statement
All the data supporting the findings are included in the manuscript.
Ethical Clearance
The study protocol was reviewed and approved by the institutional ethics committee, Faculty of Pharmacy, Ain Shams University (ENREC-ASU-2019-272). This study was conducted in accordance with the ethical principles stated in the Declaration of Helsinki.
Acknowledgments
We would like to acknowledge the microbiology laboratories of New Kasr El Aini Hospital, Cairo, Egypt for providing us with the clinical isolates. We would like to acknowledge Department of Microbiology and immunology, of both Faculty of pharmacy, Ain Shams University (ASU) and Heliopolis University for Sustainable Development, Egypt for providing support and facilities whenever needed.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. World Health Organization (WHO). List of bacteria for which new antibiotics are urgently needed; 2017. Available from: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed.
2. Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis. 2008;197:1079–1081. doi:10.1086/533452
3. Gordon NC, Png K, Wareham DW. Potent synergy and sustained bactericidal activity of a vancomycin-colistin combination versus multidrug-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54(12):5316–5322. doi:10.1128/AAC.00922-10
4. Lee CR, Lee JH, Park M, et al. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017;7:55. doi:10.3389/fcimb.2017.00055
5. Wong D, Nielsen TB, Bonomo RA, et al. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev. 2017;30:409–447. doi:10.1128/CMR.00058-16
6. Yang CH, Su PW, Moi SH, Chuang LY. Biofilm formation in Acinetobacter Baumannii: genotype-phenotype correlation. Molecules. 2019;24(10):1849. doi:10.3390/molecules24101849
7. Mangas EL, Rubio A, Alvarez-Marin R, et al. Pangenome of Acinetobacter baumannii uncovers two groups of genomes, one of them with genes involved in CRISPR/Cas defence systems associated with the absence of plasmids and exclusive genes for biofilm formation. Microb Genom. 2019;5:e000309. doi:10.1099/mgen.0.000309
8. Tang B, Gong T, Zhou X, et al. Deletion of cas3 gene in Streptococcus mutans affects biofilm formation and increases fluoride sensitivity. Arch Oral Biol. 2019;99:190–197. doi:10.1016/j.archoralbio
9. Cui L, Wang X, Huang D, et al. CRISPR-cas3 of salmonella upregulates bacterial biofilm formation and virulence to host cells by targeting quorum-sensing systems. Pathogens. 2020;9:53. doi:10.3390/pathogens9010053
10. Chen TL, Siu LK, Wu RC, et al. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin Microbiol Infect. 2007;13:801e6. doi:10.1111/j.1469-0691.2007.01744.x
11. Falah F, Shokoohizadeh L, Adabi M. Molecular identification and genotyping of Acinetobacter baumannii isolated from burn patients by PCR and ERIC-PCR. Scars Burn Heal. 2019;5:1–7. doi:10.1177/2059513119831369
12. Wang H, Chen M, Ni Y, et al. Antimicrobial resistance among clinical isolates from the Chinese Surveillance Study (CMSS) 2003–2008. Int J Antimicrobial Agents. 2010;35:227–234. doi:10.1016/j.ijantimicag.2009.11.010
13. Cantas L, Shah S, Cavaco L, et al. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front Microbiol. 2013;4:96–110. doi:10.3389/fmicb.2013.00096
14. Gootz TD, Marra A. Acinetobacter baumannii: an emerging multidrug-resistant threat. Expert Rev Anti Infect Ther. 2008;6:309–325. doi:10.1586/14787210.6.3.309
15. Coates AR, Hu Y. Novel approaches to developing new antibiotics for bacterial infections. Br J Pharmacol. 2007;152:1147–1154. doi:10.1038/sj.bjp.0707432
16. Lee NY, Wang CL, Chuang YC, et al. Combination carbapenem-sulbactam therapy for critically ill patients with multidrug-resistant Acinetobacter baumannii bacteremia: four case reports and an in vitro combination synergy study. Pharmacother. 2007;27:1506–1511. doi:10.1592/phco.27.11.1506
17. Lane D. Designer combination therapy for cancer. Nat Biotechnol. 2006;24(2):163–164. doi:10.1038/nbt0206-163
18. Li J, Fu Y, Zhang J, et al. Efficacy of tigecycline monotherapy versus combination therapy with other antimicrobials against carbapenem-resistant Acinetobacter baumannii sequence type 2 in Heilongjiang Province. Ann Palliat Med. 2019;8(5):651–659. doi:10.21037/apm.2019.11.06
19. Wiseman LR, Wagstaff AJ, Brogden RN, et al. Meropenem. Drugs. 1995;50:73–101. doi:10.2165/00003495-199550010-00007
20. Zavascki AP, Goldani LZ, Li J, Nation RL. Nation, Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother. 2007;60(6):1206–1215. doi:10.1093/jac/dkm357
21. Jean SS, Hsieh TC, Hsu CW, Lee WS, Bai KJ, Lam C. Comparison of the clinical efficacy between tigecycline plus extended-infusion imipenem and sulbactam plus imipenem against ventilator-associated pneumonia with pneumonic extensively drug-resistant Acinetobacter baumannii bacteremia, and correlation of clinical efficacy with in vitro synergy tests. J Microbiol Immunol Infect. 2016;49(6):924–933. doi:10.1016/j.jmii.2015.06.009
22. Nguyen TV, Nguyen TTH, Wang SL, et al. Preparation of chitosan nanoparticles by TPP ionic gelation combined with spray drying, and the antibacterial activity of chitosan nanoparticles and a chitosan nanoparticle–amoxicillin complex. Res Chem Intermed. 2017;43:3527–3537. doi:10.1007/s11164-016-2428-8
23. Marudova M, Zahariev N, Milenkova S, Pilicheva B, Viraneva A, Yovcheva Y. Development and in-vitro characterization of benzydamine loaded chitosan nanoparticles. J Macromolecular Symposia. 2021;395:2000279. doi:10.1002/masy.202000279
24. Parveen S, Sahoo SK. Evaluation of cytotoxicity and mechanism of apoptosis of doxorubicin using folate-decorated chitosan nanoparticles for targeted delivery to retinoblastoma. Cancer Nanotechnol. 2010;1:47–62. doi:10.1007/s12645-010-0006-0
25. Kong M, Chen XG, Xing K, Park HJ. Antimicrobial properties of chitosan and mode of action: a state of the art review. Int J Food Microbiol. 2010;144(1):51–63. doi:10.1016/j.ijfoodmicro.2010.09.012
26. Costa EM, Silva S, Vicente S, Veiga M, Tavaria F, Pintado MM. Chitosan as an effective inhibitor of multidrug resistant Acinetobacter baumannii. Carbohydr Polym. 2017;178:347–351. doi:10.1016/j.carbpol.2017.09.055
27. Berger J, Reist M, Mayer JM, Felt O, Peppas NA, Gurny R. Structure and interactions in covalently and ionically crosslinked chitosan hydrogels for biomedical applications. Eur J Pharm Biopharm. 2004;57(1):19–34. doi:10.1016/s0939-6411(03)00161-9
28. Hu B, Wang SS, Li J, Zeng XX, Huang QR. Assembly of bioactive peptide–chitosan nanocomplexes. J Phys Chem B. 2011;115(23):7515–7523. doi:10.1021/jp2013557
29. Qi L, Xu Z, Jiang X, Hu C, Zou X. Preparation and antibacterial activity of chitosan nanoparticles. Carbohydr Res. 2004;339(16):2693–2700. doi:10.1016/j.carres.2004.09.007
30. Hebeish AA, Ramadan MA, Montaser AS, Farag AM. Preparation, characterization and antibacterial activity of chitosan-g-poly acrylonitrile/silver nanocomposite. Int J Biol Macromol. 2014;68:178–184. doi:10.1016/j.ijbiomac.2014.04.028
31. Berne BJ, Pecora R. Dynamic Light Scattering: With Applications to Chemistry, Biology, and Physics. Courier Corporation; 2000. doi:10.1016/0307-4412(77)90025-5
32. Al-Onazi WA, Ali MHH. Synthesis and characterization of cerium oxide hybrid with chitosan nanoparticles for enhancing the photodegradation of Congo Red dye. J Mater Sci: Mater Electron. 2021;32:12017–12030. doi:10.1007/s10854-021-05832-7
33. Chiang MC, Kuo SC, Chen YC, Lee YT, Chen TL, Fung CP. Polymerase chain reaction assay for the detection of Acinetobacter baumannii in endotracheal aspirates from patients in the intensive care unit. J Microbiol Immunol Infect. 2011;44(2):106–110. doi:10.1016/j.jmii.2010.04.003
34. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement.
35. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi:10.1111/j.1469-0691.2011.03570.x
36. Versalovic J, Koeuth T, Lupski R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 1991;19(24):6823–6831. doi:10.1093/nar/19.24.6823
37. Tsai HC, Huang TY, Chen JS, Chen WJ, Lin CY, Hsu BM. Acinetobacter baumannii and methicillin-resistant Staphylococcus aureus in long-term care facilities in eastern Taiwan. Tzu-Chi Med J. 2019;31(4):222. doi:10.4103/tcmj.tcmj_136_18
38. Hsieh MH, Chen MY, Victor LY, Chow JW. Synergy assessed by checkerboard a critical analysis. Diagn Microbiol Infect Dis. 1993;16(4):343–349. doi:10.1016/0732-8893(93)90087-n
39. Abdelkader MM, Aboshanab KM, El-Ashry MA, Aboulwafa MM. Prevalence of MDR pathogens of bacterial meningitis in Egypt and new synergistic antibiotic combinations. PLoS One. 2017;12(2):e0171349. doi:10.1371/journal.pone.0171349
40. Alqahtani F, Aleanizy F, El Tahir E, et al. Antibacterial activity of chitosan nanoparticles against pathogenic N. gonorrhoea. Int J Nanomed. 2020;15:7877–7887. doi:10.2147/ijn.s272736
41. Agarwal M, Agarwal MK, Shrivastav N, et al. Preparation of chitosan nanoparticles and their in-vitro characterization. Int J Life Sci Scienti Res. 2018;4(2):1713–1720. doi:10.21276/ijcesr.2018.4.2.17
42. Mohammadpour Dounighi N, Eskandari R, Avadi MR, et al. Preparation and in vitro characterization of chitosan nanoparticles containing Mesobuthus eupeus scorpion venom as an antigen delivery system. J Venom Anim Toxins Incl Trop Dis. 2012;18:44–52. doi:10.1590/S1678-91992012000100006
43. Valgas C, De Souza SM, Smânia EFA. Screening methods to determine antibacterial activity of natural products. Braz J Microbiol. 2007;38:369–380. doi:10.1590/S1517-83822007000200034
44. Magaldi S, Mata-Essayag S, Hartung de Capriles C, et al. Well diffusion for antifungal susceptibility testing. Int J Infect Dis. 2004;8(1):39–45. doi:10.1016/j.ijid.2003.03.002
45. Donia M, Hamann MT. Marine natural products and their potential applications as anti-infective agents. Lancet Infect Dis. 2003;3(6):338–348. doi:10.1016/s1473-3099(03)00655-8
46. Katas H, Mohamad A, Zin NM. Physicochemical effects of chitosan-tripolyphosphate nanoparticles on antibacterial activity against gram-positive and gram-negative bacteria. J Med Sci. 2011;11:192–197. doi:10.3923/jms.2011.192.197
47. Huguet A, Pensec J, Soumet C. Resistance in Escherichia coli: variable contribution of efflux pumps with respect to different fluoroquinolones. J Appl Microbiol. 2013;114(5):1294–1299. doi:10.1111/jam.12156
48. Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91–102. doi:10.1038/nrmicro.2017.148
49. Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(10):3471–3484. doi:10.1128/aac.01464-06
50. Bergogne-Bérézin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9(2):148–165. doi:10.1128/cmr.9.2.148
51. Ying C, Li Y, Wang Y, Zheng B, Yang C. Investigation of the molecular epidemiology of Acinetobacter baumannii isolated from patients and environmental contamination. J Antibiot. 2015;68(9):562–567. doi:10.1038/ja.2015.30
52. Dalla-Costa LM, Irino K, Rodrigues J, Rivera IN, Trabulsi LR. Characterization of diarrhoeagenic Escherichia coli clones by ribotyping and ERIC-PCR. J Med Microbiol. 1998;47(3):227–234. doi:10.1099/00222615-47-3-227
53. Hammoudi D, Moubareck CA, Hakime N, et al. Spread of imipenem-resistant Acinetobacter baumannii co-expressing OXA-23 and GES-11 carbapenemases in Lebanon. Int J Infect Dis. 2015;36:56–61. doi:10.1016/j.ijid.2015.05.015
54. Mshachal MA, Abdulrahman TR, Khudair MS, Hassan JS. Molecular detection of multidrug resistance Acinetobacter baumannii from different clinical samples. Iraqi J Med Sci. 2017;15:314–323.
55. Al Marjani M, Al-Ammar M, Kadhem E. Occurrence of ESBL and MBL genes in Pseudomonas aeruginosa and Acinetobacter baumannii isolated from Baghdad, Iraq. Int J Cur Res. 2013;5:2482–2486.
56. Soudeiha MAH, Dahdouh EA, Azar E, Sarkis DK, Daoud Z. In vitro evaluation of the colistin-carbapenem combination in clinical isolates of A. baumannii using the checkerboard, Etest, and time-kill curve techniques. Front Cell Infect Microbiol. 2017;7:209. doi:10.3389/fcimb.2017.00209
57. Katip W, Uitrakul S, Oberdorfer P. A comparison of colistin versus colistin plus meropenem for the treatment of carbapenem-resistant Acinetobacter baumannii in critically ill patients: a propensity score-matched analysis. Antibiotics. 2020;9(10):647. doi:10.3390/antibiotics9100647
58. Pal S, Tak YK, Song JM. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the Gram-negative bacterium Escherichia coli. Appl Environ Microbiol. 2007;73(6):1712–1720. doi:10.1128/aem.02218-06
59. Weir E, Lawlor A, Whelan A, Regan F. The use of nanoparticles in anti-microbial materials and their characterization. Analyst. 2008;133(7):835–845. doi:10.1039/b715532h
60. Safari-Amiri M, Mortazavi-Derazkola S, Salavati-Niasari M, et al. Synthesis and characterization of Dy2O3 nanostructures: enhanced photocatalytic degradation of rhodamine B under UV irradiation. J Mater Sci Mater Electron. 2017;28:6467–6474. doi:10.1007/s10854-017-6333-8
61. Mortazavi-Derazkola S, Naimi-Jamal MR, Ghoreishi SM. Synthesis, characterization, and atenolol delivery application of functionalized mesoporous hydroxyapatite nanoparticles prepared by microwave-assisted co-precipitation method. Curr Drug Deliv. 2016;13(7):1123–1129. doi:10.2174/1567201813666160321115543
62. Shafiee Ardestani M, Bitarafan-Rajabi A, Mohammadzadeh P, et al. Synthesis and characterization of novel 99mTc-DGC nano-complexes for improvement of heart diagnostic. Bioorg Chem. 2020;96:103572. doi:10.1016/j.bioorg.2020.103572
63. Ebrahimzadeh MA, Mortazavi-Derazkola S, Zazouli MA. Eco-friendly green synthesis and characterization of novel Fe3O4/SiO2/Cu2O–Ag nanocomposites using Crataegus pentagyna fruit extract for photocatalytic degradation of organic contaminants. J Mater Sci: Mater Electron. 2019;30:10994–11004. doi:10.1007/s10854-019-01440-8
64. Xu C, Ou M, Zhou H, Yang C. Preparation and properties of bifunctional Gd2O3/GQD composite nanoparticles. J Rare Earths. 2021. doi:10.1016/j.jre.2021.06.010
65. Mani S, Balasubramanian B, Balasubramani R, et al. Synthesis and characterization of proanthocyanidin-chitosan nanoparticles: an assessment on human colorectal carcinoma HT-29 cells. J Photochem Photobiol B: Biol. 2020;210:111966. doi:10.1016/j.jphotobiol.2020.111966
66. Shirzadi-Ahodashti M, Ebrahimzadeh MA, Ghoreishi SM, Naghizadeh A, Derazkola S. Facile and eco-benign synthesis of a novel MnFe2O4@SiO2@Au magnetic nanocomposite with antibacterial properties and enhanced photocatalytic activity under UV and visible-light irradiations. Appl Organomet Chem. 2020;34(5):e5614. doi:10.1002/aoc.5614
67. Zayed MF, Eisa WH, Abd ElHameed M, Hosam AM, Zeid A. Spectroscopic investigation of chitosan-supported Cu2O/CuO nanocomposite; a separable catalyst for water-pollutants degradation. J Alloys Compd. 2020;835:155306. doi:10.1016/j.jallcom.2020.155306
68. Yang W, Fu J, Wang T, He N. Chitosan/sodium tripolyphosphate nanoparticles: preparation, characterization and application as drug carrier. J Biomed Nanotechnol. 2009;5(5):591–595. doi:10.1166/jbn.2009.1067
69. Gan Q, Wang T. Chitosan nanoparticle as protein delivery carrier–systematic examination of fabrication conditions for efficient loading and release. Colloids Surf B Biointerfaces. 2007;59(1):24–34. doi:10.1016/j.colsurfb.2007.04.009
70. Cobrado L, Azevedo MM, Silva-Dias A, Ramos JP, Pina-Vaz C, Rodrigues AG. Cerium, chitosan and hamamelitannin as novel biofilm inhibitors? J Antimicrob Chemother. 2012;67(5):1159–1162. doi:10.1093/jac/dks007
71. Pourhajibagher M, Hosseini N, Boluki E, Chiniforush N, Bahador A. Photoelimination potential of chitosan nanoparticles-indocyanine green complex against the biological activities of Acinetobacter baumannii strains: a preliminary in vitro study in burn wound infections. J Lasers Med Sci. 2020;11(2):187–192. doi:10.34172/jlms.2020.31
72. Chen YH, Wang WH, Lin SH, et al. Synergistic antibacterial effect of casein-AgNPs combined with tigecycline against Acinetobacter baumannii. Polymers. 2021;13(9):1529. doi:10.3390/polym13091529
73. Hetta HF, Al-Kadmy IMS, Khazaal SS, et al. Antibiofilm and antivirulence potential of silver nanoparticles against multidrug-resistant Acinetobacter baumannii. Sci Rep. 2021;11(1):10751. doi:10.1038/s41598-021-90208-4
74. Mureed S, Naz S, Haider A, et al. Development of multi-concentration Cu: Ag Bimetallic nanoparticles as a promising bactericidal for antibiotic-resistant bacteria as evaluated with molecular docking study. Nanoscale Res Lett. 2021;16(1):91. doi:10.1186/s11671-021-03547-6
75. Slavin YN, Ivanova K, Hoyo J, et al. Novel lignin-capped silver nanoparticles against multidrug-resistant bacteria. ACS Appl Mater Interfaces. 2021;13(19):22098–22109. doi:10.1021/acsami.0c16921
76. Abbasi A, Ghorban K, Nojoomi F, Dadmanesh M. Smaller copper oxide nanoparticles have more biological effects versus breast cancer and nosocomial infections bacteria. Asian Pac J Cancer Prev. 2021;22(3):893–902. doi:10.31557/apjcp.2021.22.3.893
77. Elnaggar YS, Elwakil BH, Elshewemi SS, El-Naggar MY, Bekhit AA, Olama ZA. Novel Siwa propolis and colistin-integrated chitosan nanoparticles: elaboration; in vitro and in vivo appraisal. Nanomedicine. 2020;15(13):1269–1284. doi:10.2217/nnm-2019-0467
78. Abdelkader A, El-Mokhtar MA, Abdelkader O, Hamad MA, Elsabahy M, El-Gazayerly ON. Ultrahigh antibacterial efficacy of meropenem-loaded chitosan nanoparticles in a septic animal model. Carbohydr Polym. 2017;174:1041–1050. doi:10.1016/j.carbpol.2017.07.030
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.