Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 12
Effects of coagulation factor VII polymorphisms on warfarin sensitivity and responsiveness in Jordanian cardiovascular patients during the initiation and maintenance phases of warfarin therapy
Authors AL-Eitan LN , Almasri AY, Al-Habahbeh SO
Received 3 October 2018
Accepted for publication 17 December 2018
Published 14 January 2019 Volume 2019:12 Pages 1—8
DOI https://doi.org/10.2147/PGPM.S189458
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin Bluth
Laith N AL-Eitan,1,2 Ayah Y Almasri,1 Sahar O Al-Habahbeh1
1Department of Applied Biological Sciences, Jordan University of Science and Technology, Irbid 22110, Jordan; 2Department of Biotechnology and Genetic Engineering, Jordan University of Science and Technology, Irbid 22110, Jordan
Purpose: This study aims to investigate the relationships between genetic polymorphisms of the coagulation factor VII (FVII) gene and warfarin responsiveness and sensitivity.
Patients and methods: The study population consisted of 417 subjects (207 Jordanian cardiovascular patients and 210 healthy individuals). Cardiovascular patients were classified into two groups: those sensitive to warfarin dosage (sensitive, moderate, and resistant) and those responsive to warfarin based on International Normalized Ratios (INRs; poor, good, and extensive responders). The HVR4 polymorphism of the FVII gene was genotyped.
Results: Our results showed that there are significant differences between patients and controls according to both genotypic and allelic frequencies (P<0.0001) in the genetic susceptibility study. Moreover, the pharmacogenetics study reported that HVR4 had no association with warfarin sensitivity or responsiveness during the initiation and maintenance phases of therapy, the only significant differences were in the INR outcome measured during the maintenance phase of therapy (P=0.012).
Conclusion: Our data suggests lacking of association between the HVR4 polymorphism in the FVII gene and warfarin sensitivity and responsiveness during the initiation and maintenance phases of therapy. It is possible that these patients carry additional mutations in genes involved in the coagulation pathway.
Keywords: HVR4 polymorphism, cardiovascular disease, pharmacogenetics study
Introduction
Warfarin (3-(α-acetonylbenzyl)-4-hydroxycoumarin) has been in commercial use for therapeutic purposes for six decades and remains the most common oral anticoagulant in use.1,2 Although it was first used in 1948 as a rat poison, it has been medically approved for human use in the USA since 1954.3
Warfarin is prescribed for the treatment and prevention of myocardial infarction (MI), ischemic stroke, venous thrombosis, atrial fibrillation, and after cardiac valve replacement procedures.4 Despite its many applications, warfarin has a narrow therapeutic range and dosage varies widely among patients. It has been found that an inadequately low dose cannot inhibit thromboembolism, while an overdose results in an increased risk of bleeding.5
Warfarin acts as an anticoagulant by inhibiting the vitamin K epoxide reductase enzyme, which is encoded for by the VKORC1. This inhibition prevents VKORC1’s ability to reproduce reduced vitamin K from its epoxide form (2,3-epoxide). Thus, vitamin K epoxide will be decreased in the liver, which, in turn, deactivates coagulation factors II, VII, IX, and X.5
More than 50 years ago, warfarin dose was determined by trial and error, with the initial dose (2–10 mg/day) based on indication and clinical factors.6 However, in 1982, the WHO’s Expert Committee on Biologic Standardization developed International Normalized Ratio (INR),7 which is defined as “the ratio of a patient’s prothrombin time to a control sample” and it is used for monitoring the effectiveness of warfarin and measures the blood coagulation pathway.8 In order to maintain treatment with warfarin efficiently and safely, INR should be kept within the target range (ie, the therapeutic range of about 2–3). Therefore, the dosage should be tailored to target INR.8
Warfarin personalization approaches have been followed. Factors such as ethnicity, comorbidities, concomitant medication, vitamin K intake, and genetic variation all play a role in patients’ sensitivity to warfarin.9
Genetic variation has been found to modulate warfarin response and sensitivity, and at least 30 genes play a role in warfarin’s anticoagulation mechanism, including VKORC1, CYP2C9, FVII, APOE, epoxide hydrolase 1, and many others.10 From among these genes, two genes have been extensively studied in various populations: CYP2C9 and VKORC1. Polymorphisms in these two genes affect the required dose and influence the initial INR responses at the start of warfarin treatment.9
Exactly 25% of clinically used medications are metabolized by CYP2C9, which is located on chromosome 10q24.2 and spans 55 kb.11,12 The genetic variability of CYP2C9 influences the treatment regimens of drugs with a narrow therapeutic index, including warfarin.13
In contrast, FVII is an inactive protease that depends on vitamin K, which, upon contact with tissue factor, is converted to its active form and then activated to factor X and factor IX. Genetic polymorphisms in the FVII gene may alter the responsiveness of warfarin during the initiation of treatment.14,15
Previous studies have investigated the association of FVII gene polymorphisms, including the R353Q polymorphism, the intron seven polymorphism (HVR4) and the insertion/deletion promoter polymorphism (–323 0/10 bp), with the risk of cardiovascular disease.16 The variable tandem repeat HVR4 polymorphism consists of three alleles (H5, H6, and H7). Some studies found a protective role for H7H7 genotypes in the context of MI in patients with cardiovascular disease,17,18 but such results could not be confirmed in some populations.19,20
Algorithms with genetic factors (CYP2C9 and VKORC1), clinical factors, and demographic factors to predict the warfarin dose could reduce the risk of overdose in the initiation of treatment.5 Still, there is no accurate way to determine the warfarin dose to lower the risk of bleeding. Therefore, it is helpful to increase the number of candidate genes responsible for the metabolism of oral anticoagulants.21,22
In this study, we examined the effects of the HVR4 polymorphism of the FVII gene and other clinical characteristics on warfarin sensitivity and responsiveness during initiation and maintenance of treatment in Jordanian cardiovascular patients. To the best of our knowledge, there are no studies conducted in the Jordanian population to assess the association of these polymorphisms with cardiovascular patients and warfarin sensitivity.
Patients and methods
Study population and data collection
The Human Research Committees at Jordan University of Science and Technology in Irbid, Jordan and the Royal Medical Services in Amman, Jordan approved this study. This study was also conducted in accordance with the Declaration of Helsinki. Patients were recruited from the outpatient anticoagulation clinic at the Queen Alia Heart Institute in Amman, Jordan after they gave their written informed consent. The study population consisted of 417 subjects: patients with cardiovascular disease, n=207 and healthy control, n=210. The healthy control subjects were defined as subjects older than 18 years who did not have any cardiovascular disorder. Totally, 207 patients met the inclusion criteria for this study, including patients 18 years or older who had been receiving warfarin for at least 3 months. Patients who did not give written informed consent, those who were pregnant, alcoholic, or taking CYP2C9 inducer medications, and those who did not visit the anticoagulation clinic regularly were excluded.
Data on demographic (age, height, weight, and gender) and lifestyle (smoking status and dietary plane) factors were collected along with the patients’ medical records. Health conditions and clinical features such as the main indication for treatment with warfarin, target INR, average weekly warfarin doses needed to achieve the target INR, and the use of concomitant medication were determined from the medical records.
Study design
In this study, patients were divided according to warfarin sensitivity as being resistant, normal, or sensitive to warfarin. Warfarin resistance is rare; but those who needed large daily doses (dose required >49 mg/week) to maintain their INR within a normal therapeutic range were called extensive metabolizers. Warfarin normal patients require intermediate dose (doses between 21 and 49 mg/week) and patients in this group were called intermediate metabolizers. Warfarin-sensitive patients need a lower dose (required dose <21 mg/week) and patients in this group were called poor metabolizers.23
Moreover, patients were divided according to warfarin responsiveness as good responders with an INR value within the target range (therapeutic range), poor responders with an INR value below the target range, and extensive responders with an INR above the target range.
Outcome measure
In this study, 207 patients were treated with warfarin in two phases; the first phase is called the initiation phase of the treatment, which is defined as the phase in which initially the INR value of the patient is unstable and flops up and down, with the doses given at that time and the measured INR values being called the initiation dose and initiation INR, respectively. Otherwise, of 207 patients, only 129 had entered the second stage, where the patient is within the target INR in at least two consecutive visits, and thus, a stabilization phase of therapy is achieved; the dose is referred to as the maintenance dose and the INR value measured during this phase as the maintenance INR.24
DNA analysis
A blood sample (5 mL) was collected for the determination of venous INR and for genotyping FVII polymorphisms.
Genomic DNA was extracted by the Gentra Pure gene Blood Kit according to the manufacturer’s protocol (Gentra System; Promega Corporation, Fitchburg, WI, USA). The concentration and purity of the extracted DNA were evaluated using the Nano-Drop ND-1000 ultraviolet–visible spectrophotometer (BioDrop, Cambridge, UK).
Gene amplification was carried out using a PCR assay. DNA samples were amplified by PCR in a final volume of 25 µL containing 1.5 µL forward primer, 1.5 µL reverse primer, 12.5 µL master mix, 9.5 µL nuclease-free water, and 2 µL genomic DNA. Sequences for the forward and reverse primers were 5-AAT GTG ACT TCC ACA CCT CC-3 and 5-GAT GTC TGT CTG TCT GTG GA-3, respectively, and taken from a previously published paper.25 PCR conditions consisted of 94°C for 11 minutes and 35 cycles of 30-second denaturation at 94°C, 30-second annealing at 55°C, and 30-second extension at 72°C. The final cycle included a 3-minute extension step at 72°C. PCR products were visualized by 3% gel electrophoresis with ethidium bromide.
Statistical analysis
To evaluate which of the selected polymorphisms of FVII (HVR4) was associated with warfarin response, several statistical analyses were performed to find the genetic associations, which included the chi-squared test, Kruskal–Wallis test, one-way ANOVA, Tukey pairwise comparison test, and the non-parametric correlation test. SPSS version 21.0 was used to perform all analyses.
Results
Baseline characteristics of the study population
A total of 207 unrelated Jordanian (Arab) cardiovascular patients treated with warfarin, including 94 female (45.41%) and 113 male (54.59%), were included in this study. The average age (± SD) was 53.86±15.47 years. The average weekly initiation dose of warfarin was 38.10 mg/week. There were 32 extensive responders (15.5%), 144 moderate responders (69.5%), and 31 poor responders (15.0%). The descriptive analysis of the demographic and clinical pattern of the study group (n=207) as well as the demographic pattern for the control group (n=210) are presented in Tables 1 and 2, respectively. There were significant correlations between the warfarin dose, age of the patient and smoking status, while no correlation was observed with the body mass index, gender, comorbidities and the indication of treatment (Table 1).
  | Table 2 Demographic characteristics of 210 controls Note: aMean SD in square brackets. Abbreviation: BMI, body mass index. |
Genotypic and allele frequencies of the FVII gene
Genotypic and allelic frequencies at FVII loci in patients and controls are shown in Table 3. The genotypic and allelic distribution for HVR4 was significantly different between patients and the controls (P<0.001).
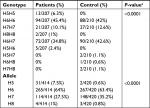  | Table 3 Allele and genotype frequencies of the FVII polymorphisms in 210 controls and 207 Jordanian cardiovascular patients Note: aFisher’s exact test with P<0.05 is considered significant. |
Association of warfarin sensitivity and FVII polymorphism
When comparing the three inclusion groups (sensitive, moderate and resistant), no significant differences in the proportions of the FVII (HVR4) genotypes were observed, during the initiation and maintenance phases of therapy with Fisher’s Exact value = 12.322 and P=0.192 (Table 4). Moreover, no significant differences in the genotypic distribution of 129 patients over the above-mentioned warfarin sensitive groups were observed during the maintenance phase of therapy (Fisher’s Exact value = 11.001, P=0.265; Table 5).
In case of the association between warfarin required dose and FVII (HVR4) genotypes, no statistically significant differences were observed during the two phases (with P=0.192 and 0.555, respectively; Table 6).
  | Table 6 Association of FVII polymorphism with variability in the dose of warfarin required Note: aOne-way ANOVA with P<0.05 is considered significant, mean doses. |
Association of warfarin responsiveness and FVII polymorphism
When comparing the three warfarin response groups (poor, good and extensive responders), no significant differences in the proportions of 207 patients between the FVII (HVR4) genotypes were observed with Fisher’s Exact value = 4.884 and P=0.920 (Table 7). In addition, no significant differences in this genotypic distribution of 129 patients over the above-mentioned warfarin response groups were observed during the maintenance phase of therapy with Fisher’s Exact value = 11.034 and P=0.296 (Table 8).
In 207 cardiovascular patients treated with warfarin, no significant differences were observed with respect to FVII (HVR4) genotypes and initiation INR values P=0.995 (Table 9). However, significant differences were found between the maintenance INR values measured for 129 patients during the stabilization period and FVII (HVR4) genotypes (P=0.012; Table 9).
  | Table 9 Association of FVII polymorphism with INR treatment outcome Note: aOne-way ANOVA with P<0.05 is considered significant, mean INR. Abbreviation: INR, International Normalized Ratio. |
Discussion
Warfarin is a widely used drug, even though it is complicated due to interindividual variability, requiring patients to visit coagulation clinics regularly and repeat INR measurements.5,22
It was shown that genetic polymorphisms in the VKORC1 and CYP2C9 genes are associated with warfarin responsiveness during initiation and maintenance of therapy.14,26,27 Moreover, numerous clinical studies of diverse populations have shown that CYP2C9 polymorphisms (* 2, * 3), VKORC1-1639G>A, and FVII-411G>T and their allelic variants are significant to determination of warfarin dosage.5,28–30
Numerous studies have confirmed that the individual variation in the warfarin dose, in addition to clinical factors, is also related to the CYP2C9 and VKORC1 genotypes. In contrast, few studies have been conducted to investigate the influence of other genes on the remaining variability, especially with regard to FVII. Polymorphisms in these genes have previously been suggested as contributors to variation in stable dose.31–33
In this study, we examined the association of FVII polymorphisms (HVR4, 37 bp repeat) and susceptibility to cardiovascular heart disease and the association between this polymorphism in addition to non-genetic factors with the response and sensitivity to warfarin during the initiation and maintenance periods of therapy in 207 Jordanian (Arabian) cardiovascular patients.
It was found that age and smoking status, which have been previously reported as causative factors,34 are correlated with the dose requirements of warfarin in our study, while the remaining factors such as BMI, gender and comorbidities are not associated with warfarin required dose (Table 1).
Iacoviello et al reported that the FVII polymorphisms are associated with the risk of familial MI. They found that the H7allelic frequency was significantly lower in patients with a thrombosis family history than in controls, so they suggested that the H7 allele protects against familial MI. Otherwise, carrying of H5 allele appears to be associated with an increased risk of familial MI.35 Accordingly, our study showed that the frequency of (H5H5/H5H6) was found only in patients (6.3%, 2.4%) and absent in controls (0%), while the frequency of (H6H7/H7H7) was significantly higher in controls (42.6%, 12.6%) than in patients (34.8%, 10.1%; P<0.0001), as shown in Table 3.
Among the three classified groups (sensitive, moderate, and resistant), we found that 30.8% of sensitive patient were homozygous for the H5H5 allele and 20% of them were heterozygous for this allele with H5H6 during initiation phase of therapy but it was not significantly different with a P=0.192 (Table 4), Moreover, the H5H5 genotype did not show any significant differences among the three groups during the maintenance phase of therapy (P=0.265), which means that there is no significant effect of FVII polymorphism on warfarin sensitivity in our population (Table 5).
In addition, we divided the patients into three groups (poor, good, and extensive) of responders according to the average INR measurements during initiation and maintenance of therapy. Our results showed no significant differences among the three groups in the FVII polymorphism during both initiation (P=0.920) and maintenance (P=0.296) and the results are shown in Tables 7 and 8.
According to our findings, there is no effect of the FVII polymorphism on warfarin dosage during the initiation and maintenance phases of therapy in a Jordanian population with P=0.192 and P=0.555, respectively (Table 6).
Significant differences exhibited in this study with regard to the INR outcome measures during the maintenance phase of therapy with a P=0.012, in contrast, there were no significant differences in regard to the initiation INR measurements with a (P=0.995; Table 9). It is possible that these patients carry additional mutations in genes involved in the coagulation pathway. Similarly, a study carried out by Shikata et al32 showed that homozygous carriers for the genotype H7H7 in the FVII gene had lower INR values than those of the genotype H6H6. Taking these results into account, mutations in vitamin K-dependent protein genes were associated with a lower INR value, indicating a reduced susceptibility to warfarin.27
Conclusion
We suggest that this study must be validated in other populations. We hope to continue and develop this study with a larger sample size, since we need to discover new factors that contribute to variability in dosing and capture additional data from rare genotyping cases. Although the use of genetic testing and dosing algorithms is currently possible in the USA, these tools are currently not available for clinical application. As personalized medicine becomes more of a reality, physicians will be able to prescribe warfarin with greater efficiency and ease.
Acknowledgments
We thank the Anticoagulant Clinic in Queen Alia Heart Institute for its generous assistance in the course of this study. This work was supported by the Deanship of Research at Jordan University of Science and Technology under the grant number (207/2017).
Disclosure
The authors report no conflicts of interest in this work.
References
Kaminsky LS, Zhang ZY. Human P450 metabolism of warfarin. Pharmacol Ther. 1997;73(1):67–74. | ||
Natarajan S, Ponde CK, Rajani RM, et al. Effect of CYP2C9 and VKORC1 genetic variations on warfarin dose requirements in Indian patients. Pharmacol Rep. 2013;65(5):1375–1382. | ||
Raviña E. The Evolution of Drug Discovery: From Traditional Medicines to Modern Drugs. Weinheim city: John Wiley & Sons; 2011. | ||
Hirsh J, Dalen JE, Anderson DR, et al. Oral anticoagulants: mechanism of action, clinical effectiveness, and optimal therapeutic range. Chest. 1998;114(5 Suppl):445S–469. | ||
Yin T, Miyata T. Warfarin dose and the pharmacogenomics of CYP2C9 and VKORC1 – rationale and perspectives. Thromb Res. 2007;120(1):1–10. | ||
Gage BF, Lesko LJ. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J Thromb Thrombolysis. 2008;25(1):45–51. | ||
Hirsh J, Dalen J, Anderson DR, et al. Oral anticoagulants: mechanism of action, clinical effectiveness, and optimal therapeutic range. Chest. 2001;119(1 Suppl):8S–21. | ||
Piatkov I, Rochester C, Jones T, Boyages S. Warfarin toxicity and individual variability – clinical case. Toxins. 2010;2(11):2584–2592. | ||
Li C, Schwarz UI, Ritchie MD, Roden DM, Stein CM, Kurnik D. Relative contribution of CYP2C9 and VKORC1 genotypes and early INR response to the prediction of warfarin sensitivity during initiation of therapy. Blood. 2009;113(17):3925–3930. | ||
Hughes DA, Pirmohamed M. Warfarin pharmacogenetics: economic considerations. Pharmacoeconomics. 2007;25(11):899–902. | ||
Rendic S. Summary of information on human CYP enzymes: human P450 metabolism data. Drug Metab Rev. 2002;34(1-2):83–448. | ||
Zhou SF, Zhou ZW, Yang LP, Cai JP. Substrates, inducers, inhibitors and structure–activity relationships of human cytochrome P450 2C9 and implications in drug development. Curr Med Chem. 2009;16(27):3480–3675. | ||
Wang D, Sun X, Gong Y, et al. CYP2C9 promoter variable number tandem repeat polymorphism regulates mRNA expression in human livers. Drug Metab Dispos. 2012;40(5):884–891. | ||
Mlynarsky L, Bejarano-Achache I, Muszkat M, Caraco Y. Factor VII R353Q genetic polymorphism is associated with altered warfarin sensitivity among CYP2C9 *1/*1 carriers. Eur J Clin Pharmacol. 2012;68(5):617–627. | ||
Greisenegger S, Weber M, Funk M, et al. Polymorphisms in the coagulation factor VII gene and risk of primary intracerebral hemorrhage. Eur J Neurol. 2007;14(10):1098–1101. | ||
Mo X, Hao Y, Yang X, Chen S, Lu X, Gu D. Association between polymorphisms in the coagulation factor VII gene and coronary heart disease risk in different ethnicities: a meta-analysis. BMC Med Genet. 2011;12(1):107. | ||
Di Castelnuovo A, D’Orazio A, Amore C, Falanga A, Donati MB, Iacoviello L. The decanucleotide insertion/deletion polymorphism in the promoter region of the coagulation factor VII gene and the risk of familial myocardial infarction. Thromb Res. 2000;98(1):9–17. | ||
Girelli D, Russo C, Ferraresi P, et al. Polymorphisms in the factor VII gene and the risk of myocardial infarction in patients with coronary artery disease. N Engl J Med. 2000;343(11):774–780. | ||
Wang XL, Wang J, McCredie RM, Wilcken DE. Polymorphisms of factor V, factor VII, and fibrinogen genes. Relevance to severity of coronary artery disease. Arterioscler Thromb Vasc Biol. 1997;17(2):246–251. | ||
Ardissino D, Mannucci PM, Merlini PA, et al. Prothrombotic genetic risk factors in young survivors of myocardial infarction. Blood. 1999;94(1):46–51. | ||
Gage BF. Pharmacogenetics-based coumarin therapy. Hematology Am Soc Hematol Educ Program. 2006;1:467–473. | ||
D’Andrea G, D’Ambrosio R, Margaglione M. Oral anticoagulants: pharmacogenetics relationship between genetic and non-genetic factors. Blood Rev. 2008;22(3):127–140. | ||
Hylek EM, D’Antonio J, Evans-Molina C, Shea C, Henault LE, Regan S. Translating the results of randomized trials into clinical practice: the challenge of warfarin candidacy among hospitalized elderly patients with atrial fibrillation. Stroke. 2006;37(4):1075–1080. | ||
Kuruvilla M, Gurk-Turner C. A review of warfarin dosing and monitoring. Proc (Bayl Univ Med Cent). 2001;14(3):305–306. | ||
Marchetti G, Gemmati D, Patracchini P, Pinotti M, Bernardi F. PCR detection of a repeat polymorphism within the F7 gene. Nucleic Acids Res. 1991;19(16):4570. | ||
Schelleman H, Chen J, Chen Z, et al. Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin Pharmacol Ther. 2008;84(3):332–339. | ||
Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med. 2013;369(24):2294–2303. | ||
Yuan HY, Chen JJ, Lee MT, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005;14(13):1745–1751. | ||
Daly AK. Pharmacogenomics of anticoagulants: steps toward personal dosage. Genome Med. 2009;1(1):10. | ||
Jonas DE, McLeod HL. Genetic and clinical factors relating to warfarin dosing. Trends Pharmacol Sci. 2009;30(7):375–386. | ||
Caldwell MD, Berg RL, Zhang KQ, et al. Evaluation of genetic factors for warfarin dose prediction. Clin Med Res. 2007;5(1):8–16. | ||
Shikata E, Ieiri I, Ishiguro S, et al. Association of pharmacokinetic (CYP2C9) and pharmacodynamic (factors II, VII, IX, and X; proteins S and C; and gamma-glutamyl carboxylase) gene variants with warfarin sensitivity. Blood. 2004;103(7):2630–2635. | ||
Kohnke H, Sörlin K, Granath G, Wadelius M. Warfarin dose related to apolipoprotein E (APOE) genotype. Eur J Clin Pharmacol. 2005;61(5–6):381–388. | ||
Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84(3):326–331. | ||
Iacoviello L, di Castelnuovo A, de Knijff P, et al. Polymorphisms in the coagulation factor VII gene and the risk of myocardial infarction. N Engl J Med. 1998;338(2):79–85. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





