Back to Journals » International Journal of Nanomedicine » Volume 13
Difunctional bacteriophage conjugated with photosensitizers for Candida albicans-targeting photodynamic inactivation
Authors Dong S, Shi H, Zhang X, Chen X, Cao D, Mao C, Gao X, Wang L
Received 12 November 2017
Accepted for publication 24 January 2018
Published 11 April 2018 Volume 2018:13 Pages 2199—2216
DOI https://doi.org/10.2147/IJN.S156815
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Linlin Sun
Shuai Dong,1,2 Hongxi Shi,1 Xintong Zhang,1,2 Xi Chen,1 Donghui Cao,2 Chuanbin Mao,3,4 Xiang Gao,1 Li Wang1
1Key Laboratory of Molecular Epigenetics of Ministry of Education, Institute of Genetics and Cytology, Northeast Normal University, 2First Hospital of Jilin University, Changchun, Jilin, 3School of Materials Science and Engineering, Zhejiang University, Hangzhou, Zhejiang, China; 4Department of Chemistry and Biochemistry, Stephenson Life Science Research Center, University of Oklahoma, Norman, OK, USA
Background: Candida albicans is the most prevalent fungal pathogen of the human microbiota, causing infections ranging from superficial infections of the skin to life-threatening systemic infections. Due to the increasing occurrence of antibiotic-resistant C. albicans strains, new approaches to control this pathogen are needed. Photodynamic inactivation is an emerging alternative to treat infections based on the interactions between visible light and photosensitisers, in which pheophorbide a (PPA) is a chlorophyll-based photosensitizer that could induce cell death after light irradiation. Due to PPA’s phototoxicity and low efficiency, the main challenge is to implement photosensitizer cell targeting and attacking.
Methods: In this study, PPA was conjugated with JM-phage by EDC/NHS crosslinking. UV-Vis spectra was used to determine the optimum conjugation percentages of PPA and JM-phage complex for photodynamic inactivation. After photodynamic inactivation, the efficacy of PPA-JM-phage was assessed by performing in vitro experiments, such as MTS assay, scanning electron microscopy, measurement of dysfunctional mitochondria, ROS accumulation, S cell arrest and apoptotic pathway.
Results: A single-chain variable-fragment phage (JM) with high affinity to MP65 was screened from human single-fold single-chain variable-fragment libraries and designed as a binding target for C. albicans cells. Subsequently, PPa was integrated into JM phage to generate a combined nanoscale material, which was called PPA-JM-phage. After photodynamic inactivation, the growth of C. albicans was inhibited by PPA-JM-phage and apoptosis was observed. Scanning electron microscopy analysis revealed shrinking and rupturing of C. albicans. We also found that depolarization of mitochondrial membrane potential was decreased and intracellular reactive oxygen species levels were elevated significantly in C. albicans inhibited by PPA-JM-phage. Additionally, PPA-JM-phage also lead to S-phase arrest, and metacaspase activation resulting from mitochondrial dysfunction was also found to be involved in C. albicans apoptosis.
Conclusion: PPa-JM-phage may induce C. albicans apoptosis through a caspase-dependent pathway and the results herein shed light on the potential application of phtototherapeutic nanostructures in fungal inactivation.
Keywords: PPA, phage, photodynamic therapy, apoptosis, metacaspase
Introduction
Candida albicans is an opportunistic fungal pathogen, which can cause superficial infections and life-threatening systemic infections in humans. Pathogenesis depends on both C. albicans virulence and host-defense systems.1 Management of infections caused by clinically relevant fungal pathogens is a challenge, due to resistance developed during the therapy process, especially in immunocompromised individuals.2 Such resistance is the main factor that makes fungal infection treatment intractable.3 Furthermore, the most commonly used antifungal azoles have posed their own risks, due to interactions with other drugs and inherent organ toxicity. Therefore, the lack of effective antifungal agents and the emergence of drug-resistant fungi strains have prompted research into novel antifungal approaches,4 among which photodynamic (PD) treatment is a promising candidate.
PD inactivation (PDI) is a kind of PD-treatment strategy based on the integration of photosensitizers (PSs) in combination with visible light, which has been implemented in oncology, dermatology, and ophthalmology,5,6 as well as other areas, including antifungal therapy. Previous studies have demonstrated that the PS methylene blue had fungicidal effects on various Candida spp. (C. albicans, C. dubliniensis, C. krusei, and C. tropicalis).7 It has been confirmed that PSs generate reactive oxygen species (ROS) to treat infections caused by C. albicans.8 Oxidative stress could lead to damage of fungal cells by increased production of various reactive species and decreased ability of the natural protective effects of the organism that are responsible for the inhibition of these reactive compounds.9
Pheophorbide A (PPA) is a chlorophyll-based second-generation PS that is localized to the mitochondria.10 It has been shown to have high potency in PDI, and is most commonly used in cancer therapy.11,12 PPA is activated by light with a wavelength of approximate 670 nm, and red light is an ideal choice for the more deeply localized targets.13 Although some studies have shown the susceptibility of Candida spp. to PDI, further work is needed to address long-standing problems in antifungal PDI treatments. First, the therapeutic effectiveness of PPA in native form is limited, due to its poor solubility in water, which leads to aggregation and inefficient PD activity. Second, PSs can only function within 0.01–0.02 μm of skin.14 Third, the lack of cell-specific targeting can decrease the site-specific concentration of PSs and in turn restrict their PD efficacy.15 Previous studies have reported that PSs in conjunction with nanoparticles further improved the efficacy of PD treatment of microbial infection.16 Wei et al reported that a complex of PPA–graphene oxide-based nanosystem-monoclonal antibody significantly enhanced mitochondria-mediated apoptosis of PDI in tumor cells.17 Therefore, a similar nanoscale complex may improve the PDI of fungi infections after integration of PSs with target proteins.
Filamentous bacteriophage (phage) is an artificially modifiable supramacromolecule (~900 nm long and ~8 nm wide) that is surrounded by a cylinder of coat proteins. Phages are suitable for synthesizing novel nanostructures and biomaterials, and have played a crucial role in advancing modern biomedicine.18 Phage-display technology has proven to be a robust method for the selection and generation of monoclonal antibodies.19 A single-chain variable fragment (scFv) consists of variable regions of heavy (VH) and light (VL) chains, which are joined together by a flexible peptide linker. The scFv fragment is the smallest unit of antibody, functioning in antigen-binding activities. In the phage format, all gene III molecules are expressed as scFv and three to five copies are displayed on the tip of bacteriophage, which allows the selection of binders with high affinity. Phage display has been applied for antibody detection in numerous scientific fields, including life science and clinical medicine. For instance, biomarker-binding peptides displayed on the surface of phages were identified by different targets, such as Sap2 and p53 antibodies.20,21 In our previous studies, phage-displaying anti-rSap2 scFvs were identified, which targeted both C. albicans cells and treated systemic infection caused by C. albicans.22 Therefore, phage-display antibody libraries can be readily screened for specific binders to the antigen proteins presented on cells by biopanning.23
The cell wall of C. albicans is a crucial factor in maintaining structural integrity, and is comprised of two main layers. The inner layer is composed of a network of β1,3-glucan molecules, and the outer layer consists of mannoproteins termed “cell-wall proteins”, accounting for approximately 40% of cell-wall mass. The 65 kDa β-glucanase mannoprotein (MP65) is an essential cell-wall mannoprotein of C. albicans, and its expression is constitutive in both yeast cells and hypha-promoting conditions.24 A previous study has shown that the MP65 gene can be applied as a diagnostic marker for systemic C. albicans and non-C. albicans infections.25 Furthermore, the mannoprotein MP65 also plays a dominant role in the morphogenesis and pathogenicity of C. albicans.26 Researchers have demonstrated that MP65 domain antibodies (DAbs) inhibit C. albicans adherence to rat vaginal epithelial cells in vivo.27 The effectiveness of monoclonal antibodies in treating Candida continues to show promise. For example, monoclonal antibodies have been shown to mediate direct antifungal effects in C. albicans, including inhibition of adherence and growth, and direct candidacidal activity.28
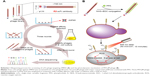
In this study, an scFv-phage (JM) that specifically recognized the mannoprotein MP65 of C. albicans was screened from phage-display antibody libraries. Then, the N-terminus of JM-phage surfaces was modified and conjugated with PPA, which was designated PPA-JM-phage. To determine the effect and functionality of this bifunctional target-specific sensitizer, we measured the in vitro effect of PDI on C. albicans in the presence of various reagents, including control and treatment groups. The depolarization of mitochondrial membrane potential (ΔΨm), ROS levels and cell-cycle arrest of C. albicans were also detected, which were involved in phototoxic effects stimulating cell death in C. albicans (Scheme 1). Furthermore, the involvement of the caspase-dependent pathway in PPA-JM-phage-induced apoptosis of C. albicans cells was investigated. These results warrant further attention for the use of PPA-JM-phage as a novel treatment for C. albicans.
Materials and methods
Screening of phage library
High-affinity scFv phages binding to recombinant MP65 protein (rMP65) were selected from the human single-fold scFv libraries I + J (Tomlinson I + J), which were purchased from MRC (Cambridge, England). We used a previously described panning protocol, with some modifications.29 Briefly, immunotubes (Thermo Fisher Scientific, Waltham, MA, USA) were coated with 100 μg/mL rMP65 in the first round, and then gradually decreased to 50 μg/mL, 25 μg/mL, and 12.5 μg/mL in the following three rounds. After blocking with 2% MBS (2% Marvel milk powder in PBS) for 2 hours at room temperature (RT), tubes were loaded with 1013 phages in 4 mL 2% MPBS and incubated for 60 minutes at RT. After thorough washing with PBST (PBS containing 0.1% Tween 20) ten times, the bound scFv-phages were eluted with 500 μL trypsin-PBS (1 mg/mL) for 10 minutes. Selected phages (250 μL) were used to infect the Escherichia coli bacteria strain TG1 for phage amplification, as previously described.30
Preparation and assessment of JM-phage
After affinity selection and panning, the positive phage was selected and amplified in TG1. Phage-infected cells were grown overnight in a shaker at 37°C. A total of 50 μL of overnight culture was used for inoculation in fresh 2× TY (1.6% tryptone, 1% yeast extract, 0.5% NaCl) at 37°C for 2 hours. Helper phages (KM13, 5×1010) were added to 10 mL culture and incubated in a 37°C water bath for 30 minutes (not shaken). Following centrifugation, cell pellets were grown overnight at 30°C in 50 mL 2× TY containing 100 μg/mL ampicillin, 50 μg/mL kanamycin, and 0.1% glucose. Phage virions were purified by precipitation with 20% PEG–NaCl solution (20% PEG6,000, 2.5 M NaCl) and centrifugation at 3,300 g for 10 minutes.
In order to assess the binding ability of phage with rMP65, postbiopanning phages were diluted serially and detected by enzyme-linked immunosorbent assay (ELISA): 100 μg/mL of rMP65 were coated in a 96-well plate overnight. Then, the plate was washed with PBS three times and blocked with 2% MPBS for 1 hour. Subsequently, 100 μL diluted phage (1:20–1:2,560) were transferred into the plate and incubated for 1 hour at RT with KM13 as a negative control. After extensive washing with PBST five times, 100 μL anti-phage g3p (pIII) and antibody (1:1,000; MoBiTec, Göttingen, Germany) was added and incubated for 1 hour. Next, the plate was washed with PBST five times and reacted with 100 μL HRP-conjugated antibody (ZSJQ-Bio, Beijing, China) for 1 hour. After washing with PBST five times, 100 μL TMB solution (Beyotime Biotechnology, Haimen, China) was added to each well and terminated with 50 μL 1 M sulphuric acid. OD492 was recorded with an ultraviolet (UV)-visible plate reader (Molecular Devices, Sunnyvale, CA, USA).
Preparation of PPA-JM-phage
The procedure for covalent immobilization of JM-phage followed the 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC)–N-hydroxysuccinimide (NHS) method.15 PPA (Frontier Scientific, Logan, UT, USA) in dimethyl sulfoxide was activated with 0.4 mg EDC (Sigma Aldrich, St Louis, MO, USA). The reaction mixture was incubated at 37°C on a Roto-Shaker for 1 hour. Unreacted EDC was quenched with 2-mercaptoethanol (Sigma Aldrich) to avoid self-conjugation of JM-phages in the next step. The reaction mixture was then incubated at 37°C on the Roto-Shaker for 1 hour. Activated PPA was then stored at −80°C until further use. For conjugation of JM-phages, a total of 2 mL phage solution containing 6×1012 virions/mL was added by serial dilution of EDC-activated PPA. Samples were incubated at RT on the Roto-Shaker overnight. They were then dialyzed in PBS at 4°C for 24 hours.
Characterization of PPA-JM-phage
The ultraviolet-visible absorption spectra of PPA-JM-phage and JM-phage were recorded on a spectrometer (Molecular Devices). Fluorescence spectra of PPA-JM-phage with different ratios of PPA to JM-phage were obtained using a microplate fluorescence reader (Molecular Devices) with excitation of 326 nm and 671 nm. In addition, to confirm that PPA was covalently attached to the JM-phages, PPA-JM-phages were subjected to sodium dodecyl sulfate (SDS) polyacrylamide-gel electrophoresis (PAGE) and imaged using an Odyssey imaging system (LI-COR Biosciences, Lincoln, NE, USA). Details can be found in the Supplementary materials. The morphology of the PPA-JM-phage was analyzed by transmission electron microscopy (TEM; JEOL, Tokyo, Japan). Samples were prepared by depositing one drop of dilute solution onto a dry copper grid that had been coated with carbon in advance. Subsequently, the grid was stained with 2% uranium acetate before TEM observation.
Confocal fluorescence imaging of C. albicans cells after incubation with phages
In order to confirm the binding specificities of PPA-JM-phage and JM-phage screened from the libraries, PPA-JM-phage and JM-phage were incubated with C. albicans cells on glass coverslips in 12-well cell-culture plates using PBS, KM13 phage, and anti-BSA phage as controls. After incubation, coverslips were washed three times with PBS and fixed in 4% paraformaldehyde for 30 minutes. Then, cells were permeabilized in 0.5% Triton X-100 for 30 minutes and blocked with 3% BSA for 1 hour. After washing with PBS three times, they were incubated with anti-phage g3p (pIII) antibody (1:1,000) for 1 hour at 4°C, then secondary fluorescein isothiocyanate (FITC)-conjugated antibody (ZSJQ-Bio) was added and incubated for 1 hour after another round of PBS rinsing, followed by DAPI (1:200) staining for 10 minutes. Finally, the stained C. albicans cells were monitored by fluorescence microscopy (BX50; Olympus, Tokyo, Japan).
Observation of C. albicans cell morphology after PDI treatment
To visualize the morphology of the C. albicans cells after PDI with PPA-JM-phage, they were examined under scanning electron microscopy (SEM; Hitachi, Tokyo, Japan). C. albicans cells were incubated in 12-well cell culture plates (Sigma Aldrich) with sterile glass coverslips at the bottom for 12 hours at 37°C. Each well was washed three times with sterile PBS and treated with experimental PPA-JM-phage, JM-phage, PPA and control groups (KM13, anti-BSA, PBS). The concentration of PPA in samples was 5 μM, and mixtures were washed three times with PBS to remove unbound phages and PPA. After 1 hour of incubation, C. albicans cells were irradiated with a 658 nm visible red laser under 50 mW/cm2 for 10 minutes. Total power output was 50 mW and spot size 1 cm2. Subsequently, disks were washed three times with PBS and fixed in 2.5% glutaradehyde for 1 hour at 4°C. After fixation, disks were rinsed three times with PBS and dehydrated in an ethanol series (30%, 50%, 70%, 80%, 90% for 7 minutes each, 100% for 10 minutes). Disks were critical point-dried prior to gold sputter-coating and analyzed by SEM.
In vitro photodynamic treatment and cell-viability assay
To verify the cytotoxicity and selectivity of PPA-JM-phage, the viability of C. albicans cells was evaluated with a cell counting kit-8 (CCK-8) assay (TransGen Biotech, Beijing, China). All experiments were conducted using C. albicans (ATCC 10231) maintained in yeast extract–peptone–dextrose (YPD) medium at 37°C overnight. After growth, cells were washed once and suspended in PBS (pH 7.4). Experimental and control groups were incubated with C. albicans cells (5×106 CFU/mL) in a sterile 96-well microplate at 37°C for 1 hour and washed three times with PBS to remove unbound phages and PPA. The following PDI treatment was identical to that described earlier. Cell viability was assessed by CCK-8 assay in the presence and absence of laser: 10 μL CCK-8 reagent was added to the 96-well microplate and incubated for 4 hours. After incubation, absorbance at 450 nm was recorded using a microplate fluorescence reader. Each assay was repeated three times.
Fluctuations in C. albicans concentration were calculated from initial CFU/mL and log10-transformed. After PDI treatment, samples from each group were tenfold serially diluted in sterile PBS. Then, 100 μL aliquots were spread on YPD solid medium and plates incubated at 37°C for 24 hour. Colony-forming units were counted, and results were log10-transformed. Three independent assays were carried out.
Cell-apoptosis study
Cell apoptosis was analyzed using a Hoechst 33342–propidium iodide (PI) double-staining apoptosis-detection kit (Beyotime Biotechnology). Following PDI treatment with experimental and control groups, cells were centrifuged at 5,000 g for 5 minutes. Pellets were rinsed twice with PBS, then incubated with 5 μL Hoechst 33342 and 5 μL PI according to the manufacturer’s instructions. Then, samples were observed under confocal microscopy with a 63× NA 1.4 oil-immersion objective.
Stress-sensitivity test
To determine PPA-JM-phage-induced alterations that could change the pathogenicity of C. albicans cells, the sensitivity of C. albicans to the stress of hydrogen peroxide and SDS was examined. Stress-sensitivity assays were carried out to evaluate response to oxidative stress and cell-wall integrity following PDI. Tenfold serial dilutions of treated cells were plated on the YPD solid medium supplemented with or without H2O2 and SDS. Plates were incubated at 37°C overnight.
Measurement of mitochondrial membrane potential
To assess changes in ΔΨm in C. albicans after PDI treatment with PPA-JM-phage, cells were stained with JC1 (Beyotime Biotechnology). Briefly, PDI-treated cells (106 CFU/mL) were washed and resuspended in PBS (pH 7.4). Untreated cells were incubated with 10 μM CCCP for 20 minutes and served as a positive control. Then, JC1 was added to the final concentration of 5 μM and the mixture incubated at 37°C for 20 minutes. Fluorescence intensity was recorded by the microplate fluorescence reader at an excitation wavelength of 490 nm and emissions at 530 nm.
Measurement of oxidative stress
ROS level was measured using an ROS-assay kit (Beyotime Biotechnology) in samples under PD treatment. Briefly, PDI-treated cells were incubated in 10 μM dichlorodihydrofluorescein diacetate (DCFH-DA) for 20 minutes at 37°C. Subsequently, cells were washed three times with PBS. The positive control was treated with 200 mM H2O2 for 10 minutes prior to the addition of the DCFH-DA probe. For all analyses, fluorescence intensity was monitored by the microplate fluorescence reader at an excitation wavelength of 488 nm and emissions at 525 nm. ROS level was examined by the ratio of (Ftext – Fblank):(Fcontrol – Fblank). Ftext was the fluorescence intensity of cells treated with PDI, Fcontrol the fluorescence intensity of PBS-treated cells, and the fluorescence intensity of wells without cells.
Cell-cycle analysis
For the analysis of the cell cycle of C. albicans after PPA-JM-phage treatment, C. albicans cells (5×106 CFU/mL) were incubated with experimental and control groups at 37°C for 1 hours. Then, mixtures were washed and irradiated with a 658 nm visible red laser light under 50 mW/cm2 for 10 minutes. Samples were centrifuged at 1,000 g for 5 minutes, washed once with cold PBS, and fixed with 75% cold ethanol overnight at 4°C. Cells were centrifuged again and washed once with cold PBS. Pellets were then incubated with a mixture of RNase A (0.1 mg/mL) and PI (0.1 mg/mL) for 30 minutes in the dark. DNA content was analyzed with flow cytometry (FACScan; Beckman Coulter, Brea, CA, USA). The percentage of the cell population in each phase was evaluated using Modfit LT 3.0.
Detection of metacaspase activation
Activation of metacaspases was measured using the CaspACE FITC-VAD-FMK in situ marker (Promega, Fitchburg, WI, USA) according to the manufacturer’s instructions. PDI-treated cells (105 CFU/mL) were washed with PBS before staining with CaspACE FITC-VAD-FMK (10 μM). After 20 minutes of incubation at 37°C, cells were washed twice with PBS and visualized with fluorescence microscopy. Fluorescence intensity was recorded by the microplate fluorescence reader at an excitation wavelength of 494 nm and emission wavelength at 518 nm.
Statistical analysis
All experiments were performed in triplicate and all data are expressed as mean ± SD. Statistical significance between experimental and control groups was assessed by two-tailed Student’s t-test or one-way analysis of variance. Statistical significance was set at P<0.05.
Results and discussion
Synthesis, characterization, and binding specificity of PPA-JM-phage
The structural features of phages make them ideal candidates for novel drug-delivery carriers: they are able to carry genomic and proteomic information in the same body and easily accommodate various target genes and proteins by genetic engineering.31 The Tomlinson human single-fold scFv I + J is a phage library that has been applied in selecting antibodies with both strong and modest affinity for targets in the past.32 In this study, we searched the I + J library for binders that recognized mannoprotein rMP65 of C. albicans. After four rounds of regular biopanning with rMP65, we obtained a unique high-affinity scFv phage clone from 40 clones, which was called JM. The amino-acid sequence of JM was submitted to DeepView to predict 3-D structures (Figure 1A). In indirect ELISA, titers of JM were determined with rMP65 at a dilution of 1:640, whereas KM13 was poorly recognized (Figure 1B). It is worth noting that the complementarity-determining regions of JM consisted of 50% hydrophilic amino acids (asparagines, cysteine, glutamine, serine, threonine, and tyrosine). Comparatively, the percentage of hydrophobic amino acids (alanine, isoleucine, leucine, phenylalanine, tryptophan, and valine) was 27% (Table S1). This indicated that JM had good hydrophilic properties. The amino-acid sequence of the selected scFv is presented in Figure 1C.
The PPA-JM-phage conjugates were synthesized by the conjugating-COOH group of the PPA to the N-terminal end of pVIII protein through EDC–NHS cross-linking (Figure 2A). Previous studies have demonstrated that phage-PSs can be used in vitro and in vivo selectively to eliminate tumors after PDI.15 Here, we extended the application of PDI to treat the pathogenesis of C. albicans. In order to determine the optimum conjugation percentages of PPA and JM-phage complex for PDI, we prepared varying ratios, calculated by the number of PPA molecules conjugated to pVIII protein on JM-phage/total pVIII protein on JM-phage (~3,900 copies) (Figure S1). As shown in Figure 2B, the characteristic absorption peaks of PPA-JM-phage in UV-visible spectra were at 405 nm and 668 nm, respectively, which were proportional to the increased percentage conjugation of PPA. Moreover, there was no absorption shift in UV-visible spectra of PPA-JM-phage before or after conjugation. Fluorescence spectroscopy was used to determine the enhancement or quenching of PPA after conjugation with JM-phage. Fluorescence-emission spectra at 671 nm showed that the maximum fluorescence was 50% after conjugation of PPA to JM-phage, likely due to energy hopping between highly ordered PPA molecules on JM-phage, as seen in Figure 2C. In contrast, fluorescence spectra emitted at 326 nm showed that the increase in percentage conjugation of PPA decreased fluorescence intensity, due to the fluorescence resonance-energy transfer between tryptophan of pVIII protein and PPA, as shown in Figure 2D. Furthermore, PPA-JM-phage was also found to be fluorescent under 700 nm wavelength-pass filter on SDS-PAGE gels (Figure S2). Together, these results supported the covalent conjugation of PPA to the pVIII protein of JM-phage.
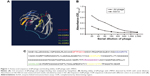
TEM was performed to observe the morphology of phages before and after modification. Unmodified phages were very flexible and brighter, while conjugated phages were darker, which further confirmed that PPA was successfully conjugated onto the phage (Figure 2E and F). To investigate the targeting efficiency of JM-phage and PPA-JM-phage, confocal fluorescence microscopy observation was carried out (Figure 3). After incubation with PPA-JM-phage and JM-phage, C. albicans cells showed green fluorescence, while no fluorescence signal was detected in control groups, eg, KM13 (phage did not display any scFv on its surface) and anti-BSA phage (phage display anti-BSA scFv on its surface). The results suggested that PPA-JM-phage and JM-phage recognized C. albicans cells specifically.
PPA-JM-phage destroyed the surface integrity of C. albicans cells
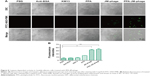
SEM was employed to determine surface-morphology changes in C. albicans cells. Before the SEM experiment, we examined the PDI efficacy of PPA with different ratios on phage and hybrid concentrations. PPA-JM-phage (50%) at 5 μM concentration was chosen as the suitable combination for optimal activity (Figure S3). As shown in Figure 4A–F, C. albicans cells incubated with control groups showed intact cell walls after PDI treatment, and there were no obvious morphological changes in yeast-type cells, whereas the PPA-JM-phage-treated group showed a detrimental effect: cells were mostly ruptured and shrunken, and leaking of yeast-cell content was observed. Other studies have suggested that this type of cell damage may be caused by interference by the cytoplasmatic membrane, which could increase permeability and cause damage to intracellular targets.33–35 These effects occurred because of free radicals and singlet oxygen, which were the products of PDI, reacting with several cellular structures and different metabolic pathways.36 Yeast cells treated with JM-phage exhibited similar symptoms as described earlier, but to a noticeably lesser extent. These morphological deformations suggested that C. albicans mannoprotein mediated direct antifungal effects.28
PPA-JM-phage induced apoptosis/necrosis under laser irradiation
To investigate the antifungal effect of PPA-JM-phage, a cell-viability assay was performed. In the dark-toxicity assay there was no significant cell death in either experimental or control groups, suggesting that KM13 itself does not cause cytotoxicity (Figure 4G). In the phototoxicity assay, the proportion of viable C. albicans decreased to 48% after treatment with PPA-JM phage, whereas PPA caused no measurable cell death after washing three times, indicating that PPA did not target C. albicans cells specifically. JM-phage alone showed slight toxicity to C. albicans cells, and scFv-phage may have had relatively weaker antifungal ability.
Following PDI, C. albican cells treated with PPA-JM-phage and showed a reduction of 0.92 log10 (CFU/mL) (Figure 4H). Statistically significant (P<0.05) differences between PPA-JM-phage and control groups were observed. PPA-JM-phage integrated both JM-phage and chemicals into a unique complex, meaning the phage can be synthesized automatically in a bacterial system. This decreases the costs of reagents and improves cell-wall targeting by JM-phage, which is also beneficial for the specificity of PPA-JM-phage. Therefore, the concept and results based on PPA-JM-phage (the combination of chemicals and phage) have the potential to be developed into novel and effective antifungi drugs.
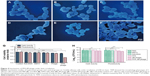
Apoptotic responses have been described in C. albicans after exposure to UV irradiation, oxidative stress, and weak acid stress.37 By integrating the excellent properties of antibody-based therapy and PDI into a phage-PS, PPA-JM-phage was expected to induce apoptosis/necrosis upon light irradiation more efficiently. The apoptotic effects of PPA-JM-phage were determined by the Hoechst 33342–PI double-staining method, which are apoptosis- and necrosis-specific dyes, respectively (Figure 5). It was observed that cells treated with control groups stained with blue fluorescent dye (Hoechst+/PI−) did not show any necrosis or apoptosis. In the PPA group, extensive blue fluorescence (Hoechst+/PI−) was observed with minor red fluorescence signals (Hoechst+/PI+). In contrast, the proportion of red fluorescence (Hoechst+/PI+) was significantly strengthened in PPA-JM-phage- and JM-phage-treated groups, suggesting both early apoptotic and necrotic death signals in C. albicans cells. These results were supported by the fact that the Hoechst stain binds to condensed chromatin in apoptotic cells more brightly than normal cells, and PI binds to DNA only in membrane-damaged dead cells.
PPA-JM-phage increased C. albicans sensitivity to H2O2 and SDS
H2O2 is a kind of ROS that can damage the DNA and proteins of microbial cells. However, it cannot diffuse freely across cell membranes. The cell wall is a vital structure, and plays a crucial role in cellular protection, growth, and adherence. SDS is a cell-wall stressor that can perturb normal cell-wall functions. A previous study reported that PDI resulted in impaired resistance to H2O2 oxidative stress and that C. albicans cells were sensitive to SDS.7 In this study, we found that the growth of C. albicans cells treated with PPA-JM-phage and phage were inhibited, even without direct H2O2/SDS treatment. However, PPA-JM-phage exhibited increased sensitivity to the stress conditions compared with control groups, which showed no obvious effects on growth inhibition of C. albicans cells under either stress condition tested (Figure 6). In general, PPA-JM-phage may be able to reduce ROS resistance and affect cell-wall structure in C. albicans.
PPA-JM-phage influenced mitochondrial membrane potential
Apoptosis is a normal physiological process that leads to cell death in organisms and is crucial for normal cell growth, development, and homeostasis.38 It can be induced by many forms of cellular stresses, such as irradiation and cytotoxic drugs. Mitochondrial dysfunction is regarded as the onset of apoptosis.39 The dissipation of ΔΨm is a key cellular event in the regulation of apoptosis. As such, the effect of PPA-JM-phage on mitochondria was tested by JC1, which can selectively enter mitochondria. In healthy cells with high ΔΨm, it formed J-aggregates that exhibited intense red fluorescence, while in apoptotic cells with low ΔΨm, it remained in the monomeric form, showing green fluorescence. Figure 7A shows the ratios of red:green fluorescence representing ΔΨm. Percentages of PPA-JM-phage-treated and JM-phage-treated cells were 5.3% and 10.6%, respectively, similar to positive cells (5.3%). These results indicate that PPA-JM-phage and JM-phage caused mitochondrial dysfunction in C. albicans by decreasing the ΔΨm. The decreasing ΔΨm represented inhibition of the proton-pumping function of the respiratory chain, which can lead to decreased ATP production and cell death.40
ROS production was elevated in PPA-JM-phage-treated C. albicans cells
Oxidative stress is considered a key mechanism in cellular defense, and is involved in mitochondrial membrane permeability, cell-death induction, and ROS formation. Elevated endogenous ROS production is regarded as another apoptotic feature, which is followed by depolarization of ΔΨm.41,42 ROS not only can damage a wide range of biomacromolecules but also plays a physiological role in the regulation of apoptosis.43,44 ROS is essential for almost all apoptotic pathways, and acts as a primary trigger of apoptosis.45
In this study, ROS levels in PDI-treated cells were monitored by DCFH-DA. The fluorescence emitted by the cells was measured by spectrofluorometry at excitation and emission wavelengths of 488 nm and 525 nm, respectively. As shown in Figure 7B, ROS levels in PPA-JM-phage-treated cells increased significantly, almost equally to the positive control. High ROS levels were also observed in JM-phage-treated cells, though to a lesser extent than PPA-JM-phage. No increase in ROS levels was observed in the control groups. These results suggested that PPA-JM-phage induced apoptosis through the activation of ROS-mediated cell-death in C. albicans cells, which supports previous reports that ROS accumulation induces apoptosis in C. albicans.46
PPA-JM-phage inhibited cell proliferation of C. albicans through confinement of cell cycle in S phase
Excessive amounts of ROS can trigger a series of downstream cellular events that lead to disorganization of DNA and cell death.35 We assessed the effect of PPA-JM-phage on cell proliferation. Cell-cycle arrest is closely related to apoptosis/necrotic death.11 To provide further evidence for inhibition of the cell cycle by PPA-JM-phage treatment, DNA cell-cycle analysis was conducted by flow cytometry. As shown in Figure 7C, the PPA-JM-phage group after PDI induced a significant S phase of cell-cycle arrest. Moreover, slight S-phase cell-cycle arrest in phage group was also detected, whereas no statistical difference in cell-cycle distribution was found in control groups. We also examined the effects of ROS production on C. albicans cells using NAC as an ROS scavenger. NAC treatment decreased ROS production on PDI-treated cells (Figure S4A). As expected, there was no difference in cell-cycle distribution among NAC-pretreated groups (Figure S4C). These results indicated that ROS can influence cell-cycle arrest. In conclusion, cell-cycle analysis showed that PPA-JM-phage caused S-phase cell-cycle arrest in C. albicans, which resulted in the inhibition of DNA synthesis. This could have been caused by the speed of G1–S phase transition being faster and more cells being promoted from the G1 phase into the S phase. Conversely, a delay in mitotic entry is essential for cells to repair any DNA damage that may have accumulated in the S phase.47
PPA-JM-phage activated metacaspase activity in C. albicans
Fungus apoptosis comprises many biochemical pathways involving a suite of regulators and effectors. In this study, caspase pathways were investigated. Caspase activation has been recognized as an important process associated with apoptosis.48 In C. albicans, both caspase-dependent and caspase-independent cell-death pathways were utilized. Although caspases are not present in fungi, researchers have identified orthologues of the mammalian caspase family, termed “metacaspases” in fungi and plants.
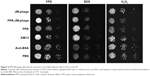
Metacaspases in C. albicans are important effector molecules in apoptosis and activated in the early stages of apoptosis. The activation of metacaspases was detected with the CaspACE FITC-VAD-FMK in situ marker, and green fluorescence revealed activated intracellular metacaspases in C. albicans cells. Indeed, cells treated with PPA-JM-phage and JM-phage showed strong and weak green fluorescence, respectively, whereas no fluorescence was observed in the control groups (Figure 8A). Furthermore, the fluorescence intensities detected by the microplate fluorescence reader were consistent with these fluorescence results (Figure 8B). Meanwhile, the fluorescence intensity of NAC-pretreated cells showed no difference between control groups and experimental groups (Figure S4B), which demonstrated that PDI treatment did not trigger metacaspase activation in C. albicans cells pretreated by NAC. Therefore, the assessment of the effect of PPA-JM-phage on metacaspase activity showed that apoptosis in C. albicans may be metacaspase-dependent and was caused by ROS production.
Conclusion
In this study, PPA was conjugated with JM-phage, which targeted the cell-wall mannoprotein MP65 of C. albicans. PPA-JM-phage combined two treatment modalities – PDI by PPA and targeting C. albicans by phage – and also had the function of antibody-based therapy. The improved feature of this bifunctional nanodrug is that PPA accumulation in the cells of C. albicans was enhanced significantly. A measurable therapeutic effect of PPA-JM-phage on C. albicans was observed. PPA-JM-phage inhibited the growth and damaged the surface morphology of C. albicans. It also triggered apoptotic features in C. albicans, namely dysfunctional mitochondria, ROS accumulation, and S-phase cell arrest. Furthermore, our study demonstrates that the apoptotic pathway in C. albicans triggered by PPA-JM-phage may be metacaspase-dependent. Therefore, it can be concluded that the combination of PSs–PPA, targeting agents, and phage, can improve the therapeutic capabilities of the newly synthesized bifunctional antifungal drugs, which will broaden the applicability of nanophototherapeutics.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81373231, 51673168), the Department of Science and Technology of Jilin Province, China (20160101216JC), and the Zhejiang Provincial Natural Science Foundation of China (LZ16E030001).
Disclosure
The authors report no conflicts of interest in this work.
References
Gualco L, Debbia EA, Bandettini R, et al. Antifungal resistance in Candida spp. isolated in Italy between 2002 and 2005 from children and adults. Int J Antimicrob Agents. 2007;29(2):179–184. | ||
Pfaller MA, Diekema DJ, Jones RN, Messer SA, Hollis RJ. Trends in antifungal susceptibility of Candida spp. isolated from pediatric and adult patients with bloodstream infections: SENTRY antimicrobial surveillance program, 1997 to 2000. J Clin Microbiol. 2002;40(3):852–856. | ||
Pappas PG, Kauffman CA, Andes D, et al. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(5):503–535. | ||
Dai TH, Fuchs BB, Coleman JJ, et al. Concepts and principles of photodynamic therapy as an alternative antifungal discovery platform. Front Microbiol. 2012;3:120. | ||
Costa A, Rasteiro VM, Pereira CA, Rossoni RD, Junqueira JC, Jorge AO. The effects of rose bengal- and erythrosine-mediated photodynamic therapy on Candida albicans. Mycoses. 2012;55(1):56–63. | ||
Rezusta A, Lopez-Chicon P, Paz-Cristobal MP, et al. In vitro fungicidal photodynamic effect of hypericin on Candida species. Photochem Photobiol. 2012;88(3):613–619. | ||
Kato IT, Prates RA, Sabino CP, et al. Antimicrobial photodynamic inactivation inhibits Candida albicans virulence factors and reduces in vivo pathogenicity. Antimicrob Agents Chemother. 2013;57(1):445–451. | ||
Moreira LM, dos Santos FV, Lyon JP, Maftoum-Costa M, Pacheco-Soares C, da Silva NS. Photodynamic therapy: porphyrins and phthalocyanines as photosensitizers. Aust J Chem. 2008;61(10):741–754. | ||
Tang PM, Liu XZ, Zhang DM, Fong WP, Fung KP. Pheophorbide a based photodynamic therapy induces apoptosis via mitochondrial-mediated pathway in human uterine carcinosarcoma. Cancer Biol Ther. 2009;8(6):533–539. | ||
Kessel D, Antolovich M, Smith KM. The role of the peripheral benzodiazepine receptor in the apoptotic response to photodynamic therapy. Photochem Photobiol. 2001;74(2):346–349. | ||
Zhang HS, Zhang M, Yu LH, Zhao Y, He NW, Yang XB. Antitumor activities of quercetin and quercetin-5′,8-disulfonate in human colon and breast cancer cell lines. Food Chem Toxicol. 2012;50(5):1589–1599. | ||
Robey RW, Steadman K, Polgar O, Bates SE. ABCG2-mediated transport of photosensitizers: potential impact on photodynamic therapy. Cancer Biol Ther. 2005;4(2):195–202. | ||
Munin E, Giroldo LM, Alves LP, Costa MS. Study of germ tube formation by Candida albicans after photodynamic antimicrobial chemotherapy (PACT). J Photochem Photobiol B. 2007;88(1):16–20. | ||
Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23(9):1105–1116. | ||
Gandra N, Abbineni G, Qu XW, Huai YY, Wang L, Mao CB. Bacteriophage bionanowire as a carrier for both cancer-targeting peptides and photosensitizers and its use in selective cancer cell killing by photodynamic therapy. Small. 2013;9(2):215–221. | ||
Bhaumik J, Mittal AK, Banerjee A, Chisti Y, Banerjee UC. Applications of phototheranostic nanoagents in photodynamic therapy. Nano Res. 2014;8(5):1373–1394. | ||
Wei YC, Zhou FF, Zhang D, Chen Q, Xing D. A graphene oxide based smart drug delivery system for tumor mitochondria-targeting photodynamic therapy. Nanoscale. 2016;8(6):3530–3538. | ||
Cao B, Yang M, Mao C. Phage as a genetically modifiable supramacromolecule in chemistry, materials and medicine. Acc Chem Res. 2016;49(6):1111–1120. | ||
Du B, Qian M, Zhou ZL, et al. In vitro panning of a targeting peptide to hepatocarcinoma from a phage display peptide library. Biochem Biophys Res Commun. 2006;342(3):956–962. | ||
Pan P, Wang Y, Zhu Y, et al. Nontoxic virus nanofibers improve the detection sensitivity for the anti-p53 antibody, a biomarker in cancer patients. Nano Res. 2015;8(11):3562–3570. | ||
Wang Y, Ju Z, Cao B, et al. Ultrasensitive rapid detection of human serum antibody biomarkers by biomarker-capturing viral nanofibers. ACS Nano. 2015;9(4):4475–4483. | ||
Dong S, Shi H, Cao D, et al. Novel nanoscale bacteriophage-based single-domain antibodies for the therapy of systemic infection caused by Candida albicans. Sci Rep. 2016;6:32256. | ||
Sohn K, Urban C, Brunner H, Rupp S. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol Microbiol. 2003;47(1):89–102. | ||
Stamati I, Kuimova MK, Lion M, Yahioglu G, Phillips D, Deonarain MP. Novel photosensitisers derived from pyropheophorbide-A: uptake by cells and photodynamic efficiency in vitro. Photochem Photobiol Sci. 2010;9(7):1033–1041. | ||
Arancia S, Sandini S, Cassone A, de Bernardis F. Use of 65 kDa mannoprotein gene primers in PCR methods for the identification of five medically important Candida species. Mol Cell Probes. 2009;23(5):218–226. | ||
Sandini S, la Valle R, de Bernardis F, Macri C, Cassone A. The 65 kDa mannoprotein gene of Candida albicans encodes a putative β-glucanase adhesin required for hyphal morphogenesis and experimental pathogenicity. Cell Microbiol. 2007;9(5):1223–1238. | ||
de Bernardis F, Liu HQ, O’Mahony R, et al. Human domain antibodies against virulence traits of Candida albicans inhibit fungus adherence to vaginal epithelium and protect against experimental vaginal candidiasis. J Infect Dis. 2007;195(1):149–157. | ||
Moragues MD, Omaetxebarria MJ, Elguezabal N, et al. A monoclonal antibody directed against a Candida albicans cell wall mannoprotein exerts three anti-C. albicans activities. Infect Immun. 2003;71(9):5273–5279. | ||
Lee CM, Iorno N, Sierro F, Christ D. Selection of human antibody fragments by phage display. Nat Protoc. 2007;2(11):3001–3008. | ||
Garet E, Cabado AG, Vieites JM, Gonzalez-Fernandez A. Rapid isolation of single-chain antibodies by phage display technology directed against one of the most potent marine toxins: palytoxin. Toxicon. 2010;55(8):1519–1526. | ||
Farr R, Choi DS, Lee SW. Phage-based nanomaterials for biomedical applications. Acta Biomater. 2014;10(4):1741–1750. | ||
Barderas R, Shochat S, Altschuh D, Meloen R, Casal JI. A fast mutagenesis procedure to recover soluble and functional scFvs from synthetic and semisynthetic antibody libraries. J Immunol Methods. 2006;312(1):182–189. | ||
Giroldo LM, Felipe MP, de Oliveira MA, Munin E, Alves LP, Costa MS. Photodynamic antimicrobial chemotherapy (PACT) with methylene blue increases membrane permeability in Candida albicans. Lasers Med Sci. 2009;24(1):109–112. | ||
Cormick MP, Alvarez MG, Rovera M, Durantini EN. Photodynamic inactivation of Candida albicans sensitized by tri- and tetra-cationic porphyrin derivatives. Eur J Med Chem. 2009;44(4):1592–1599. | ||
Lambrechts SA, Aalders MC, Van MJ. Mechanistic study of the photodynamic inactivation of Candida albicans by a cationic porphyrin. Antimicrob Agents Chemother. 2005;49(5):2026–2034. | ||
Paardekooper M, van den Broek PJ, de Bruijne AW, Elferink JG, Dubbelman TM, van Steveninck J. Photodynamic treatment of yeast cells with the dye toluidine blue: all-or-none loss of plasma membrane barrier properties. Biochim Biophys Acta. 1992;1108(1):86–90. | ||
Ramsdale M. Programmed cell death in pathogenic fungi. Biochim Biophys Acta. 2008;1783(7):1369–1380. | ||
Kerr JF, Winterford CM, Harmon BV. Apoptosis: its significance in cancer and cancer therapy. Cancer. 1994;73(8):2013–2026. | ||
Estaquier J, Vallette F, Vayssiere JL, Mignotte B. The mitochondrial pathways of apoptosis. Adv Exp Med Biol. 2012;942:157–183. | ||
Krysko DV, Roels F, Leybaert L, d’Herde K. Mitochondrial transmembrane potential changes support the concept of mitochondrial heterogeneity during apoptosis. J Histochem Cytochem. 2001;49(10):1277–1284. | ||
Moradasferreira P, Costa V, Piper P, Mager W. The molecular defences against reactive oxygen species in yeast. Mol Microbiol. 1996;19(4):651–658. | ||
Pozniakovsky AI, Knorre DA, Markova OV, Hyman AA, Skulachev VP, Severin FF. Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J Cell Biol. 2005;168(2):257–269. | ||
Perrone GG, Tan SX, Dawes IW. Reactive oxygen species and yeast apoptosis. Biochim Biophys Acta. 2008;1783(7):1354–1368. | ||
Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem. 2000;275(35):27393–27398. | ||
Phillips AJ, Crowe JD, Ramsdale M. Ras pathway signaling accelerates programmed cell death in the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A. 2006;103(3):726–731. | ||
Shirazi F, Kontoyiannis DP. Micafungin triggers caspase-dependent apoptosis in Candida albicans and Candida parapsilosis biofilms, including caspofungin non-susceptible isolates. Virulence. 2015;6(4):385–394. | ||
Chastain PD, Brylawski BP, Zhou YC, et al. DNA damage checkpoint responses in the S phase of synchronized diploid human fibroblasts. Photochem Photobiol. 2015;91(1):109–116. | ||
Alnemri ES, Livingston DJ, Nicholson DW, et al. Human ICE/CED-3 protease nomenclature. Cell. 1996;87(2):171. |
Supplementary materials
Assessment of NAC-induced changes in ROS production, cell cycle, and metacaspase activation
For ROS quenching, N-acetylcysteine (NAC; Sigma Aldrich, St Louis, MO, USA) was administered to Candida albicans cells to achieve a final concentration of 5 mM for 10 minutes at 28°C. Then, cells underwent photodynamic treatment and were incubated in 10 μM dichlorodihydrofluorescein diacetate (DCFH-DA) for 20 minutes at 37°C. Subsequently, cells were washed three times with PBS. The positive control was treated with 200 mM H2O2 for 10 minutes prior to addition of the DCFH-DA probe. For all analyses, fluorescence intensity was monitored by a microplate fluorescence reader at an excitation wavelength of 488 nm and emissions at 525 nm. ROS level was examined by the ratio of (Ftext – Fblank):(Fcontrol – Fblank). Ftext was the fluorescence intensity of cells treated with photodynamic inactivation, Fcontrol the fluorescence intensity of PBS-treated cells, and Fblank was fluorescence intensity of wells without cells.
C. albicans cells (5×106 CFU/mL) were pretreated with NAC to achieve a final concentration of 5 mM for 10 minutes at 28°C and incubated with experimental and control groups at 37°C for 1 hour. Then, mixtures were washed and irradiated with a 658 nm visible red laser light under 50 mW/cm2 for 10 minutes. Samples were centrifuged at 1,000 g for 5 minutes, washed once with cold PBS, and fixed with 75% cold ethanol overnight at 4°C. Cells were centrifuged again and washed once with cold PBS. The pellets were then incubated with a mixture of RNase A (0.1 mg/mL) and propidium iodide (0.1 mg/mL) for 30 minutes in the dark. DNA content was analyzed with flow cytometry (FACScan; Beckman Coulter, Brea, CA, USA). The percentage of the cell population in each phase was evaluated using Modfit LT 3.0.
C. albicans cells were pretreated with NAC to achieve a final concentration of 5 mM for 10 minutes at 28°C. Then, 105 CFU/mL C. albicans cells were treated with photodynamic inactivation and washed with PBS before staining with CaspACE fluorescein isothiocyanate-VAD-FMK (10 μM). After 20 minutes of incubation at 37°C, cells were washed twice with PBS and visualized by fluorescence microscopy. Fluorescence intensity was recorded by the microplate fluorescence reader at an excitation wavelength of 494 nm and emissions at 518 nm.
  | Table S1 Characteristics of amino acids in CDRs |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.