Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 13
Differences of Microbiomes Found in Non-Inflammatory and Inflammatory Lesions of Acne Vulgaris
Authors Jusuf NK , Putra IB , Sari L
Received 20 July 2020
Accepted for publication 17 September 2020
Published 22 October 2020 Volume 2020:13 Pages 773—780
DOI https://doi.org/10.2147/CCID.S272334
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
Nelva Karmila Jusuf, Imam Budi Putra, Lovena Sari
Department of Dermatology and Venerology, Faculty of Medicine, Universitas Sumatera Utara, Medan, Indonesia
Correspondence: Nelva Karmila Jusuf
Department of Dermatology and Venerology, Faculty of Medicine, Universitas Sumatera Utara, Medan, North Sumatera 20995, Indonesia
Tel +62 811651324
Email [email protected]
Purpose: Bacterial activity and inflammation both influence acne vulgaris (AV) formation. Cutibacterium acnes is considered as an actor involved in inflammation of AV. Besides Cutibacterium acnes, other microbiomes found in AV may also play a role in the pathogenesis. This research was conducted to overview microbiomes found in non-inflammatory and inflammatory lesions of AV.
Patients and Methods: An observational descriptive study with cross-sectional approach was designed. Sample collection was performed with 40 subjects with AV. In every patient, both non-inflammatory (closed comedone) and inflammatory (pustule) lesion samples were collected by swab. Afterward, bacterial culture was performed, continued by bacterial identification.
Results: In non-inflammatory lesions, the growth of nine bacterial species was observed from 40 samples. In an anaerobic culture, Cutibacterium acnes (17,5%) was identified. In aerobic cultures, different bacterial species were found including Staphylococcus epidermis (52.5%), Staphylococcus hominis (12.5%), Staphylococcus haemolyticus (7.5%), Micrococcus luteus (7.5%), Leuconostoc mesentroides (7.5%), Staphylococcus aureus (5%), Kocuria varians (5%), and Staphylococcus vitulinus (2.5%). In inflammatory lesions, nine bacterial species were found, in which was the anaerobic culture we identified Cutibacterium acnes (25.0%). Aerobic cultures have revealed the growth colonies of Staphylococcus epidermidis (42.5%), Staphylococcus hominis (22.5%), Staphylococcus aureus (12.5%), Staphylococcus haemolyticus (10.0%), Leuconostoc mesentroides (5.0%), Staphylococcus cohnii (2.5%), Staphylococcus arlettae (2.5%), and Dermacoccus nishinomyaensis (2.5%). Two mixed bacterial growths were observed in non-inflammatory lesions, while four mixed bacterial growths were found in inflammatory lesions.
Conclusion: Differences in bacterial isolates were observed both in non-inflammatory and inflammatory lesions of AV.
Keywords: acne vulgaris, microbiome, non-inflammatory, inflammatory
Introduction
Acne vulgaris (AV) is an inflammation of pilosebaceous unit which commonly occurs in adolescence. Clinical manifestation of AV is pleomorphic lesions which are found in the form of non-inflammatory lesions (open and closed comedone) and the inflammatory lesions (papule, pustule, and nodule) with various extent and severity.1
Colonization of Propionibacterium acnes (P. acnes), which is now renamed as Cutibacterium acnes (C. acnes), in the sebaceous glands and follicular keratinocyte contributes to the process of inflammation that leads to the formation of acne.1–3 Cutibacterium acnes triggers the keratinocyte and phagocyte cells to produce pro-inflammatory cytokines such as interleukin (IL)-1, IL-1β, IL-12, and tumor necrosis factor (TNF)-α, which induces vigorous inflammation.4 Cutibacterium acnes also perform a stimulation to produce antimicrobial peptide and degradative matrix metalloproteinases (MMPs) that can trigger the inflammation.5 A recent study has shown that the phylotypes IA-2, IB01, and IC of C. acnes are related to the presence of AV.4 Some studies have also reported that the other bacterial species, including Staphylococcus epidermidis (S. epidermidis), S. aureus, Micrococcus sp., Klebsiella sp., Escherichia coli, Enterobacter sp., and Proteus sp., were observed in the AV lesions.6,7 These findings have revealed the contributions of other bacterial species in the progression of AV pathogenesis other than C. acnes.8 In addition to investigating the role of microbiome in the formation of AV, this study was conducted to understand the microbiome profile in both non-inflammatory and inflammatory AV.
Patients and Methods
Sample Collection
A descriptive study with cross-sectional design was carried out by recruiting outpatients who visited the Dermatology and Venereology Clinic at Universitas Sumatera Utara Hospital between November and December 2019. The inclusion criteria were patients who had been diagnosed with acne vulgaris, aged over 18 years-of-old, and were willing to sign the informed consent. The exclusion criteria were patients who were pregnant or breast-feeding, or those who had consumed certain systemic and topical antibiotics in the past 4 weeks. Subjects who had AV were classified into several groups, based on the Lehmann AV severity scale, ie, mild, moderate, and severe AV. Samples were collected from 40 AV patients. In order to collect the samples, both non-inflammatory (closed comedone) and inflammatory lesions (pustule) were collected from each subject. As a control group, a swab sample was collected from the skin area in the face without acne lesion in each patient.
Research Protocol
Demographical data were collected to obtain certain demographic characteristics including age, gender, contact number, as well as the addresses. All participants had the study explained to them, and their willingness to participate in this study was obtained after receiving the informed consent. After receiving the consent, samples were collected from both non-inflammatory (closed comedone) and inflammatory (pustule) lesions from every subject. These specimens were collected via sterilized acne extractors (No name, Indonesia), which was followed by sterilized swabbing sample into growth culture media and moistening with nutrient broth (Oxoid Product, PT. Dipa Puspa Labs, Jakarta, Indonesia). Samples from skin areas on the face without acne lesions were also taken by swab, and were compiled as the control samples. Every specimen was then divided into two groups which were used as culture media for the growth of anaerobic and aerobic bacterial species, in which every culture media was prepared by adding blood agar (Oxoid Product) and Brucella blood agar (Oxoid Product,). The investigation of Gram positive and negative bacteria was carried out by impression smears taken on a transparent slide for Grams staining (Color Gram Biomeruex product, PT. Enseval Medika Prima, Jakarta, Indonesia). Afterward, samples were prepared for inoculation, which was continuously followed by incubation step both in the anaerobic and aerobic condition at 37°C for 24–48 hours. Inside the anaerobic tube, AnaeroGen® Compact (Anaerogen Thermo Scientific, PT. Dipa Puspa Labs, Jakarta, Indonesia) was placed to isolate the anaerobic bacterial species, whereas to isolate the aerobic bacterial species, no AnaeroGen® Compact was used. The bacterial species identification was performed with Vitek®2 compact (Biomeruex product, PT. Enseval Medika Prima, Jakarta, Indonesia).
The Vitek® 2 is an automated microbial identification system. With its colorimetric reagent cards, and associated hardware and software advances, this study used a technology platform for phenotypic identification methods. This accommodates the same colorimetric reagent cards that are incubated and interpreted automatically. The reagent cards have 64 wells that can each contain an individual test substrate. Substrates measure various metabolic activities such as acidification, alkalinization, enzyme hydrolysis, and growth in the presence of inhibitory substances. An optically clear film present on both sides of the card allows for the appropriate level of oxygen transmission while maintaining a sealed vessel that prevents contact with the organism-substrate admixtures. Identification cards are inoculated with microorganism suspensions using an integrated vacuum apparatus. A test tube containing the microorganism suspension is placed into a special rack (cassette) and the identification card is placed in the neighboring slot while inserting the transfer tube into the corresponding suspension tube. The filled cassette is placed manually into a vacuum chamber station. After the vacuum is applied and air is re-introduced into the station, the organism suspension is forced through the transfer tube into micro-channels that fill all the test wells. Inoculated cards are passed by a mechanism, which cuts off the transfer tube and seals the card prior to loading into the carousel incubator. All card types are incubated on-line at 35.5+1.0ºC. Each card was removed from the carousel incubator once every 15 minutes, transported to the optical system for reaction readings, and then returned to the incubator until the next read time. Data were collected at 15 minute intervals during the entire incubation period. A transmittance optical system allows interpretation of test reactions using different wavelengths in the visible spectrum. During incubation, each test reaction is read every 15 minutes to measure either turbidity or colored products of substrate metabolism. The databases of the Vitek® 2 identification products are constructed with large strain sets of well-characterized microorganisms tested under various culture conditions. These strains are derived from a variety of clinical and industrial sources as well as from public and university culture collections.9 The card was automatically filled by a vacuum device, sealed and inserted into the Vitek® 2 reader-incubator module (incubation temperature, 35.5°C), and subjected to a kinetic fluorescence measurement every 15 minutes. The results were interpreted by the test panel database, and final results were obtained automatically. All cards used were automatically discarded into a waste container.10
Ethical Clearance
We declare that the procedures were followed according to the regulations established by the Clinical Research and Ethics Committee and to the Declaration of Helsinki of the World Medical Association. This research protocol has been approved by the Ethical Committee of the Faculty of Medicine, Universitas Sumatera Utara, with given number 807/TGL/KEPK FK USU-RSUP HAM/2019.
Statistical Analysis
To determine the differences among the obtained data, statistical analysis was performed via descriptive analysis. The distribution of anaerobic and aerobic bacterial species was also statistically analyzed to find the differences which are based on the non-inflammatory and inflammatory lesions. The result was a frequency for anaerobic and aerobic bacteria distribution of acne vulgaris non-inflammatory and inflammatory lesions.
Results
The subject characteristics are displayed in Table 1. It can be seen that the most populated characteristic was in subjects aged between 17–25 years old (72.5%), and 11 subjects were 26–35 years old (27.5%). Most of the subjects were female (65%, 26 subjects), while there were 14 males (35%). There were 14 subjects who had mild AV, and both moderate and severe AV accounted for 13 subjects, each.
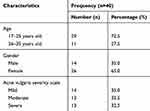 |
Table 1 Demographical Characteristics of Samples |
The microbiome distribution on the non-inflammatory lesions is displayed in Table 2 and Figure 1. There were 40 samples taken from 40 subjects with non-inflammatory lesions of AV. From these samples, nine growth of bacterial species were observed. In anaerobic culture, we identified C. acnes on seven samples (17,5%). In anaerobic cultures were observed the growth of S. epidermidis on 21 samples (52.5%), Staphylococcus hominis (S. hominis) on five samples (12.5%), Staphylococcus haemolyticus (S. haemolyticus), Micrococcus luteus (M. luteus), and Leuconostoc mesentroides (L. mesentroides) were each seen on three samples (7.5%), while other species such as S. aureus and Kocuria varians were each seen on two samples (5.0%), and Staphylococcus vitulinus (S. vitulinus) on one sample (2.5%). The mixed growths of anaerobic and aerobic bacterial species that were observed on non-inflammatory lesions were two types of mixed growth. The first mixed growth was C. acnes and S. epidermidis, on six samples (15.0%), and the second was C. acnes and S. hominis, on one sample (2.5%).
 |
Table 2 Microbiome Distributions of Non-Inflammatory Acne Vulgaris Lesions |
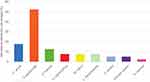 |
Figure 1 Microbiome distributions of non-inflammatory acne vulgaris lesions. |
The microbiome distribution on inflammatory lesions is shown in Table 3 and Figure 2. The samples collection was carried out from 40 subjects with AV, and the number of samples taken was 40. On these lesions, nine bacterial species were found, in which the anaerobic culture was observed to have growth of C. acnes on 10 samples (25%). On the other hand the aerobic cultures were observed to have certain bacterial species including S. epidermidis on 17 samples (42.5%), S. hominis on nine samples (22.5%), S. aureus on five samples (12.5%), S. haemolyticus on four samples (10.0%), L. mesentroides on two samples (5%), while other species such as Staphylococcus cohnii (S. cohnii), Staphylococcus arlettae (S. arlettae), and Dermacoccus nishinomyaensis (D. nishinomyaensis) were each seen on one sample (2.5%). The mixed growths of anaerobic and aerobic bacterial species that were observed on non-inflammatory lesions were four types mixed growth. The first mixed growth was C. acnes and S. epidermidis on five samples (12.5%), the second was C. acnes and S. hominis on two samples (5.0%), the third was C. acnes and S. aureus on two samples (5.0%), and the last one was C. acnes and S. haemolyticus on one sample (2.5%).
 |
Table 3 Microbiome Distributions of Inflammatory Acne Vulgaris Lesions |
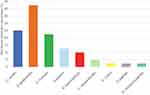 |
Figure 2 Microbiome distributions of inflammatory acne vulgaris lesions. |
The distribution of microbiome skin area on the face without acne lesion is highlighted in Table 4 and Figure 3. These samples were collected from 40 subjects, which resulted in 40 samples. Based on these samples, 10 bacterial species were found to be growing, in which no C. acnes and other anaerobic bacterial growth was observed. Thus, aerobic cultures that were observed in these samples were S. epidermidis on 25 samples (62.5%), S. hominis on three samples (7.5%), S. aureus, S. haemolyticus, S. arlettae, and Staphylococcus warneri (S. warneri) were seen, each on two samples (5.0%). While other species such as Staphylococcus lugdunensis (S. lugdunensis), Staphylococcus capitis (S. capitis), M. luteus, and D. nishinomyaensis were each seen on one sample (2.5%).
 |
Table 4 Microbiome Distributions of Skin Area Without Acne Lesion |
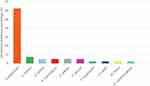 |
Figure 3 Microbiome distributions of skin area on the face without acne vulgaris lesions. |
In this study, six exact bacterial isolates that were observed both on non-inflammatory and inflammatory lesions were C. acnes, S. epidermidis, S. hominis, S. haemolyticus, L. mesentroides, and S. aureus. Moreover, in non-inflammatory and inflammatory AV lesions, six different bacterial isolates were found. On the non-inflammatory lesions, M. luteus, Kocuria varians, and S. vitulinus were found, whereas on the inflammatory lesions, S. chonii, S. arlettae, and D. nishinomyaensis were observed.
The mixed growth bacterial species were found more in inflammatory lesions (10 samples) than those in non-inflammatory lesions (7 samples). The skin area of the face without acne lesions was found to have no mixed growth bacteria.
Discussion
In this study, the results showed that the S. epidermidis is the predominant aerobic bacterial isolate, while the C. acnes is the predominant anaerobic bacterial isolate, both in non-inflammatory and inflammatory lesions.
The bacterial species of non-inflammatory lesions that were observed in this study were C. acnes, S. epidermidis, S. hominis, S. haemolyticus, Micrococcus luteus, L. mesentroides, S. aureus, Kocuria varians, and S. vitulinus. The common bacterial species that were found dominantly were S. epidermidis and C. acnes. These results have confirmed that the only bacterial species which were commonly found were not only the C. acnes, the other bacterial species were also observed in the AV lesions. A study conducted by Syahrial et al,11 who have identified the bacterial species in comedone lesions, has reported that the most common bacteria were C. acnes for 37.2% and S. epidermidis for 30.2%. Another study by Sitohang et al12 also confirmed that the majority bacterial species in comedone lesions was S. epidermidis, with 50.5%, while 11.0% of C. acnes was found.
In this study, the bacterial species observed from the inflammatory AV lesions were C. acnes, S. epidermidis, S. hominis, S. aureus, S. haemolyticus, L. mesentroides, S. cohnii, S. arlettae, and D. nishinomyaensis. In this lesion, S. epidermidis and C. acnes were the two top common bacterial species that were found. These results are in accordance to the study by Srikanth et al,13 who observed in pustules of AV lesions that the C. acnes was observed as the most common anaerobic bacterial species and S. epidermidis was the most common aerobic bacteria. Moon et al14 also reported similar findings that S. epidermidis (36%) and C. acnes (30%) dominated the types of bacterial colonies in papule and pustule lesions of AV patients. In a study by Brook et al,15 the most common aerobic bacteria found in pustule of AV patients was S. epidermidis (46.2%). A study conducted by Leyden et al.16 also found that C. acnes in infants was found to be more common in inflammation lesions (80%) than comedone (25%), while in preteenagers, 95% of C. acnes was found in inflammation lesions with 0% in comedone lesions.
In this study, the bacterial species that were observed on non-inflammatory lesions as well as on inflammatory AV-lesions had similarity, particularly in term of the species. In both lesions, it was collectively found that bacterial isolates of C. acnes, S. epidermidis, S. hominis, S. haemolyticus, L. mesentroides, and S. aureus with different amounts of isolates. On the other hand, it was also found that bacterial species that were found on the non-inflammatory lesions were differently observed on the inflammatory lesions. On the non-inflammatory lesions, the bacterial isolates were M. luteus, Kocuria varians, and S. vitulinus, whereas these three species were not observed on the inflammatory lesions. Whilst in the inflammatory lesions, bacterial isolates of S. cohnii, S. arlettae, and D. nishinomyaensis were observed, in which none of these three species were observed on the non-inflammatory lesions.
In this study, we also observed the differences in species and the growth of bacterial isolates that were observed on the skin area without acne lesions. It was found that the domination of S. epidermidis growth in aerobic culture was higher than the other bacteria, whereas no C. acnes or anaerobic bacterial growth was found in the anaerobic culture. These findings have suggested similar results to those that were observed by Dreno et al,2 in France, in which the predomination of Staphylococcus growth was observed on the skin area where AV lesions were absent.
Several samples in this study have suggested the presence of mixed growth bacteria, which were observed on the non-inflammatory and inflammatory AV-lesions with both anaerobic and aerobic bacteria. The differences in species mixed growth bacteria on the non-inflammatory and inflammatory lesions were observed, in which two mixed growth of anaerobic and aerobic bacterial were found in the non-inflammatory lesions. These species were C. acnes and S. epidermidis as the first mixed, whereas the second mixed growth bacterial were C. acnes and S. hominis. On the inflammatory lesions, four mixed growth bacterial species were found, in which all of the mixed growth bacterial species were observed to be anaerobic and aerobic species; ie, first, C. acnes and S. epidermidis, the next is C. acnes and S. hominis, then is C. acnes and S. aureus, and finally is C. acnes and S. haemolyticus. The amounts of bacterial mixed growth were observed to be higher in the inflammatory lesions rather than in the non-inflammatory sides.
Cutibacterium acnes is considered to have important contributions in the process of pathogenesis of acne.3 These anaerobic bacteria produce a lot of types of enzymes, including lipase, protease, and any other extracellular enzymes, and this bacteria also secretes the chemotactic factors that can trigger the inflammation due to the stimulations on polymorphonuclear leucocyte, lymphocyte, and macrophages.1,17 Cutibacterium acnes also produces free fatty acids from triglycerides, which induce the comedogenesis.1 This bacteria also produces a biofilm which leads to C. acnes adhering to the follicular wall and binding to corneocytes to produce microcomedone. This condition explains the contribution of C. acnes in the formation of microcomedo as well as the inflammation on the AV.17 Our results have also confirmed higher amounts of C. acnes colonies on the inflammatory lesions compared to those on the non-inflammatory lesions, in which no C. acnes colonies were observed on the skin area without acne lesions. A previous study has reported that a healthy follicle subject was dominated by the C. acnes strain CC18, whereas C. acnes phylogeny I-1a particularly related to clonal complex of CC19 and CC3 were observed on the follicular area of patients with acne lesions. These results have shown the potential effects of inflammation caused by C. acnes strain on the skin, which may influence the development and severity of acne vulgaris.4
The other contribution of other bacterial species in the pathogenesis of AV remains controversial.8 According to Pathatk et al,18 the C. acnes and S. epidermidis are common bacterial species that were found in 70–82% of AV patients.. Staphylococcus epidermidis produces a biofilm that creates anaerobic conditions, in which these conditions are effective for the growth of C. acnes. Staphylococcus epidermidis also contributes in the production of virulent factors such as lipase and haemolysin delta that can be considered as the main factors of the AV formation.8,19 Staphylococcus epidermidis was the abundant commensal bacteria found in human skin, and one study has suggested the amounts of S. epidermidis can be equivalent or higher than C. acnes in some follicles. Therefore, while S. epidermidis is one of the most dominant bacterial species on the skin surface, its contribution to health and follicular disease remains poorly understood.20,21 The result of this study is expected to provide evidence for developing more suitable treatment guidelines for acne vulgaris in the future.
Conclusions
Differences in bacterial isolates were observed both in non-inflammatory and inflammatory lesions of AV. In this study C. acnes was found more in inflammatory lesions than non-inflammatory lesions. It is also known that not only does C. acnes play role in the inflammatory process of AV, but other bacteria such as S. epidermidis were also abundant in inflammatory lesions of AV.
Acknowledgments
We would like to express thanks to Universitas Sumatera Utara Hospital.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Goh C, Cheng C, Agak G, et al. Acneiform Disorder. In: Kang S, Amagai M, Bruckner AL, et al., editors. Fitzpatrick’s Dermatology in General Medicine.
2. Dreno B, Martin R, Moyal D, Henley JB, Khammari A, Seite S. Skin microbiome and acne vulgaris: staphylococcus, a new actor in acne. Exp Dermatol. 2017;26(9):798–803. doi:10.1111/exd.13296
3. Dreno B, Pecastaings S, Corver S, Veraldi S, Khammari A, Roques C. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J Eur Acad Dermatol Venereol. 2018;32(2):5–14. doi:10.1111/jdv.15043
4. Keshari S, Kumar M, Balasubramaniam A, et al. Prospects of acne vulgaris targetting secreted virulences factors of Cutibacterium acnes. Expert Rev Vaccines. 2019;18(5):433–437.
5. McLaughlin J, Watterson S, Layton AM, et al. Propionibacterium acnes and acne vulgaris: new insights from the integration of population genetic, multi-omic, biochemical and host-microbe studies. Microorganisms. 2019;7(128):1–29.
6. Hassanzadeh P, Bahmani M, Mehrabani D. Bacterial resistance to antibiotics in acne vulgaris: an in vitro study. Indian J Dermatol. 2008;53(3):122. doi:10.4103/0019-5154.43213
7. Neubert U, Jansen T, Plewig G. Bacteriologic and immunologic aspects of gram‐negative folliculitis: a study of 46 patients. Int J Dermatol. 1999;38(4):270–274. doi:10.1046/j.1365-4362.1999.00688.x
8. Kumar B, Pathak R, Mary PB, Jha D, Sardana K, Gautam HK. New insights into acne pathogenesis: exploring the role of acne-associated microbial populations. Dermatologica Sinica. 2016;34(2):67–73. doi:10.1016/j.dsi.2015.12.004
9. Pincus DH. Microbial identification using biomerieux vitek 2 system. Encycl Rapid Microbiol Methods. 2006;1:1–32.
10. Ligozzi M, Bernini C, Bonora MG, Fatima M, Zuliani J, Fontana R. Evaluation of the vitek 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J Clin Microbiol. 2002;40(5):1681–1686. doi:10.1128/JCM.40.5.1681-1686.2002
11. Syahrial MA, Nasution D, Jusuf NK, Lubis SE Pola Resistensi Propionibacterium acnes Terhadap Antibiotika Oral Pada Pasien Akne Vulgaris di RSUP H. Adam Malik Medan. [unpublished disertation]. Medan: Universitas Sumatera Utara; 2009
12. Sitohang IBS, Fathan H, Effendi E, Wahid M. The susceptibility of pathogens associated with acne vulgaris to antibiotics. Med J Indones. 2019;28(1):21–27. doi:10.13181/mji.v28i1.2735
13. Srikanth M, Kalyani CS, Mohan N, Sridhar K, Padmaja IJ. Bacteriology of acne. J Evol Med Dent Sci 2015;4(19):3267–3274. doi:10.14260/jemds/2015/473
14. Moon SH, Roh HS, Kim YH, Kim JE, Ko JY, Ro YS. Antibiotic resistance of microbial strains isolated from Korean acne patients. J Dermatol. 2012;39(10):833–837.
15. Brook I, Frazier EH, Cox ME, Yeager JK. The aerobic and anaerobic microbiology of pustular acne lesions. Anaerobe. 1995;1(6):305–307. doi:10.1006/anae.1995.1031
16. Leyden JJ, McGinley KJ, Vowels B. Propionibacterium acnes colonization in acne and nonacne. Dermatology. 1998;196(1):55–58. doi:10.1159/000017868
17. Beylot C, Auffret N, Poli F, et al. Propionibacterium acnes: an update on its role in the pathogenesis of acne. J Eur Acad Dermatol Venereol. 2014;28(3):271–278. doi:10.1111/jdv.12224
18. Pathak R, Kasama N, Kumar R, Gautama HK. Staphylococcus epidermidis in human skin microbiome associated with acne: a cause of disease or defence? Res J Biothecnol. 2013;8:79–82.
19. Vuong C, Voyich JM, Fischer ER, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–275.
20. Neill AM, Gallo RL. Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome. 2018;6:1–16.
21. Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo R. The microbiome extends to subepidermal compartments od normal skin. Nat Commun. 2013;4(1):1–8. doi:10.1038/ncomms2441
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
