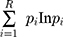Back to Journals » Clinical and Experimental Gastroenterology » Volume 13
Circulating α4β7+ Memory T Cells in Pediatric IBD Patients Express a Polyclonal T Cell Receptor Repertoire
Authors Gamliel A, Werner L, Pinsker M, Salamon N, Weiss B, Shouval DS
Received 10 July 2020
Accepted for publication 21 August 2020
Published 2 October 2020 Volume 2020:13 Pages 439—447
DOI https://doi.org/10.2147/CEG.S271565
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Wing-Kin Syn
Adir Gamliel,1,2 Lael Werner,1,2 Marina Pinsker,1,3 Naomi Salamon,1 Batia Weiss,1,2 Dror S Shouval1,2
1Pediatric Gastroenterology Unit, Edmond and Lily Safra Children’s Hospital, Sheba Medical Center, Ramat Gan, Israel; 2Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel; 3Institute of Nanotechnology and Advanced Materials, Bar Ilan University, Ramat Gan, Israel
Correspondence: Dror S Shouval
Pediatric Gastroenterology Unit, Edmond and Lily Safra Children’s Hospital, Sheba Medical Center, Ramat Gan, Israel
Tel +972-3-5305006
Email [email protected]
Background: The integrin α 4β 7 is highly expressed on activated T cells and is thought to direct homing of lymphocytes to the intestine. Since ulcerative colitis (UC) and Crohn’s disease (CD) are characterized by mucosal oligoclonal T cells’ expansion, we aimed to assess whether similar repertoire features are identified in circulating gut-specific memory T cells.
Methods: Memory CD3+ T cells were isolated from blood samples of control subjects and patients with active UC or CD and then FACS-sorted into α 4β 7+ and α 4β 7− populations. DNA was extracted from each subset and subjected to next-generation sequencing of the TCRβ. Different repertoire characteristics were compared between α 4β 7+ and α 4β 7− subsets for each subject, and between groups.
Results: The percentages of memory T cells and α 4β 7+ cells were comparable between groups. α 4β 7+ memory T cells displayed a polyclonal distribution, in control subjects and in UC or CD patients, with similar indices of diversity. Strikingly, the clonal overlap between α 4β 7+ and α 4β 7− T cells for each subject in all three groups was high, ranging between 20%– 50%. We were unable to identify shared T cell clones that were specific to one of the groups.
Conclusion: α 4β 7+ memory T cells exhibited a polyclonal repertoire in both control subjects and patients with active inflammatory bowel disease, with high rates of overlap with α 4β 7− memory T cells. Our study, along with additional recent reports, may suggest that the suppression of intestinal inflammation by vedolizumab is independent of the drug’s effect on T cell migration to the gut.
Keywords: IBD, TCR, integrin, α 4β 7, T cells, immune repertoire
Introduction
Integrins are cell surface glycoprotein receptors that bind to adhesion molecules and mediate homing of leukocytes to peripheral sites such as the intestine.1 More than 20 different integrins have been identified that vary according to their expression pattern and specificity of ligand binding. These processes are further strengthened by the expression of different chemokine receptors, such as CCR9, that stabilize the interaction between the immune cells and the vessel walls during the extravasation process in peripheral sites. Drugs that interfere with this process have been developed for patients with inflammatory bowel disease (IBD) or multiple sclerosis, based on selectivity of the integrin.2,3
The α4β7 integrin complex binds mucosal addressin cell adhesion molecule 1 (MAdCAM‐1), expressed exclusively on intestinal endothelial cells.4 Ligation of α4β7 and MAdCAM-1 leads to leukocyte extravasation into intestinal high endothelial venules and is therefore considered gut-selective. Understanding this process led to the development of vedolizumab, a monoclonal antibody targeting the α4β7 integrin. Vedolizumab has been shown to be effective for induction and maintenance of remission in Crohn’s disease (CD) and ulcerative colitis (UC) in multiple studies.5–9 Its main mechanism of action has been considered to be the blockade of activated T cell migration to the gut, as α4β7 is upregulated on activated cells.4 However, emerging data have challenged this dogma by suggesting that other immune cells, and specifically innate immune populations, also express α4β7,10–13 and question whether vedolizumab alleviates intestinal inflammation solely by blocking migration of T cells.
The adaptive immune arm contains trillions of different T cell clones. The marked diversity in T cell receptors (TCRs) is formed following a complex rearrangement process of different gene segments (VDJ recombination).14 There are four types of TCR protein monomers: α, β, γ and δ; more than 95% of T cells express α/β chains, encoded by genes in the TCRA and TCRB loci. The antigenic specificity of T cells occurs via generation and rearrangement that involves recombination of variable (Vβ), diversity (Dβ) and joining (Jβ) genes, accompanied by deletion and insertion of random nucleotides, generating trillions of unique TCRs. Each TCR recognizes a unique antigen, and ligation can trigger proliferation, differentiation and/or cytokine secretion. Next-generation sequencing (NGS) platforms, developed in the last decade, facilitate detailed assessment of TCR repertoire patterns at the nucleotide or amino acid level.15 A restricted TCR (also referred to as oligoclonal expansion) was demonstrated in various autoimmune disorders including multiple sclerosis,16 rheumatoid arthritis17 and psoriasis.18 Few NGS repertoire studies in IBD revealed oligoclonality in intestinal samples of patients with active CD and UC.19–23 We were able to show that newly-diagnosed, pediatric UC patients have marked alterations in TCRB repertoire patterns in inflamed rectum, compared with controls, characterized by oligoclonal expansion and decreased diversity.24
There are no data on TCR repertoire patterns of circulating memory T cells, and specifically those subsets with upregulated α4β7 expression, among IBD patients. Since T cells are important in mediating the mucosal inflammatory response in IBD, one could expect that cells expressing α4β7 would include a high percentage of disease-associated clonotypes. We aimed to examine repertoire features of α4β7+ memory T cells in IBD patients vs control subjects, and specifically compare these features between α4β7+ and α4β7− populations.
Methods
Sorting and DNA Isolation
The study was conducted in accordance with the Declaration of Helsinki and approved by the local ethics committee of Sheba Medical Center. Informed written consent was obtained from parents of participating subjects before experiments were conducted. Blood samples were obtained in EDTA-containing tubes from control subjects and patients with UC or CD. Disease activity in IBD patients was determined based on the pediatric ulcerative colitis activity index (PUCAI), the pediatric Crohn’s disease activity index (pCDAI) or endoscopic features. Samples were subjected to lymphoprep gradient centrifugation (Stemcell, MA, USA) for isolation of mononuclear cells. Next, negative selection of CD3+ cells was conducted using MACS beads (Miltenyi Biotec, CA, USA). Cells were stained with CD3-FITC, CD45RO-APC, (both from Miltenyi Biotec) and α4β7-PE (using vedolizumab as the antibody). Conjugation of vedolizumab to R-Phycoerythrin (R-PE) was performed using the SiteClick™ antibody labeling kit (Thermo Scientific, MA, USA). Antibody carbohydrate domain modification, azide attachment to the antibody, and conjugation with DIBO-modified label were performed according to manufacturer’s instructions. CD3+ cells were subjected to FACS sorting into two sub-populations: memory T cells (CD3+CD45RO+) that are either α4β7+ or α4β7− (Supplementary Figure 1). Purity was >95%. Genomic DNA was extracted from blood using a commercially available kit (Wizard kit, Promega, WA, USA), according to the manufacturer’s instructions.
Next-Generation Sequencing
Up to 2 µg DNA were used for TRB library generation of various V and J gene segments of the rearranged complementary determining region (CDR3β, ImmunoSeq TRB Survey Service, Adaptive Biotechnologies, Seattle, USA). To ensure equal depth of sequencing, we used the Survey level (up to 500,000 reads per sample). The resulting libraries were subjected to high-throughput sequencing using Illumina technology. Samples were sequenced in two separate batches: first, controls and patients with UC and second, patients with CD. The number of sequences in the second run on the illumina was significantly lower compared to the first run (Supplementary Table 1), but was still high, enabling a thorough analysis of immune repertoire patterns in the CD group as well.
TCRβ Repertoire Analysis
ImmunoSeq software was used for determination of diversity parameters and overlapping clones. Clonality was calculated as 1-normalized Shannon’s entropy. This measures how evenly receptor sequences are distributed among a set of T cells, with values ranging from 0 to 1. Values near 1 represent samples with one or a few predominant clones (monoclonal or oligoclonal samples) dominating the repertoire. Clonality values near 0 represent polyclonal samples. Simpson’s D is the sum over all observed rearrangements of the square fractional abundances of each rearrangement. Shannon’s H, which measures the overall diversity in a given population, and takes into account the number of unique sequences (richness of the repertoire) and how evenly the sequences are distributed, was calculated using the following formula:
R = Total templates
i = Unique rearrangements
pi = Proportion of the total sequences belonging to the “i”th unique rearrangement.
Graphical presentation of the repertoire was presented using hierarchical tree maps using the Treemap software (www.treemap.com).
Since the number of total templates differed between the groups, and to enable a valid comparison of different repertoire characteristics, we used the 10,000 most frequent unique clones for all analyses except for overlap between α4β7+ and α4β7− populations for each subject.
Statistical Analysis
Values are expressed as mean ± standard error of the mean (SEM). Unpaired Student’s t-test was used to test for statistical significance. Significance was determined if P value was ≤0.05. For differential V-gene, and J-family usage, Bonferroni's correction for multiple comparisons was performed.
Results
Study Population
Blood samples were obtained from 5 control subjects, 6 patients with UC and 5 patients with CD (Table 1). Control subjects were patients referred for endoscopic evaluation of abdominal pain or diarrhea, that had a normal macroscopic and histologic esophagogastroduodenoscopy or colonoscopy, without past or present history of IBD or other immune-mediated disorder (eg, celiac, diabetes, etc.). Among patients with UC, three patients had mild disease (based on the PUCAI score) and two experienced moderate disease activity. An additional patient was in clinical remission, but colonoscopy demonstrated moderate colitis (Mayo score 2). In the cohort of patients with CD, three patients had moderate disease activity (based on the pCDAI score), two had mild activity and one was in clinical remission, but with endoscopic and histologic evidence of inflammation of the ileocecal region, with large ulcers and cobblestoning. The median age of the subjects in the control group was 15.3 years (interquartile range [IQR] 13.5–16.4), compared with 16.8 years (IQR 15.0–17.9) and 15.8 years (IQR 15.1–17.5) in the UC and CD groups, respectively.
 |
Table 1 Demographic and Clinical Phenotype of Studied Patients |
Immunophenotyping of Memory T Cells
We first analyzed the frequency of different immune populations in the studied groups. The frequency of CD3+CD45RO+ memory T cells was comparable between groups (43.8±5.5%, 32.2±5.4% and 47.0±6.0% among control, UC and CD groups, respectively, Figure 1A). Moreover, the frequency of α4β7+ cells among memory T cells was also similar (33.6±7.0%, 36.0±7.2% and 37.4±4.1%, among control, UC and CD groups, respectively, Figure 1B).
Assessment of TCRβ Repertoire Features
In order to characterize the landscape of memory T cells, NGS of the TCRβ region was performed on α4β7+ and α4β7− memory T cells. The mean number of total templates and unique templates in each of the groups is presented in Supplementary Table 1. We first conducted treemap analysis of the most frequent 10,000 clones. In these studies, each square represents a unique clone and the size indicates how frequent it is. As can be seen in representative images in Figure 2A, a polyclonal distribution of T cells was observed in α4β7− and α4β7+ memory T cells, in control subjects, patients with active UC and CD. To further characterize the immune repertoire in these populations, various indices of diversity were calculated. The cumulative percent of the top 100 most frequent clones was comparable between α4β7− and α4β7+ populations in all studied groups (Figure 2B). Moreover, Shannon’s H, Simpson’s D and clonality were also similar between sub-populations in controls, UC and CD patients, and also comparable between the groups (Figure 2C–E). These findings indicate that the TCRβ repertoire of α4β7+ memory T cells is polyclonal, similar to the α4β7− population, and that comparable diversity features are identified in these immune subsets in controls vs patients with active UC or CD.
Identification of Overlapping Clones
We next assessed whether there were overlapping clones between α4β7− and α4β7+ memory T cells in each subject, and whether specific clonotypes could be identified solely in one of the groups. Strikingly, the degree of clonal sharing between α4β7− and α4β7+ sub-populations in each of the groups was high, ranging from 24%–50%, 29%–46% and 24%–50% in the control, UC and CD groups, respectively (Figure 3A and B). Comparison of specific clonotypes showed that among the 10 most frequent clones in each immune subset, for each subject, the mean number of shared clones was 2.4, 3.7 and 2.6 in the control, UC and CD groups, respectively (Supplementary Table 2).
We next analyzed whether we could identify α4β7− or α4β7+ memory T clones that were unique to each group. However, such specific clones that characterize solely one group could not be identified (data not shown). In addition, analysis of TRBV and TRBJ gene usage from each group showed similar frequencies, in both α4β7− or α4β7+ subpopulations (Figure 4), except for TCRBV25 which was significantly lower in both UC and CD patients compared with controls, similar to previous observations.25
Discussion
Next-generation sequencing is a powerful tool to study adaptive immune function in different diseases. Using this methodology, we and others have demonstrated oligoclonal expansion of T cells in the inflamed gut of patients with IBD.19–24 Since these T cells migrate to the gut, one could also expect to identify specific clonotypes in the blood of these patients, at least in those cells expressing α4β7. Nevertheless, we showed a polyclonal distribution of T cell clonotypes in circulating α4β7+ and α4β7− memory T cells in control subjects and in patients with active IBD. Moreover, we showed that the overlap between α4β7+ and α4β7− subsets was high, ranging between 20%–50%, highlighting that expression of this integrin is not confined to specific clones that are thought to be gut-specific.
Numerous studies have demonstrated that vedolizumab is effective in the treatment of patients with active IBD.5–9 Additional trials have also shown efficacy of new gut-specific anti-integrins in both UC and CD, such as etrolizumab,26 targeting β7, abrilumab,27 targeting α4β7 and AJM300,28 targeting α4. Since α4β7 was shown to be upregulated in memory T cells, and binds MADCAM1 expressed exclusively on intestinal endothelial cells,4 anti-α4β7 drugs are thought to exert their anti-inflammatory effects through blocking migration of T cells to the gut,4 which could explain the relatively long duration needed to demonstrate clinical and endoscopic efficacy. However, data from different mouse and human studies in recent years suggest that targeting α4β7 or β7 may affect other immune populations beside T cells. Villablanca et al reported that β7 expression on bone marrow precursors was required to facilitate reconstitution of small bowel tolerogenic retinoic acid-producing dendritic cells (DCs) and macrophages.10 In addition, Schippers et al identified that β7 expression on inflammatory monocytes was required for their migration to the gut, in mice.11 Drugs targeting α4β7 or β7 may affect migration of these innate subsets as well.
Two additional studies shed more light on the importance of α4β7 expression on monocytes in IBD. Zeissig et al compared systemic and mucosal immune profiles of 18 IBD patients treated with vedolizumab vs 20 patients treated with anti-TNFα12 While they were not able to show differences in the frequency or phenotype of mucosal T cells in patients treated with vedolizumab, before and after induction timeframe, they did identify marked changes in macrophage abundance, phenotype and pattern recognition receptor expression.12 Moreover, the intestinal TCRα and TCRβ repertoires pre- and post-vedolizumab therapy were comparable, even after 14 weeks. Another study showed that α4β7 was expressed on CD14+CD16high non-classical monocytes and mediated their migration to the gut, where they develop into CD163+ wound healing macrophages.13 Using Rag1−/- mice they showed that anti-α4β7 inhibited colonic wound healing in a T cell-independent manner.21
Overall, the previously mentioned studies suggest that vedolizumab may affect gut homing of multiple immune cell populations, and that its effects on innate cells might be more important than previously appreciated. Our findings also question the importance of α4β7 for guiding T cell migration to the gut, since a high degree of overlap was identified between α4β7+ and α4β7− memory T cells. Interestingly, the IBD group in Sheba Medical Center showed that vedolizumab therapy completely blocked α4β7 expression on circulating T cells, independent of patient’s response status or vedolizumab serum levels,29 suggesting that the integrin blockade on T cells may not be sufficient on its own for patient’s response to treatment. It is possible that surface expression of α4β7 on T cells is completely blocked by vedolizumab, as shown by that group,29 but there are still free molecules on innate cells and therefore full treatment efficacy is not achieved. Another alternative explanation is that vedolizumab may affect the function of mucosal T cells directly, irrespective of its effect on cell migration.
The overlap between circulating α4β7+ and α4β7− memory T cells can be explained in several ways. It is possible that different factors up- or down-regulate α4β7’s expression in the same cell and that levels of expression vary. Moreover, migration of T cells is influenced by expression of different chemokine receptors,4 which were not examined here. Finally, additional integrins might be important as well for gut-specific migration, including αEβ7.30
Our study has several limitations, including the small sample size in each of the groups and relatively mild clinical disease activity, especially in the UC group. However, the data of polyclonal repertoire features and high degree of clonal overlap were consistent in all subjects from the three groups. In addition, different therapies and especially immunosuppressive regimens can affect repertoire features by inhibiting proliferation of T cells. In our UC cohort one out of six subjects was receiving prednisone and azathioprine, while the rest were on mesalazine or treatment naïve. Among the patients with CD, all were on medications, including prednisone and azathioprine or biologics. It is possible that these drugs affected repertoire features, and further studies among treatment-naïve patients should address this point. Finally, we did not sequence the TCRα region, although it is well established that the most variable region is the CDR3β of the TCRβ. Nevertheless, this is the first study looking at immune repertoire profiles in specific T cell subsets in IBD patients. We used vedolizumab-conjugated antibody to sort α4β7+ cells, and therefore believe the identity of the subpopulations was correct. Moreover, our study enabled a comparison between α4β7+ and α4β7− populations, with similar results showing high degree of clonal overlap in all groups. Further studies should assess whether similar profiles are identified in mucosal α4β7+ and α4β7− memory T cells, and compare them to circulating T cells.
In conclusion, we showed a polyclonal distribution of α4β7+ memory T cells in IBD patients, similar to control subjects, along with a high degree of clonal overlap between α4β7+ and α4β7− memory T cells. While it is clear that vedolizumab is an effective drug for patients with active IBD, its mechanism of action might be broader than blocking T cell migration to the gut. Additional studies are required to characterize the dynamics of various integrins on specific immune populations in the intestine (small vs large bowel) and blood, and define these expression profiles in steady state, during active inflammation and in mucosal healing.
Disclosure
This study was funded by a research grant from Takeda. Dr Dror Shouval reports grants from Takeda, during the conduct of the study. The authors report no other potential conflicts of interest for this work.
References
1. Zundler S, Becker E, Schulze LL, Neurath MF. Immune cell trafficking and retention in inflammatory bowel disease: mechanistic insights and therapeutic advances. Gut. 2019;68:1688–1700. doi:10.1136/gutjnl-2018-317977
2. Lamb CA, O’Byrne S, Keir ME, Butcher EC. Gut-selective integrin-targeted therapies for inflammatory bowel disease. J Crohns Colitis. 2018;12:S653–S668. doi:10.1093/ecco-jcc/jjy060
3. Rice GP, Hartung HP, Calabresi PA. Anti-alpha4 integrin therapy for multiple sclerosis: mechanisms and rationale. Neurology. 2005;64:1336–1342. doi:10.1212/01.WNL.0000158329.30470.D0
4. Habtezion A, Nguyen LP, Hadeiba H, Butcher EC. Leukocyte trafficking to the small intestine and colon. Gastroenterology. 2016;150:340–354. doi:10.1053/j.gastro.2015.10.046
5. Sandborn WJ, Feagan BG, Rutgeerts P, et al. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med. 2013;369:711–721. doi:10.1056/NEJMoa1215739
6. Feagan BG, Rutgeerts P, Sands BE, et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2013;369:699–710. doi:10.1056/NEJMoa1215734
7. Kopylov U, Avni-Biron I, Ron Y, et al. Effectiveness and safety of vedolizumab for maintenance treatment in inflammatory bowel disease-The Israeli real world experience. Dig Liver Dis. 2019;51:68–74. doi:10.1016/j.dld.2018.07.040
8. Danese S, Sandborn WJ, Colombel J-F, et al. Endoscopic, radiologic, and histologic healing with vedolizumab in patients with active crohn’s disease. Gastroenterology. 2019;157:1007–1018 e1007. doi:10.1053/j.gastro.2019.06.038
9. Sandborn WJ, Colombel JF, Panaccione R, et al. Deep remission with vedolizumab in patients with moderately to severely active ulcerative colitis: a GEMINI 1 post hoc analysis. J Crohns Colitis. 2019;13:172–181. doi:10.1093/ecco-jcc/jjy149
10. Villablanca EJ, De Calisto J, Torregrosa Paredes P, et al. Beta7 integrins are required to give rise to intestinal mononuclear phagocytes with tolerogenic potential. Gut. 2014;63:1431–1440. doi:10.1136/gutjnl-2013-305386
11. Schippers A, Muschaweck M, Clahsen T, et al. Beta7-Integrin exacerbates experimental DSS-induced colitis in mice by directing inflammatory monocytes into the colon. Mucosal Immunol. 2016;9:527–538. doi:10.1038/mi.2015.82
12. Zeissig S, Rosati E, Dowds CM, et al. Vedolizumab is associated with changes in innate rather than adaptive immunity in patients with inflammatory bowel disease. Gut. 2019;68:25–39. doi:10.1136/gutjnl-2018-316023
13. Schleier L, Wiendl M, Heidbreder K, et al. Non-classical monocyte homing to the gut via alpha4beta7 integrin mediates macrophage-dependent intestinal wound healing. Gut. 2019. doi:10.1136/gutjnl-2018-316772
14. Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4:123–132. doi:10.1038/nri1292
15. Nielsen SCA, Boyd SD. Human adaptive immune receptor repertoire analysis-past, present, and future. Immunol Rev. 2018;284:9–23. doi:10.1111/imr.12667
16. Lossius A, Johansen JN, Vartdal F, Holmoy T. High-throughput sequencing of immune repertoires in multiple sclerosis. Ann Clin Transl Neurol. 2016;3:295–306. doi:10.1002/acn3.295
17. Henderson LA, Volpi S, Frugoni F, et al. Next-generation sequencing reveals restriction and clonotypic expansion of treg cells in juvenile idiopathic arthritis. Arthritis Rheum. 2016;68:1758–1768. doi:10.1002/art.39606
18. Harden JL, Hamm D, Gulati N, Lowes MA, Krueger JG. Deep sequencing of the T-cell receptor repertoire demonstrates polyclonal T-cell infiltrates in psoriasis. F1000Res. 2015;4:460. doi:10.12688/f1000research.6756.1
19. Chapman CG, Yamaguchi R, Tamura K, et al. Characterization of T-cell receptor repertoire in inflamed tissues of patients with crohn’s disease through deep sequencing. Inflamm Bowel Dis. 2016;22:1275–1285. doi:10.1097/MIB.0000000000000752
20. Doorenspleet ME, Westera L, Peters CP, et al. Profoundly expanded T-cell clones in the inflamed and uninflamed intestine of patients with crohn’s disease. J Crohns Colitis. 2017;11:831–839. doi:10.1093/ecco-jcc/jjx012
21. Allez M, Auzolle C, Ngollo M, et al. T cell clonal expansions in ileal Crohn’s disease are associated with smoking behaviour and postoperative recurrence. Gut. 2019;68:1961–1970. doi:10.1136/gutjnl-2018-317878
22. Gunaltay S, Repsilber D, Helenius G, et al. Oligoclonal T-cell receptor repertoire in colonic biopsies of patients with microscopic colitis and ulcerative colitis. Inflamm Bowel Dis. 2017;23:932–945. doi:10.1097/MIB.0000000000001127
23. Saravanarajan K, Douglas AR, Ismail MS, et al. Genomic profiling of intestinal T-cell receptor repertoires in inflammatory bowel disease. Genes Immun. 2020;21:109–118. doi:10.1038/s41435-020-0092-x
24. Werner L, Nunberg MY, Rechavi E, et al. Altered T cell receptor beta repertoire patterns in pediatric ulcerative colitis. Clin Exp Immunol. 2019;196:1–11. doi:10.1111/cei.13247
25. Rosati E, Pogorelyy MV, Dowds CM, et al. Identification of disease-associated traits and clonotypes in the T cell receptor repertoire of monozygotic twins affected by inflammatory bowel diseases. J Crohns Colitis. 2020. doi:10.1093/ecco-jcc/jjz210
26. Vermeire S, O’Byrne S, Keir M, et al. Etrolizumab as induction therapy for ulcerative colitis: a randomised, controlled, Phase 2 trial. Lancet. 2014;384:309–318. doi:10.1016/S0140-6736(14)60661-9
27. Sandborn WJ, Cyrille M, Hansen MB, et al. Efficacy and safety of abrilumab in a randomized, placebo-controlled trial for moderate-to-severe ulcerative colitis. Gastroenterology. 2019;156:946–957 e918. doi:10.1053/j.gastro.2018.11.035
28. Yoshimura N, Watanabe M, Motoya S, et al. Safety and efficacy of AJM300, an oral antagonist of alpha4 integrin, in induction therapy for patients with active ulcerative colitis. Gastroenterology. 2015;149:1775–1783 e1772. doi:10.1053/j.gastro.2015.08.044
29. Ungar B, Kopylov U, Yavzori M, et al. Association of vedolizumab level, anti-drug antibodies, and alpha4 beta7 occupancy with response in patients with inflammatory bowel diseases. Clin Gastroenterol Hepatol. 2018;16:697–705 e697. doi:10.1016/j.cgh.2017.11.050
30. Dotan I, Allez M, Danese S, et al. The role of integrins in the pathogenesis of inflammatory bowel disease: approved and investigational anti-integrin therapies. Med Res Rev. 2020;40:245–262. doi:10.1002/med.21601
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.