Back to Journals » International Journal of Nanomedicine » Volume 13
Biosynthesis of Au, Ag and Au–Ag bimetallic nanoparticles using protein extracts of Deinococcus radiodurans and evaluation of their cytotoxicity
Authors Li JL , Tian B, Li T, Dai S, Weng YL, Lu JJ, Xu XL, Jin Y, Pang RJ, Hua YJ
Received 26 October 2017
Accepted for publication 24 January 2018
Published 9 March 2018 Volume 2018:13 Pages 1411—1424
DOI https://doi.org/10.2147/IJN.S149079
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Linlin Sun
Jiulong Li,1 Bing Tian,1 Tao Li,1 Shang Dai,1 Yulan Weng,1 Jianjiang Lu,2 Xiaolin Xu,2 Ye Jin,1 Renjiang Pang,1 Yuejin Hua1
1Key Laboratory for Nuclear-Agricultural Sciences of Chinese Ministry of Agriculture and Zhejiang Province, Institute of Nuclear-Agricultural Sciences, Zhejiang University, Hangzhou, People’s Republic of China; 2Key Laboratory for Green Processing of Chemical Engineering of Xinjiang Bingtuan, School of Chemistry and Chemical Engineering, Shihezi University, Xinjiang, People’s Republic of China
Background: Biosynthesis of noble metallic nanoparticles (NPs) has attracted significant interest due to their environmental friendly and biocompatible properties.
Methods: In this study, we investigated syntheses of Au, Ag and Au–Ag bimetallic NPs using protein extracts of Deinococcus radiodurans, which demonstrated powerful metal-reducing ability. The obtained NPs were characterized and analyzed by various spectroscopy techniques.
Results: The D. radiodurans protein extract-mediated silver nanoparticles (Drp-AgNPs) were preferably monodispersed and stably distributed compared to D. radiodurans protein extract-mediated gold nanoparticles (Drp-AuNPs). Drp-AgNPs and Drp-AuNPs exhibited spherical morphology with average sizes of 37.13±5.97 nm and 51.72±7.38 nm and zeta potential values of -18.31±1.39 mV and -15.17±1.24 mV at pH 7, respectively. The release efficiencies of Drp-AuNPs and Drp-AgNPs measured at 24 h were 3.99% and 18.20%, respectively. During the synthesis process, Au(III) was reduced to Au(I) and further to Au(0) and Ag(I) was reduced to Ag(0) by interactions with the hydroxyl, amine, carboxyl, phospho or sulfhydryl groups of proteins and subsequently stabilized by these groups. Some characteristics of Drp-AuNPs were different from those of Drp-AgNPs, which could be attributed to the interaction of the NPs with different binding groups of proteins. The Drp-AgNPs could be further formed into Au–Ag bimetallic NPs via galvanic replacement reaction. Drp-AuNPs and Au–Ag bimetallic NPs showed low cytotoxicity against MCF-10A cells due to the lower level of intracellular reactive oxygen species (ROS) generation than that of Drp-AgNPs.
Conclusions: These results are crucial to understand the biosynthetic mechanism and properties of noble metallic NPs using the protein extracts of bacteria. The biocompatible Au or Au–Ag bimetallic NPs are applicable in biosensing, bioimaging and biomedicine.
Keywords: extreme bacterium, green synthesis, noble metallic nanoparticles, reactive oxygen species
Introduction
The materials composed of nanoparticles (NPs) produce fascinating and diverse properties including size-related electronic, optical, thermal and catalytic properties as a result of their exceptional size and surface area to volume ratio compared with their bulk counterparts.1–3 With the advancements in the field of nanotechnology, noble metallic NPs including gold nanoparticles (AuNPs), silver nanoparticles (AgNPs) or Au–Ag bimetallic NPs have demonstrated wide application potentials in antimicrobial agents,4,5 diagnosis,6 therapy,6,7 biosensing,8 drug delivery,9,10 and industrial catalysis.11 It is vital to prepare nontoxic and safe NPs to use in these applications.12
Green synthesis of metal NPs deserves emerging and inestimable merit due to its ecofriendly, energy-efficient, sustainable and biocompatible potentials. Biosynthesis of NPs using biomolecules from microorganisms has been proposed because the microorganisms are easily available and cultured and as well there is effective production of them on a large scale without seasonal and geographic effects, compared to other organisms including plants.13–15 The biomolecules such as proteins and DNA can be used as reactants, templates and capping agents in the synthesis of nanomaterials.2,16,17 The synthesized NPs are bioconjugated to peptides, proteins or DNA, which endows the NPs with additional bioactive properties.2 For example, the bacterial extract of Marinobacter lipolyticus that produced lipase could be used to synthesize the silver nanomaterials for anti-candidal activities.18 In particular, the use of proteins or peptides as the reactants of noble metals has received extraordinary attention as they have distinct recognition, reduction and adsorption abilities of the metal precursors or NPs and provide a reservoir for the exploration of NP fabrication.2,19–22 Protein extracts of fungus have been recently applied to synthesize AuNPs or AgNPs.23,24 However, the biosynthetic mechanism of metal NPs using proteins from bacteria has been rarely investigated.25,26
Deinococcus radiodurans, which is a nonpathogenic bacterium known for its extreme resistance to radiation and oxidants and which has numerous reducing metabolites, was used for transformation or detoxification of heavy metals under in situ oxidative stresses.27–29 Recently, the synthesis of AgNPs and AuNPs via the cultures of D. radiodurans with silver and gold ions was investigated.30,31 Moreover, surface layer protein lattices of D. radiodurans exhibited a biotemplating effect on the preformed AuNPs, into ordered arrays.32 The intracellular proteins containing metal capturing and reducing groups might provide a reducing microenvironment for the formation of metal NPs. In the previous work,31 we proposed the involvement of proteins in the biosynthesis of AuNPs by the cultures of D. radiodurans incubated with HAuCl4 solution. To date, the biosynthetic mechanism and evaluation of cytotoxicity of Au, Ag or Au–Ag bimetallic NPs synthesized using protein extracts of D. radiodurans remains unexplored.
Herein, we report a facile method for biosynthesis of Au, Ag and Au–Ag bimetallic NPs using the protein extracts of D. radiodurans as a reductant and capping agent in aqueous solution without any external energy. The characteristics and comparison of D. radiodurans protein extract-mediated gold nanoparticles (Drp-AuNPs) and D. radiodurans protein extract-mediated silver nanoparticles (Drp-AgNPs) were demonstrated using ultraviolet and visible (UV/Vis) absorption spectroscopy, electron microscopy, X-ray diffraction (XRD) and dynamic light scattering (DLS). The underlying mechanisms and differences in the formation of Drp-AuNPs and Drp-AgNPs were investigated using Fourier-transform infrared spectroscopy (FTIR) and high-performance X-ray photoelectron spectroscopy (XPS). The biosynthetic Au–Ag bimetallic NPs formed on the as-synthesized Drp-AgNPs with addition of Au(III) were characterized. The cytotoxicity of these metallic NPs was determined using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt (MTS) assay and reactive oxygen species (ROS) assay.
Materials and methods
Bacterial cultures and chemicals
D. radiodurans (ATCC13939) was cultured aerobically in an orbital shaker at 220 rpm and 30°C in the tryptone glucose yeast (TGY) medium (0.5% tryptone, 0.1% glucose, 0.3% yeast extract, w/v). Chloroauric acid (HAuCl4·3H2O) and silver nitrate (AgNO3) were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Ultrapure water with Milli-Q grade (18.25 MΩ) was used for solution preparations and washing procedures. All the reagents used in this study were of analytical grade. The Au(III) and Ag(I) solutions used were prepared by dissolving HAuCl4·3H2O and AgNO3 in ultrapure water, respectively. The pH of the working solution was tuned by nitric acid and/or sodium hydroxide.
Preparation of protein extracts
D. radiodurans cells (OD600 nm=1.0) were harvested by centrifugation at 8,000× g for 10 min and then thoroughly washed with phosphate buffer solution (0.01 M, pH 7.2). Protein extracts were prepared by sonication and centrifugation at 15,000× g for 30 min at 4°C to remove cell debris. The supernatant was collected and further treated with a final concentration of 80% (w/v) solid ammonium sulfate by gently stirring for 24 h at 4°C.33 The obtained protein precipitate was collected by centrifugation at 10,000× g for 20 min at 4°C and then dissolved in water and dialyzed by using a cellulose acetate membrane (molecular weight [MW] cutoff 14,000 Da) with stirring overnight at 4°C. Following dialysis, the protein extracts including proteins from the cell envelope and cytoplasm were collected for the synthesis of NPs. Protein concentration was quantitated using the BCA Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA).
Synthesis of Drp-AuNPs and Drp-AgNPs
For the synthesis of NPs, 2 mg/mL protein extracts were reacted with a final concentration of 1 mM gold or silver ion solution at ambient conditions (pH 7, 25°C).34 The formation of AuNPs or AgNPs in the suspension was monitored by the color changes of the reaction mixtures and measured by absorption spectrum using a UV/Vis absorption spectrometer (SpectraMax M5; Molecular Devices LLC, Sunnyvale, CA, USA).35 Appropriate controls such as protein extracts, 1 mM Au(III) or Ag(I), were measured for comparison purposes. To remove the unreacted molecules or ions, the NPs were centrifuged, washed and dialyzed repeatedly as described earlier. Then, the protein-synthesized NPs were filtered using 0.22 μm syringe filters and lyophilized.
Characterization of Drp-AuNPs and Drp-AgNPs
The scanning electron microscope (SEM; Hitachi Model SU8010; Hitachi Ltd., Tokyo, Japan) and transmission electron microscope (TEM, Hitachi Model H-7650) were used to identify the morphology, size and distribution of the synthesized NPs as described previously.31,33 The particle size of NPs was measured using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The energy-dispersive X-ray spectroscopy (EDS) was recorded by focusing the electron beam onto a region of the sample surface to perform elemental analysis of the test samples in an area scan mode. To measure the XRD pattern of prepared NPs, a X-ray diffractometer (X’Pert PRO; PANalytical, Almelo, the Netherlands) was used with Cu Kα1 radiation of wavelength λ=1.540 Å. The scanning was performed in the region of the 2θ angle from 20° to 90° with a step of 0.02° and a 2-s time constant for each step. The particle size and zeta potential of NPs were measured using a laser Doppler anemometry (Zetasizer Nano ZS; Malvern Instruments, Malvern, UK) with a wavelength of 632.8 nm and an He–Ne laser beam at 25°C. An electric field of 150 mV was applied to monitor the electrophoretic velocity of the particles.31 Three independent samples were run at the same time, and the representative data were presented.
For FTIR analysis, the purified NPs were crushed with KBr in a mortar at the ratio of 1:100. The pressed pellet was covered with a clip and immediately analyzed by using Nicolet 5700 FTIR spectrometer (Thermo Fisher Scientific) over the 4,000–400 cm−1 regions at a resolution of 2 cm−1 over 1,800 scans.
The prepared NPs were mounted on a stainless steel holder, and an XPS analysis was performed by the high-performance X-ray photoelectron spectrometer (Escalab 250Xi; Thermo Fisher Scientific) using monochromatic Al·Kα radiation of energy 1,486.6 eV. In order to determine core-level binding energies, the C1s peak at 284.8 eV was used as the charge reference, and XPS spectra were analyzed by XPSPEAK41 software.
To investigate the production of noble metal NPs using pure intracellular protein, we selected the CrtI (DR0861) and the Dps2 (DRB0092) to incubate with Au(III) or Ag(I). These proteins were expressed and purified in vitro as described previously.36,37 A total of 100 μL of 1 mg/mL of the respective proteins were incubated with 1 mM Au(III) or Ag(I) solution at ambient conditions. The production of NPs was monitored for the color and the absorption spectrum changes of the suspension. The capping agents on the washed NPs were separated by boiling with 1% sodium dodecyl sulfate (SDS) solution for 10 min, followed by centrifugation at 8,000× g for 10 min, and then, the supernatants were collected. The samples were further analyzed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).23,24
Synthesis and confirmation of Au–Ag bimetallic NPs
A total of 1 mM Au(III) ion solution was added into the prepared aqueous solution of Drp-AgNPs at pH 7 and 25°C. Then, the suspensions at incubation times of 0, 1 and 60 min were monitored for their color changes and measured by UV/Vis absorption spectroscopy. Subsequently, the synthesized NPs were analyzed using the SEM–EDS.
Release kinetics of NPs
The metal release from NPs was determined as previously described.4 Briefly, the NPs were placed in a dialysis bag that was suspended in an ultrapure water solution (40 mL). Then, a continuous release of the NPs was measured over a period of 48 h using inductively coupled plasma-optical emission spectroscopy (ICP-OES; Optima 8000DV; PerkinElmer Inc., Waltham, MA, USA). The release rate was calculated using the equation: % of cumulative metal released = (Mw/Md) ×100, where Mw is the metal content in the water and Md is the total metal content in the dialysis bag.
Cytotoxicity assays
The human breast epithelial cell line MCF-10A purchased commercially from CTCC Bioscience Inc. (Wuxi, China) was utilized in the MTS assay to examine the in vitro cytotoxic effect of prepared NPs. MCF-10A cells were cultured in the growth medium consisting of high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 mg/mL of streptomycin under a humidified environment (37°C, 5% CO2 and 95% O2). The cultured cells at a density of 5×103 cells/well were seeded into a 96-well plate and exposed to 0, 2.5, 5, 12.5 and 25 μg/mL NPs. All the mixtures were incubated under identical conditions for 24 h. Then, the collected cells were incubated with the addition of 20% of MTS for another 2 h. The absorbance of the cell suspensions was measured at 490 nm using the microplate spectrophotometer (SpectraMax M5). The untreated cells were considered as 100% viable.
Intracellular ROS measurement
The dye CM-H2DCFDA was used as ROS probe to evaluate the intracellular ROS generation.38 MCF-10A cells were treated with or without 25 μg/mL NPs for 24 h. The samples were then washed twice with PBS and incubated with CM-H2DCFDA at 37°C for 30 min in the dark. Then, the cells were washed twice with PBS. The fluorescence intensity was monitored as the rate of oxidation of the dye in the cells, and untreated cells were used as a standard to calculate the level of ROS generation.
Statistical analysis
The data were processed by OriginPro 8.0 (OriginLab Co., Northampton, MA, USA) and expressed as mean ± standard deviation. The analysis of variance was followed by Student’s t-test. P-value <0.05 was considered as statistically significant.
Results and discussion
Biosynthesis of Drp–AuNPs and Drp–AgNPs
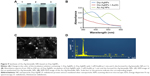
The protein extracts from the whole cell of D. radiodurans were used to investigate the biosynthesis of noble metal NPs. The color changes in reaction mixtures and formations of the characteristic absorption peaks were used to indicate the synthesis of Drp-AuNPs or Drp-AgNPs through the reduction of Au(III) or Ag(I) by protein extracts of D. radiodurans. As the biosynthesis proceeded, the color of suspensions containing protein extracts and Au(III) changed to purple at 6 h (Figure 1A). The absorption spectra of the suspensions exhibited a characteristic absorption peak at 545 nm (Figure 1B), owing to the surface plasmon resonance (SPR) of AuNPs.31 As for the formation of the Drp-AgNPs from the reaction of Ag(I) and protein extracts, a stable brown color together with the significant characteristic SPR absorption at 414 nm was observed within ~24 h (Figure 1C and D). The controls (metal ion solution or protein extracts alone) had neither corresponding color changes nor characteristic absorptions, indicating that the protein extracts were involved in the formations of NPs. UV/Vis kinetics for the formation of NPs exhibited that the characteristic absorbance at 545 nm for Drp-AuNPs increased with reaction time and remained unchanged from 6 to 24 h (Figure S1A) and the characteristic absorbance at 414 nm for Drp-AgNPs remained unchanged from 24 to 48 h (Figure S1B), indicating that Drp-AuNP and Drp-AgNP biosynthetic processes reached equilibrium by 6 h and 24 h, respectively. Moreover, the synthesized Drp-AuNPs and Drp-AgNPs exhibited no significant changes in color or characteristic absorption peak at room temperature within 3 months, indicating the relative stability of these NPs. The proteins could be used as a stabilizing agent during the biosynthetic process.
Characterization of Drp-AuNPs and Drp-AgNPs
As shown in the TEM image (Figure 2A), the Drp-AuNPs were nearly monodispersed with spherical morphology, while some of them were in agglomeration which might be ascribed to the self-assembly of protein-adsorbed Drp-AuNPs.15,39,40 In the SEM image, the accumulated Drp-AuNPs were also observed (Figure 2B). The average size of Drp-AuNPs was 51.72±7.38 nm. The as-prepared Drp-AuNPs were confirmed by a model of area profile analysis of EDS (Figure 2C–E), which revealed the presence of characteristic peaks for Au together with trace peaks for C, N, O, P and S. The strong peak at ~2.1 keV due to the characteristic SPR of AuNPs corresponded with the formation of Drp-AuNPs.41 The signals of C, O, S and P in Figure 2C could have arisen from the protein extracts. Figure 2D–E demonstrates the overlapped distribution of N with that of Au in the Drp-AuNPs, indicating that some proteins bound on the surface of NPs as a capping and stabilizing agent.
For the Drp-AgNPs, the NPs with spherical morphology were nearly monodispersed without agglomeration (Figure 3A and B). The average size of Drp-AgNPs was 37.13±5.97 nm. The analysis of as-prepared Drp-AgNPs using EDS revealed the presence of a characteristic peak for Ag and trace peaks for C, N, O, P and S (Figure 3C). The intensive peak near 3.0 keV corresponded with the formation of Drp-AgNPs according to the characteristic SPR of AgNPs.42 Figure 3D–E shows the distribution of N with that of Ag in the Drp-AgNPs, suggesting that some proteins could bind to the surface of NPs as a capping and stabilizing agent. The elemental distribution of Ag (Figure 3D) was more uniform compared to Au (Figure 2D).
The XRD analysis provided information about crystalline structures of the Drp-AuNPs and Drp-AgNPs. In Figure 4A, the XRD spectra show distinct diffraction peaks at 2θ of 38.12°, 44.32°, 64.54°, 77.54° and 81.64°, indexing the [1 1 1], [2 0 0], [2 2 0], [3 1 1] and [2 2 2] planes, respectively, of the face-centered cubic Au structure. The strong diffraction peak at 38.12° was attributed to the main reflection of X-rays from the [1 1 1] crystal plane of Drp-AuNPs. Considering the [1 1 1] peak, the average crystallite size was estimated to be 56.84 nm using the Debye–Scherrer equation as reported.31 In the XRD spectra of Drp-AgNPs (Figure 4B), the characteristic diffraction peaks at 2θ values of 32.20°, 37.93°, 46.24°, 66.51° and 76.72°, respectively, corresponded to the [1 0 1], [1 1 1], [2 0 0], [2 2 0] and [3 1 1] planes of the face-centered cubic crystal structure, demonstrating that the synthesized Drp-AgNPs were crystalline in nature (JCPDS file nos 84-0713 and 04-0783).43,44 The unidentified peaks at 27.78°, 54.86° and 57.42° could be ascribed to the protein residues used for Drp-AgNPs formation, which played a role in stabilizing the NPs.43
The average hydrodynamic diameter of the Drp-AuNPs at the incubation time of 6 h was found to be 94.63±1.97 nm with a polydispersity index (PDI) of 0.19±0.04, while the value of Drp-AgNPs at the incubation time of 24 h was 78.00±1.65 nm with a near-monodispersed particle size distribution (PSD) of 0.11±0.03, as measured using DLS (Figure 4C and D). The differences in size between TEM and DLS could be due to proteins that acted as a capping agent and formed a covering on the surface of NPs. The zeta potential value of Drp-AuNPs was −15.17±1.24 mV at pH=7, suggesting that electrostatic repulsion occurred between the Drp-AuNPs and facilitated the relative stability of these biosynthesized NPs.27 The zeta potential value of Drp-AgNPs was −18.31±1.39 mV, indicating that the Drp-AgNPs were more stable compared to that of Drp-AuNPs.
Synthetic mechanisms of Drp–AuNPs and Drp–AgNPs
The functional groups of protein(s) involved in the reduction of metal ions and formation of NPs were investigated by FTIR spectroscopy (Figure 5). The intense and distinct absorption bands of protein extracts at 3,381 and 860 cm−1 could be referred to the stretching and out-of-plane bending vibration of O−H and/or N−H groups, respectively. On the formation of Drp-AuNPs and Drp-AgNPs, the peaks shifted to 3,419 and 880 cm−1, respectively, and the band at 1,992 cm−1 (NH3+ stretching vibration) disappeared. The N−H and O−H groups were involved in the formation of NPs. The bands at 1,656 and 1,544 cm−1 corresponded to the amide I and II bands of the proteins.45 The amide I band shifted to 1,645 cm−1, while the amide II band was not obviously changed. Intriguingly, a shifted band for the amide III at 1,236 cm−1 appeared when the NPs were synthesized. These results indicated that amino groups of protein extracts contributed to synthesizing and capping NPs. The band at 1,387 cm−1 was assigned to the COO− symmetric stretch of proteins, which was with carboxyl side groups in the amino acid residues. It shifted to 1,404 cm−1 on completion of the reduction of Au(III), but did not change in the reduction of Ag(I), indicating the special interaction of carboxylate groups with gold ions. The band and out-of-plane deformation vibration of COOH group at 947 cm−1 disappeared on the formation of NPs, which was attributed to the anisotropic growth of NPs with the help of carboxyl groups.46 The bands at 1,153 and 1,072 cm−1 corresponded to P−OH and P−O−C stretching, respectively, suggesting the presence of phosphate group in the proteins.23 The band at 1,153 cm−1 changed to 1,149 and 1,120 cm−1 in the syntheses of Drp-AuNPs and Drp-AgNPs, respectively. Shifting of the P−O−C band to 1,085, 1,047 and 1,045 cm−1 suggested the interaction of phosphoprotein with the Drp-AuNPs and Drp-AgNPs, respectively. These results clearly indicated that hydroxyl, amine, carboxyl and phospho groups of proteins could participate in the reduction of metal ions and subsequent stabilization of Drp-AuNPs and Drp-AgNPs, and the carboxyl groups from proteins might be responsible for the growth of Drp-AgNPs.46
The speciation of Au or Ag in the reducing process was studied using the XPS technique. As shown in Figure 6A and B, the presence of C1s, O1s, N1s, P2p, S2p and Au4f core levels was detected in the wide scan spectrum of Drp-AuNPs, and peaks of C1s, O1s, N1s, P2p, S2p and Ag3d were found in the spectrum of Drp-AgNPs. The core-level spectra of C1s, O1s, N1s, P2p and S2p were also present in the monomer protein extracts (Figure S2A). These results indicated that the protein functional groups, for example, carboxyl, amine, hydroxyl and phospho groups, could provide reducing and capping sites for formation of NPs, which is consistent with the results of the FTIR assay. The presence of S2p was ascribed to the cysteines or reduced glutathione of proteins binding to the surface of NPs.47,48 The Au4f spectrum in Figure 6C with doublet peaks corresponding to Au4f7/2 and Au4f5/2, at 83.7 and 87.4 eV could be ascribed to Au(0). The shift of ~0.8 eV in the binding energy of the Au4f core level compared with the reported binding energy of the Au4f core level spectrum could be due to protein-capping effects on AuNPs, which is consistent with the previous report.49 The distinct asymmetry in the peaks at binding energies of 84.6 and 88.2 eV could be attributed to Au(I), indicating that the AuNPs were formed through intermediate Au(I) species. Au(I) was usually present on the surface of AuNPs coated with protein.50 Figure 6D shows the Ag3d spectrum of Drp-AgNPs, which could be attributed to Ag3d5/2 and Ag3d3/2, binding energies at ~367.6 and ~373.6 eV, respectively.51 The peaks exhibited an apparent shift of ~0.7 eV in the binding energy in comparison to the reported core level of Ag3d spectrum, which was probably due to the interactions between AgNPs and proteins.44
Figure 7A shows that the carboxylate (O=C−OH) and amide carbon (O=C−N) peaks of the Drp-AuNPs appeared at 289.2 eV, corresponding to that peak of protein extracts at 288.7 eV (Figure S2B). The N1s core level centered at 399.6 and 401.3 eV corresponded to amine and amide groups of the Drp-AuNPs, respectively (Figure 7B), but the peak at 402.5 eV for the protonation of the amino group of the protein extracts as shown in Figure S2C was not detected. The binding energy peaks for the carboxylate group of proteins at 289.2 eV and the amine group of proteins at 401.3 eV were not detected following reaction with Ag(I) ions (Figure 7D and E), suggesting that the synthesis of Drp-AgNPs was different from that of Drp-AuNPs. The O1s core levels of Drp-AgNPs and Drp-AuNPs that centered at 532.1 eV had a 0.3 eV higher energy compared to that of proteins (Figures 7C and F and S2D). The shifted or changed peaks of C1s, N1s and O1s suggested that conjugation of proteins with NPs occurred in the reducing process.
The FTIR and XPS results demonstrated that gold and silver ions could initially bind to proteins, and then, the Au(III) was reduced to Au(I) and further to Au(0); the Ag(I) was reduced to Ag(0) through the hydroxyl, amine, carboxyl, phospho and sulfhydryl groups of the proteins from D. radiodurans, subsequently forming the Drp-AuNPs and Drp-AgNPs. Binding of Drp-AuNPs and Drp-AgNPs with the proteins led to the formation of relatively stable conjugates.48 The conjugation process could be used to facilitate the post-synthetic surface modification with functional ligands.52 The different characteristics of Drp-AuNPs and Drp-AgNPs could be attributed to the interaction of the NPs with different binding groups, including amide and carboxylate groups from proteins.
Synthesis of AuNPs and AgNPs by selective proteins
Our previous results demonstrated the probable presence of proteins of D. radiodurans on the surface of AuNPs.31 We separated the proteins bound to AuNPs by SDS-PAGE. Two protein bands (~55 and ~35 kDa) were observed (Figure S3), confirming the presence of proteins on the surface of AuNPs synthesized by D. radiodurans. The intensive gel slice of ~35 kDa was collected and identified by mass spectrometry (Table S1). A total of 26 abundant proteins including S-layer proteins were identified on the AuNPs’ surface.
Here, we wonder if any intracellular protein has the biosynthetic ability of AuNPs and AgNPs. We selected the CrtI and the Dps2 to investigate the different roles of proteins in synthesizing AuNPs and AgNPs. The purified CrtI was able to synthesize both AuNPs and AgNPs (Figure 8A and B). The CrtI (~60 kDa) was eluted from the AuNPs and AgNPs and identified by SDS-PAGE analysis (Figure S4A), suggesting that the CrtI could be used as the reductant and capping agents for these NPs. However, the Dps2 could biosynthesize AgNPs rather than AuNPs (Figures 8C and D and S4B). The different roles of the Dps2 in reducing Au(III) and Ag(I) might be due to the following: first, the Dps2, which functions as a sequester of intracellular Fe2+ ion and stores iron ions in the form of Fe3+,53 could not adsorb or interact with the trivalent Au ions; however, it did reduce the monovalent Ag ions; second, Dps2 only contains three tryptophan and six tyrosine molecules that have the ability of donating electrons from their NH or OH groups (Table S2)54,55 and thus could not have enough potential to synthesize the AuNPs, considering the relative higher reduction potential of gold (EθAu(III)/Au(0)=1.50 V) than that of silver (EθAg(I)/Ag(0)=0.799 V).56,57 Thus, the metal NP’s biosynthetic ability with different proteins might be determined by their composition or reducing capability.
Biogenerated noble metal NPs of different organisms might have different properties due to the distinct composition of biomolecules between species. NPs based on some green agents like polysaccharides could be a safe and efficient nonviral gene vector to human cells.58,59 The use of proteins from microorganisms as the reactants and capping agents of noble metals could have some advantages or benefits over other green agents like polysaccharides. First, the noble metal strongly binds to specific amino acids, amine and other metal groups and these complexes are often stronger than with polysaccharide groups. This characteristic could contribute to the small release of the metal from the Drp-NPs.60 Second, the controlled synthetic process of NPs could be rationally realized because the adsorption ability of proteins composed of a wide selection of amino acids is adjustable. The synthesis of NPs by proteins can be performed at room temperature in aqueous solution; however, the polysaccharide-based synthesis of NPs needs a high temperature to increase the solubility of polysaccharides, which could lead to the reduction of metal precursors prior to complexation between polysaccharides and precursors.2
Synthesis and confirmation of Au–Ag bimetallic NPs
Following the incubation of Drp-AgNPs with 1 mM Au(III), the color of the Drp-AgNPs’ aqueous solution changed from yellow to purple within 1 h, together with a shift in the characteristic absorption peak from 414 to 550 nm (Figure 9A and B), indicating that the Au–Ag bimetallic NPs were formed. The formation of Au–Ag bimetallic NPs with a spherical morphology was also confirmed by the SEM–EDS, which revealed the presence of peaks for Au and Ag as well as the trace peaks for C, N, O, P and S of proteins bound to the NPs (Figure 9C and D). The synthesis could be attributed to the galvanic replacement reaction via the oxidation of Drp-AgNPs by Au(III) ions.26 The rapid reaction time compared to that of Drp-AuNPs (6 h) could be ascribed to the presence of proteins coating on the surface of Drp-AgNPs. The formation of Au–Ag bimetallic NPs using protein extracts of D. radiodurans provided a new way to achieve large-scale biosynthesis of noble bimetallic NPs.
Cytotoxicity analyses of synthesized NPs
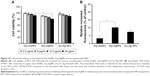
Metal release of synthesized NPs was measured over the period of the study (48 h). In all, 18.20% and 3.99% release of Drp-AgNPs and Drp-AuNPs was detected at 24 h incubation, respectively (Figure S5). For the Au–Ag bimetallic NPs, 3.51% Au release and 12.99% Ag release were determined at 24 h, respectively. The release of Drp-AgNPs was lower than the previously reported AgNPs (~25%).61 The low release of Drp-AgNPs could be attributed to the strong binding affinity of AgNPs to proteins and this resulted in the formation of large NPs.60
MTS assay using the human breast epithelial cell line MCF-10A was utilized to examine the cytotoxic effect of prepared NPs. Figure 10A shows that cell viability of MCF-10A decreased with the increase in the concentration of Drp-AuNPs, Drp-AgNPs and Au–Ag bimetallic NPs. At the concentration of 25 μg/mL, the cell viabilities following the treatment of Drp-AuNPs, Drp-AgNPs and Au–Ag bimetallic NPs were 91.02%, 79.39% and 88.81%, respectively. In comparison, the Drp-AuNPs and Au–Ag bimetallic NPs had a lower inhibitory effect on the cells than the Drp-AgNPs at a higher concentration of NPs. This result was supported by the analysis of intracellular ROS generation upon exposure of the NPs to the cells (Figure 10B). The slight cytotoxicity of Drp-AgNPs could be attributed to the release of Ag and more ROS generation.38,62 The NPs synthesized by proteins were generally assured to be biocompatible due to the presence of the stabilizing agent.12 Therefore, the Drp-AuNPs and Au–Ag bimetallic NPs with cell viability much beyond 80% in normal human cells could have advantageous applications in biosensors, bioimaging, drug delivery and nanomedicine. Further production and application of the Drp-NPs will be performed in our future research.
Conclusion
We demonstrated the facile biosynthesis of AuNPs, AgNPs and Au–Ag bimetallic NPs using protein extracts of D. radiodurans in an aqueous solution at ambient conditions. The hydroxyl, amine, carboxyl, phospho or sulfhydryl groups from proteins played a vital role in the reduction of Au(III) to Au(I) and further to Au(0), and Ag(I) to Ag(0), as well as the stabilization of the formed NPs. The synthetic Drp-AuNPs and Drp-AgNPs exhibited different characteristics, which could be ascribed to the interaction of the NPs with different binding groups of proteins. The Au–Ag bimetallic NPs could be biosynthesized based on the as-synthesized Drp-AgNPs with the addition of Au(III). The Drp-AuNPs and Au–Ag bimetallic NPs demonstrated relatively low cytotoxicity against MCF-10A compared to the Drp-AgNPs. Understanding the characteristics and biosynthetic mechanisms of noble metallic NPs mediated by D. radiodurans proteins highlights the wide and advantageous applicability of the NPs to future “nanofactories” and biomedicine.
Acknowledgment
This work was supported by grants from the National Natural Science Foundation of China (31670083, 31370119, 31210103904 and 31570058).
Disclosure
The authors report no conflicts of interest in this work.
References
Ling D, Hackett MJ, Hyeon T. Surface ligands in synthesis, modification, assembly and biomedical applications of nanoparticles. Nano Today. 2014;9(4):457–477. | ||
Duan H, Wang D, Li Y. Green chemistry for nanoparticle synthesis. Chem Soc Rev. 2015;44(16):5778–5792. | ||
Dickerson MB, Sandhage KH, Naik RR. Protein- and peptide-directed syntheses of inorganic materials. Chem Rev. 2008;108(11):4935–4978. | ||
Oves M, Khan MS, Zaidi A, et al. Antibacterial and cytotoxic efficacy of extracellular silver nanoparticles biofabricated from chromium reducing novel OS4 strain of Stenotrophomonas maltophilia. PLoS One. 2013;8(3):e59140. | ||
Qayyum S, Oves M, Khan AU. Obliteration of bacterial growth and biofilm through ROS generation by facilely synthesized green silver nanoparticles. PLoS One. 2017;12(8):e01813638. | ||
Llevot A, Astruc D. Applications of vectorized gold nanoparticles to the diagnosis and therapy of cancer. Chem Soc Rev. 2012;41(1):242–257. | ||
Zhou W, Gao X, Liu D, Chen X. Gold nanoparticles for in vitro diagnostics. Chem Rev. 2015;115(19):10575–10636. | ||
You CC, Miranda OR, Gider B, et al. Detection and identification of proteins using nanoparticle-fluorescent polymer ‘chemical nose’ sensors. Nat Nanotechnol. 2007;2(5):318–323. | ||
Wang F, Wang YC, Dou S, Xiong MH, Sun TM, Wang J. Doxorubicin-tethered responsive gold nanoparticles facilitate intracellular drug delivery for overcoming multidrug resistance in cancer cells. ACS Nano. 2011;5(5):3679–3692. | ||
Tiwari PM, Vig K, Dennis VA, Singh SR. Functionalized gold nanoparticles and their biomedical applications. Nanomaterials (Basel). 2011;1(1):31–63. | ||
Heddle JG. Gold nanoparticle-biological molecule interactions and catalysis. Catalysts. 2013;3(3):683–708. | ||
Ansari MA, Shukla AK, Oves M, Khan HM. Electron microscopic ultrastructural study on the toxicological effects of AgNPs on the liver, kidney and spleen tissues of albino mice. Environ Toxicol Phar. 2016;44:30–43. | ||
Gwynne P. There’s gold in them there bugs. Nature. 2013;495(7440):S12–S13. | ||
Kerr RA. Microbiology – bacteria help grow gold nuggets from dirt. Science. 2006;313(5784):159. | ||
Iravani S. Green synthesis of metal nanoparticles using plants. Green Chem. 2011;13(10):2638–2650. | ||
Berti L, Burley GA. Nucleic acid and nucleotide-mediated synthesis of inorganic nanoparticles. Nat Nanotechnol. 2008;3(2):81–87. | ||
Tan YN, Lee JY, Wang DI. Uncovering the design rules for peptide synthesis of metal nanoparticles. J Am Chem Soc. 2010;132(16):5677–5686. | ||
Oves M, Qari HA, Felemban NM, Khan MZ, Rehan ZA, Ismail IMI. Marinobacter lipolyticus from Red Sea for lipase production and modulation of silver nanomaterials for anti-candidal activities. IET Nanobiotechnol. 2017;11(4):403–410. | ||
Boyer C, Bulmus V, Liu J, Davis TP, And MHS, Barnerkowollik C. Well-defined protein-polymer conjugates via in situ raft polymerization. J Am Chem Soc. 2007;129(22):7145–7154. | ||
Fach M, Radi L, Wich PR. Nanoparticle assembly of surface-modified proteins. J Am Chem Soc. 2016;138(45):14820–14823. | ||
Daniel MC, Tsvetkova IB, Quinkert ZT, et al. Role of surface charge density in nanoparticle-templated assembly of bromovirus protein cages. ACS Nano. 2010;4(7):3853–3860. | ||
Naik RR, Stringer SJ, Agarwal G, Jones SE, Stone MO. Biomimetic synthesis and patterning of silver nanoparticles. Nat Mater. 2002;1(3):169–172. | ||
Das SK, Dickinson C, Lafir F, Brougham DF, Marsili E. Synthesis, characterization and catalytic activity of gold nanoparticles biosynthesized with Rhizopus oryzae protein extract. Green Chem. 2012;14(5):1322–1334. | ||
Jain N, Bhargava A, Majumdar S, Tarafdar JC, Panwar J. Extracellular biosynthesis and characterization of silver nanoparticles using Aspergillus flavusNJP08: a mechanism perspective. Nanoscale. 2011;3(2):635–641. | ||
Korbekandi H, Iravani S, Abbasi S. Production of nanoparticles using organisms. Crit Rev Biotechnol. 2009;29(4):279–306. | ||
Jang H, Kim Y, Huh H, Min D. Facile synthesis and intraparticle self-catalytic oxidation of dextran-coated hollow Au–Ag nanoshell and its application for chemo-thermotherapy. ACS Nano. 2014;8(1):467–475. | ||
Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev. 2011;75(1):133–191. | ||
Daly MJ. Engineering radiation-resistant bacteria for environmental biotechnology. Curr Opin Biotech. 2000;11(3):280–285. | ||
Cox MM, Battista JR. Deinococcus radiodurans – the consummate survivor. Nat Rev Microbiol. 2005;3(11):882–892. | ||
Kulkarni RR, Shaiwale NS, Deobagkar DN, Deobagkar DD. Synthesis and extracellular accumulation of silver nanoparticles by employing radiation-resistant Deinococcus radiodurans, their characterization, and determination of bioactivity. Int J Nanomedicine. 2015;10:963–974. | ||
Li J, Li Q, Ma X, et al. Biosynthesis of gold nanoparticles by the extreme bacterium Deinococcus radiodurans and an evaluation of their antibacterial properties. Int J Nanomedicine. 2016;11:5931–5944. | ||
Hall SR, Shenton W, Engelhardt H, Mann S. Site-specific organization of gold nanoparticles by biomolecular templating. Chemphyschem. 2001;2(3):184–186. | ||
Das SK, Liang J, Schmidt M, Laffir F, Marsili E. Biomineralization mechanism of gold by zygomycete fungi Rhizopus oryzae. ACS Nano. 2012;6(7):6165–6173. | ||
Yang J, Sargent EH, Kelley SO, Ying JY. A general phase-transfer protocol for metal ions and its application in nanocrystal synthesis. Nat Mater. 2009;8(8):683–689. | ||
Albanese A, Chan WC. Effect of gold nanoparticle aggregation on cell uptake and toxicity. ACS Nano. 2011;5(7):5478–5489. | ||
Xu Z, Tian B, Sun Z, Lin J, Hua Y. Identification and functional analysis of a phytoene desaturase gene from the extremely radioresistant bacterium Deinococcus radiodurans. Microbiology. 2007;153(5):1642–1652. | ||
Cuypers MG, Mitchell EP, Romão CV, Mcsweeney SM. The crystal structure of the Dps2 from Deinococcus radiodurans reveals an unusual pore profile with a non-specific metal binding site. J Mol Biol. 2007;371(3):787–799. | ||
AshaRani PV, Mun GLK, Hande MP, Valiyaveettil S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano. 2009;3(2):279–290. | ||
Zhang D, Neumann O, Wang H, et al. Gold nanoparticles can induce the formation of protein-based aggregates at physiological pH. Nano Lett. 2009;9(2):666–671. | ||
Feliu N, Docter D, Heine M, et al. In vivo degeneration and the fate of inorganic nanoparticles. Chem Soc Rev. 2010;4(1):15–18. | ||
Das SK, Das AR, Guha AK. Microbial synthesis of multishaped gold nanostructures. Small. 2010;6(9):1012–1021. | ||
Kumar A, Vemula PK, Ajayan PM, John G. Silver-nanoparticle-embedded antimicrobial paints based on vegetable oil. Nat Mater. 2008;7(3):236–241. | ||
Wen L, Zeng P, Zhang L, Huang W, Wang H, Chen G. Symbiosis theory-directed green synthesis of silver nanoparticles and their application in infected wound healing. Int J Nanomedicine. 2016;11:2757–2767. | ||
Luo Y, Shen S, Luo J, Wang X, Sun R. Green synthesis of silver nanoparticles in xylan solution via Tollens reaction and their detection for Hg(2+). Nanoscale. 2015;7(2):690–700. | ||
Steffen’s Homepage [webpage on the Internet]. Spectroscopic Tools. Available from: http://www.science-and-fun.de/tools/. Accessed October 11, 2016. | ||
Xie J, Lee JY, Wang DI, Ting YP. Silver nanoplates: from biological to biomimetic synthesis. ACS Nano. 2007;1(5):429–439. | ||
Liu F, Cui Y, Wang L, et al. Temperature-responsive Poly(N-isopropylacrylamide) modified gold nanoparticle–protein conjugates for bioactivity modulation. ACS Appl Mater Inter. 2015;7(21):11547–11554. | ||
Yang X, Yang M, Pang B, Vara M, Xia Y. Gold nanomaterials at work in biomedicine. Chem Rev. 2015;115(19):10410–10488. | ||
Singh DK, Jagannathan R, Khandelwal P, Abraham PM, Poddar P. In situ synthesis and surface functionalization of gold nanoparticles with curcumin and their antioxidant properties: an experimental and density functional theory investigation. Nanoscale. 2013;5(5):1882–1893. | ||
Xie J, Zheng Y, Ying JY. Protein-directed synthesis of highly fluorescent gold nanoclusters. J Am Chem Soc. 2009;131(3):888–889. | ||
Lin D, Wu H, Zhang R, Pan W. Enhanced photocatalysis of electrospun Ag-ZnO heterostructured nanofibers. Chem Mater. 2009;21(15):3479–3484. | ||
Xie J, Lee JY, Wang DIC, Ting YP. Identification of active biomolecules in the high-yield synthesis of single-crystalline gold nanoplates in algal solutions. Small. 2007;3(4):672–682. | ||
Santos SP, Mitchell EP, Franquelim HG, Castanho MA, Abreu IA, Romao CV. Dps from Deinococcus radiodurans: oligomeric forms of Dps1 with distinct cellular functions and Dps2 involved in metal storage. FEBS J. 2015;282(22):4307–4327. | ||
Si S, Mandal TK. Tryptophan-based peptides to synthesize gold and silver nanoparticles: a mechanistic and kinetic study. Chemistry. 2007;13(11):3160–3168. | ||
Si S, Bhattacharjee RR, Banerjee A, Mandal TK. A mechanistic and kinetic study of the formation of metal nanoparticles by using synthetic tyrosine-based oligopeptides. Chemistry. 2006;12(4):1256–1265. | ||
Selvakannan PR, Swami A, Srisathiyanarayanan D, et al. Synthesis of aqueous Au core-Ag shell nanoparticles using tyrosine as a pH-dependent reducing agent and assembling phase-transferred silver nanoparticles at the air-water interface. Langmuir. 2004;20(18):7825–7836. | ||
Yin Y, Yu S, Liu J, Jiang G. Thermal and photoinduced reduction of ionic Au(III) to elemental Au nanoparticles by dissolved organic matter in water: possible source of naturally occurring Au nanoparticles. Environ Sci Technol. 2014;48(5):2671–2679. | ||
Deng W, Fu M, Cao Y, et al. Angelica sinensis polysaccharide nanoparticles as novel non-viral carriers for gene delivery to mesenchymal stem cells. Nanomedicine. 2013;9(8):1181–1191. | ||
Yu Q, Cao J, Chen B, et al. Efficient gene delivery to human umbilical cord mesenchymal stem cells by cationized Porphyra yezoensis polysaccharide nanoparticles. Int J Nanomedicine. 2015;10:7097–7107. | ||
Levak M, Buric P, Sikiric MD, et al. Effect of protein corona on silver nanoparticle stabilization and ion release kinetics in artificial seawater. Environ Sci Technol. 2017;51(3):1259–1266. | ||
Lu D, Liu Q, Zhang T, Cai Y, Yin Y, Jiang G. Stable silver isotope fractionation in the natural transformation process of silver nanoparticles. Nat Nanotechnol. 2016;11(8):682–686. | ||
Li T, Albee B, Alemayehu M, et al. Comparative toxicity study of Ag, Au, and Ag-Au bimetallic nanoparticles on Daphnia magna. Anal Bioanal Chem. 2010;398(2):689–700. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.