Back to Journals » International Journal of Nanomedicine » Volume 12
An annotated corpus with nanomedicine and pharmacokinetic parameters
Authors Lewinski NA, Jimenez I, McInnes BT
Received 15 March 2017
Accepted for publication 21 July 2017
Published 12 October 2017 Volume 2017:12 Pages 7519—7527
DOI https://doi.org/10.2147/IJN.S137117
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Nastassja A Lewinski,1 Ivan Jimenez,1 Bridget T McInnes2
1Department of Chemical and Life Science Engineering, Virginia Commonwealth University, Richmond, VA, 2Department of Computer Science, Virginia Commonwealth University, Richmond, VA, USA
Abstract: A vast amount of data on nanomedicines is being generated and published, and natural language processing (NLP) approaches can automate the extraction of unstructured text-based data. Annotated corpora are a key resource for NLP and information extraction methods which employ machine learning. Although corpora are available for pharmaceuticals, resources for nanomedicines and nanotechnology are still limited. To foster nanotechnology text mining (NanoNLP) efforts, we have constructed a corpus of annotated drug product inserts taken from the US Food and Drug Administration’s Drugs@FDA online database. In this work, we present the development of the Engineered Nanomedicine Database corpus to support the evaluation of nanomedicine entity extraction. The data were manually annotated for 21 entity mentions consisting of nanomedicine physicochemical characterization, exposure, and biologic response information of 41 Food and Drug Administration-approved nanomedicines. We evaluate the reliability of the manual annotations and demonstrate the use of the corpus by evaluating two state-of-the-art named entity extraction systems, OpenNLP and Stanford NER. The annotated corpus is available open source and, based on these results, guidelines and suggestions for future development of additional nanomedicine corpora are provided.
Keywords: nanotechnology, informatics, natural language processing, text mining, corpora
Introduction
Nanotechnology is enabling new strategies to detect and treat disease through multifunctional (eg, targeted, activatable, diagnostic, and therapeutic) drug design. Formulating a drug as a nanomedicine can also improve its therapeutic index by changing its stability, pharmacokinetics, and toxicity. For several decades, researchers have primarily designed new nanomedicines based on an empirical approach. However, the number of possible nanomedicine formulations continues to increase exponentially as new nanomaterials, surface coatings, bioconjugates, and drug combinations are developed. As a consequence, experimentally assessing all possible nanomedicine formulations for efficacy and safety is not feasible or realistic. There is a critical need to automatically extract information and synthesize knowledge and trends in nanomedicine research to rationally prioritize testing and development.
Natural Language Processing (NLP) approaches can semi-automate the process of converting text-based unstructured data (eg, full-text articles) to structured data (eg, tables). Interest in applying NLP techniques to nanotechnology has increased over the years, with a number of systems developed for nanomedicine information extraction and nanotechnology patent mining.1 Most NLP algorithms rely on annotated corpora for both training and evaluation of the system. Despite development of several NanoNLP systems, only one nanotechnology-related corpus has been described in the literature.2 Motivated by the need for a nanomedicine corpus, we present the Engineered Nanomedicine Database (END).
The main objective of this paper is to propose a framework for creating an annotated corpus for nanomedicine entity extraction. Toward this objective, we validate a manually annotated corpus of US Food and Drug Administration (FDA)-approved nanomedicines from drug product inserts collected from the Drugs@FDA Database.3 The extracted entities consist of nanoparticle physicochemical properties, exposure parameters, and biologic response information for 41 drugs. We evaluate the precision, recall, and F-measure between expert and non-expert generated annotations and evaluate the performance of two state-of-the-art named entity extraction systems applied to the corpus. To promote future development of nanomedicine corpora and entity extraction systems, we provide the expert annotated corpus as open source (http://rampages.us/nanoinformatics/resources).
The remainder of this paper is as follows. First, we describe related work associated with entity extraction. Second, we describe our method in developing the END dataset. Third, statistical analysis of the annotation process and the contents of the completed END corpus are presented. Finally, use of the corpus for creation of a named entity recognition (NER) system tailored to extracting nanomedicine data is discussed.
Materials and methods
Dataset
The documents selected for annotation were drug product labels for 41 nanomedicines that are currently approved for clinical use by the FDA. The labels were obtained from the Drugs@FDA online database. This document type was chosen for two reasons: compared to nanomedicines in the pipeline, FDA-approved nanomedicines are 1) expected to have the most number of unique publications in the literature; 2) already being prescribed by physicians, and therefore, mentions could be contained in clinical notes. The list of nanomedicines chosen was based on review articles.4,5 The annotated drug product labels described nanomedicines consisting of liposomes, micelles, polymer conjugates, protein conjugates, and nanoparticles, which are listed in Table 1. The extracted entities relate to nanoparticle physicochemical properties, exposure, pharmacokinetics, and biologic response in addition to description information. A complete list of these entities is shown in Table 2.
  | Table 1 US FDA-approved nanomedicines from the year 1975 to 2013 |
  | Table 2 Extracted nanomedicine entities |
Annotation process
To develop an entity extractor, a training set composed of relevant texts that have been manually annotated by domain experts is required. Manually annotated corpora are traditionally prepared by the NLP community through collective shared tasks. When conducted by individuals, it has be reported to take more than 10 hours to annotate a single research paper.2 Recruiting professional nanomedicine researchers to donate this time is challenging; therefore, we hypothesized that a potential solution could be to train non-expert (student) annotators. It has been suggested that, with training, annotation tasks can be crowdsourced to non-experts to build annotated corpora of biomedical literature.6,7 In this work, we assessed the quality of annotations generated by non-experts to those of a domain expert on texts describing nanomedicines. The annotators included three undergraduate research assistants and one professor from the Virginia Commonwealth University Department of Chemical and Life Science and Engineering. The research assistants were entering their junior year and were given training in annotation as outlined in the following section. The General Architecture for Text Engineering (GATE)8 open source, annotation, and development environment for computational language processing was used to manually annotate the drug product labels. An example of annotation is presented in Figure 1. The entity annotation guidelines, which include entity definitions and annotation rules (Table 3), were developed to reduce potential interpretation differences between annotators.
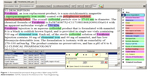  | Figure 1 Annotated ferumoxytol drug product label using GATE. |
Annotator training
The following procedure was employed when training the students:
- “Preannotation guideline discussion phase:” In this phase, orthography and grammar rules, multiword entity rules, and definitions of entities were discussed. All students completed this phase.
- “Pilot annotation phase:” In this phase, the annotators were trained on six entities (active ingredient, dose, indication, nanoparticle, route of administration, trade name). Their annotations were compared with the expert’s annotations, and all differences were discussed with them. All students completed this phase.
- “Annotation phase:” In this phase, all 21 entities were given to the annotators. Orthography and grammar rules, multiword entity rules, and definitions of entities were again discussed. The annotators were also informed that if they had any questions, they were to ask the expert annotator. All questions and responses were circulated among all three of the annotators. One student fully completed and two students partially completed this phase.
Results
Characteristics of corpus
Table 4 shows a high-level breakdown of the expert-annotated drug label inserts within the END corpus. Within the 41 drug label inserts, there are 28,276 sentences and 465,890 words and a total of 22,033 annotations. On average, each drug label contains 690 (SD 496) sentences, 11,363 (SD 5,897) words, and 537 (SD 310) annotations. The largest class of nanomedicines, liposomes, was also examined to determine if restricting to a subset could be representative of the corpus overall. Table 5 shows the number of annotated mentions, the number of unique mentions, and the number of labels containing mentions for each entity. Not all labels contained a mention for all 21 entity types. The number of mentions across the drug labels varied from 6,689 (adverse reaction) to 7 (particle diameter). The unique types of mentions also varied, with the largest number annotated for adverse reaction where 1,773 of the 6,689 mentions were unique.
  | Table 4 Summary of corpus text structure |
  | Table 5 Statistics on the 21 annotated entities |
Evaluation of non-expert annotations
We evaluated the annotation agreement using precision, recall, and F-measure, which were calculated using the GATE framework. Precision measures the number of correctly identified entities as a percentage of the number of items identified. Recall measures the number of correctly identified entities as a percentage of the total number of correct entities. F-measure is the harmonic mean between precision and recall. In this work, we compare the student annotators to the expert annotator (Table 6) and the student annotators to other student annotators (Table 7). We do not report the inter-annotator agreement (eg, Cohen’s Kappa) because the concept of a nonentity is not defined, and therefore, the number of words contained in the nonentity is not known.
  | Table 7 Annotation agreement between student annotators |
Demonstration of intended use of the corpus
We conducted an evaluation of the END corpus on two state-of-the-art NER systems. We evaluated the Apache OpenNLP Toolkit and Stanford NER. OpenNLP was created for processing general English natural language text and includes the NameFinder entity recognizer which uses a Maximum Entropy supervised learning algorithm to identify named entities in unstructured text.9 Stanford NER is a Java-based Named Entity Recognizer that has previously been used to automatically identify general English entities (eg, person and company names) and biomedical entities (eg, gene and protein names) from natural language text.10 Stanford NER uses Conditional Random Fields, also known as CRFClassifier.11
To evaluate the previously developed entity extractors, we performed 10-fold cross validation on each of the entity extraction models developed for the project for those entities that had at least 45 instances in our dataset. Each instance contains 10 words to the right and left of the entity. Table 8 shows the F1 score of the OpenNLP and Stanford NER entity extractors, and the number of instances in the current training data. The results show that for most entities, the Stanford NER system obtains a higher F1 score than the OpenNLP entity extractor for all of the entities evaluated. The Stanford NER system results show that it is able to identify some nano-entities very accurately (eg, molecular weight), but others poorly (eg, core composition). As expected, in general, entities that had a lower number of instances tended to have poorer results than those with more instances, although this is not the case for active ingredient.
The results indicate that further investigation is required into developing entity extraction methods for nanomedicines.
Current state-of-the-art named entity extraction systems, such as Stanford NER and OpenNLP, primarily utilize punctuation, lexical information (eg, previous word), morphologic information (eg, prefix), and orthographic information (eg, capitalization) as features into a machine learning algorithm.12 Although these types of features have been shown to perform well for general English entities (eg, People, Locations, and Organizations), they have been shown to be less useful within the biomedical domain.13 Analysis of the features utilized by Stanford NER and OpenNLP shows that Stanford NER incorporates more contextual information as features than OpenNLP. Our hypothesis is that the incorporation of this additional contextual information may be responsible for Stanford NER systems higher performance.
Discussion
In this work, we created and evaluated an annotated corpus with nanomedicine and pharmacokinetic parameters. FDA-approved nanomedicines were chosen due to the larger number of publications describing these nanomedicines, compared to those still in the development pipeline. We discovered in our chosen document type (ie, drug product labels) that a limited number of mentions specific to the physicochemical properties of nanomedicines are included. Of the 16 minimum characterization parameters extracted by other groups, only 5 (core composition, particle diameter, molecular weight, surface charge, surface chemistry) were contained in the drug product labels.1 Particle diameter had the lowest number of entities due to being mentioned one to two times in only 6 of the 41 labels. Similarly, only 15 out of the 41 labels described the surface coating. Future work will include full papers from the primary literature to develop larger training sets that include data on additional characterization parameters as well as relation annotations. The inclusion of relation annotations will facilitate linking the descriptors, that is, physicochemical characterization, exposure, and biologic response information, with the associated drug.
The 41 nanomedicines included in this corpus were chosen based on published literature reviews.4,5 More recent reviews identify additional nanomedicines and their drug product labels will serve as a test set when evaluating our developed entity extractor.14,15 The nanomedicines in the END corpus are all nanostructured compounds used for the treatment or prevention of disease. We recognize that the definition of nanomedicine is still not fully established in the research community. Erring on the side of inclusion, the END corpus contains conjugate formulations, although PEGylated proteins are not always recognized as nanomedicines.16 Despite some limitations, the END corpus can serve as a controlled dataset for developing entity extractors for nanomedicine.
Overall agreement between the student annotators and the expert annotator was relatively good in light of the wide experience gap. Low inter-annotator agreement primarily correlated with the complexity of the entity definition and the length of description in the drug product insert. For example, a common misannotation for dose often included values for the dosage form, which does not always equal the administered dose. Misannotations for nanoparticle were due to confusion with the active ingredient. Contributing factors to the confusion include the label listing the generic name of the nanoparticle as the active ingredient and the student annotators’ limited experience identifying components of nanostructures. This limited experience also resulted in low inter-annotator agreement for surface coating. The low precision of surface coating is skewed due to misannotations contained in one label (Zevalin). The low recall is more representative, since the missing annotations for surface coating were consistent across the labels. Out of all entities, adverse reaction had the lowest recall. This was attributed to adverse reaction having the largest number of unique entities and being described in several sections of the label. Compared to a study reporting inter-annotator agreement between crowdsourced annotations and expert annotations, our overall F-measure across all entities was lower (0.69 vs 0.76).6,7 We believe this is in part due to the inclusion of more complex concepts in our entity set compared to the biomedical entities, disease and symptom, which more people are familiar with from life experience.
The agreement across students was higher for the 10 entities that all students completed across all 41 labels. The trends observed in the student–student and expert–student agreement were similar. For example, the entities that presented the most difficulty and consequently resulted in the highest number of misannotations between students were active ingredient, dose, and nanoparticle. The reasons for the high number of false positives are the same to those described above. The student–student agreement for indication revealed an opposite result, low precision and high recall, compared to the expert–student agreement, high precision and low recall. This was due to the higher number of misannotations when comparing between students, compared to missing annotations when comparing between the expert and students. Overall, the performance of the students for entities that could describe any drug (eg, trade name, US patent) was better than for entities specific to nanomedicines (ie, nanoparticle, particle diameter).
Limitations
Several limitations must be discussed to facilitate the interpretation of the results of this study. First, the current dataset consists of entities and their context extracted from FDA drug labels. It is still unclear if the context describing the entities within the labels is similar to that within the primary literature. The next step is to utilize this framework for a large-scale data creation study that focuses on primary literature. During this time, we will compare using the drug label dataset as test data for automatically extracting entities from labels not in the training set as well as different article types (eg, preclinical vs clinical).
Second, the size of the dataset is relatively small. However, we believe that this dataset and the framework developed for its creation can be used to facilitate 1) the development of additional larger-scale datasets for nanoparticle entity extraction and 2) the evaluation of current state-of-the-art NLP methods on secondary bio-focused tasks. For example, the pharmacokinetic parameters and biologic response entities are not solely relevant to nanomedicines (eg, plasma half-life, adverse effect). This dataset may be leveraged to aid in developing systems to extract this information across a wide variety of different types of medications.
Third, many nanomedicines in the pipeline are not represented by the formulations included in the set based on the current FDA-approved nanomedicines. The language describing these more sophisticated nanomedicine complexes may differ from the FDA-approved nanomedicines based on older technology. In addition, these new nanomedicines may receive FDA approval in the future, and future work will include active learning to better cover the complete dataset.17
Lessons learned
For this project, we used GATE to manually annotate the drug product labels.8 The annotation was conducted using the Windows and Mac operating systems. This caused some compatibility issues. Future annotations will be conducted using a single operating system to avoid these difficulties. In addition, manual annotation is a time-consuming and sometimes tiresome process. Future work will include active learning to focus manual annotation efforts on entities that need more instances to improve the entity recognition algorithm.
Analysis of current state-of-the-art named entity extraction systems showed that they are not applicable for each of the different nanomedicine parameters extracted by the system. Analysis of the parameters also showed that not all parameters may need a machine learning component to identify them within the text. For example, out of the 1,192 mentions of Route of Administration, only 20 were unique. Given the low performance of identifying this entity by the NER systems, incorporating simple rules and a dictionary for a hybrid machine learning/rule-based approach may improve the overall results of the system.
Conclusion
In this paper, we proposed a framework for creating an annotated corpus for nanomedicine entity extraction. We validated our framework by annotating a corpus of FDA-approved nanomedicines from drug product inserts collected from the Drugs@FDA Database. We annotated the nanoparticle physicochemical properties, exposure parameters, and biologic response information and evaluated the reliability of the human ratings. Based on these results, we provided guidelines and suggestions for future development of additional nanomedicine corpora. We provided both the annotated corpus and the statistical software for their analysis as open source. Furthermore, we demonstrated the use of the proposed framework by evaluating two state-of-the-art named entity extraction systems on the corpus. In the future, we plan to extend this corpus to include the preclinical and clinical trial literature.
Acknowledgments
The authors thank Marley Hodson and Tanin Izadi for serving as annotators and Gabriel Jones for assisting with the open-source NER evaluation. This work was supported by the startup funds provided to Dr Lewinski and Dr McInnes by the School of Engineering at Virginia Commonwealth University.
This work was presented at the 8th International Nanotoxicology Congress as a poster presentation with interim findings. The poster’s abstract was published in the conference’s program available at http://www.nanotoxcongress.net/images/8th-International-Nanotoxicology-Congress.pdf.
Author contributions
NAL and BTM conceived the project idea, led the project, conducted data analysis, and wrote the manuscript. IJ performed annotations and assisted with data analysis. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Lewinski NA, McInnes BT. Using natural language processing techniques to inform research on nanotechnology. Beilstein J Nanotechnol. 2015;6:1439–1449. | ||
Dieb TM, Yoshioka M, Hara S. NaDev: an annotated corpus to support information extraction from research papers on nanocrystal devices. J Inform Process. 2016;24(3):554–564. | ||
Drugs@FDA Database. Available from: http://www.accessdata.fda.gov/scripts/cder/drugsatfda/. Accessed February 8, 2015. | ||
Schütz CA, Juillerat-Jeanneret L, Mueller H, Lynch I, Riediker M; NanoImpactNet Consortium. Therapeutic nanoparticles in clinics and under clinical evaluation. Nanomedicine (Lond). 2013;8(3):449–467. | ||
Weissig V, Pettinger TK, Murdock N. Nanopharmaceuticals (part 1): products on the market. Int J Nanomedicine. 2014;9:4357–4373. | ||
Good BM, Nanis M, Wu C, Su AI. Microtask crowdsourcing for disease mention annotation in PubMed abstracts. Paper presented at: Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing; 2015:282–293. | ||
Tsueng G, Nanis S, Fouquier J, Good B, Su A. Citizen science for mining the biomedical literature. Citiz Sci Theory Pract. 2016;1(2):14. | ||
Cunningham H. GATE, a General Architecture for Text Engineering. Comput Humanit. 2002;36(2):223–254. | ||
Baldridge J, Morton T, Bierner G. The opennlp maximum entropy package. Tech Rep Source Forge. 2002. Available from: https://sourceforge.net/projects/maxent/. Accessed October 2, 2017. | ||
Manning CD, Surdeanu M, Bauer J, Finkel JR, Bethard S, McClosky D. The stanford corenlp natural language processing toolkit. Paper presented at: ACL (System Demonstrations); 2014. Available from: https://nlp.stanford.edu/pubs/StanfordCoreNlp2014.pdf. Accessed October 2, 2017. | ||
Sutton C, McCallum A. An introduction to conditional random fields. Foundations and Trends® in Machine Learning. 2012;4(4):267–373. | ||
Nadeau D, Sekine S. A survey of named entity recognition and classification. Lingvisticae Investigationes. 2007;30(1):3–26. | ||
Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Paper presented at: Pacific Symposium on Biocomputing. 2008:652–663. | ||
Bobo D, Robinson KJ, Islam J, Thurecht KJ, Corrie SR. Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm Res. 2016;33(10):2373–2387. | ||
D’Mello SR, Cruz CN, Chen ML, Kapoor M, Lee SL, Tyner KM. The evolving landscape of drug products containing nanomaterials in the United States. Nat Nanotechnol. 2017;12(6):523–529. | ||
Anselmo AC, Mitragotri S. Nanoparticles in the clinic. Bioeng Transl Med. 2016;1(1):10–29. | ||
Chen Y, Lasko TA, Mei Q, Denny JC, Xu H. A study of active learning methods for named entity recognition in clinical text. J Biomed Inform. 2015;58:11–18. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



