Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 17
The Association Between Mitochondrial tRNAGlu Variants and Hearing Loss: A Case-Control Study
Authors Yu X, Li S, Guo Q, Leng J, Ding Y
Received 20 September 2023
Accepted for publication 16 March 2024
Published 28 March 2024 Volume 2024:17 Pages 77—89
DOI https://doi.org/10.2147/PGPM.S441281
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Xuejiao Yu,1 Sheng Li,2 Qinxian Guo,3 Jianhang Leng,3 Yu Ding3
1Department of Clinical Laboratory, Quzhou People’s Hospital, the Quzhou Affiliated Hospital of Wenzhou Medical University, Quzhou, Zhejiang Province, 324000, People’s Republic of China; 2Department of Otolaryngology, Quzhou People’s Hospital, the Quzhou Affiliated Hospital of Wenzhou Medical University, Quzhou, Zhejiang Province, 324000, People’s Republic of China; 3Central Laboratory, Hangzhou First People’s Hospital, Hangzhou, Zhejiang Province, 310006, People’s Republic of China
Correspondence: Xuejiao Yu, Department of Clinical Laboratory, Quzhou People’s Hospital, the Quzhou Affiliated Hospital of Wenzhou Medical University, Quzhou, People’s Republic of China, Email [email protected] Yu Ding, Central Laboratory, Hangzhou First People’s Hospital, Hangzhou City, Zhejiang Province, 310006, People’s Republic of China, Email [email protected]
Purpose: This study aimed to examine the frequencies of mt-tRNAGlu variants in 180 pediatric patients with non-syndromic hearing loss (NSHL) and 100 controls.
Methods: Sanger sequencing was performed to screen for mt-tRNAGlu variants. These mitochondrial DNA (mtDNA) pathogenic mutations were further assessed using phylogenetic conservation and haplogroup analyses. We also traced the origins of the family history of probands carrying potential pathogenic mtDNA mutations. Mitochondrial functions including mtDNA content, ATP and reactive oxygen species (ROS) were examined in cells derived from patients carrying the mt-tRNAGlu A14692G and CO1/tRNASer(UCN) G7444A variants and controls.
Results: We identified four possible pathogenic variants: m.T14709C, m.A14683G, m.A14692G and m.A14693G, which were found in NSHL patients but not in controls. Genetic counseling suggested that one child with the m.A14692G variant had a family history of NSHL. Sequence analysis of mtDNA suggested the presence of the CO1/tRNASer(UCN) G7444A and mt-tRNAGlu A14692G variants. Molecular analysis suggested that, compared with the controls, patients with these variants exhibited much lower mtDNA copy numbers, ATP production, whereas ROS levels increased (p< 0.05 for all), suggesting that the m.A14692G and m.G7444A variants led to mitochondrial dysfunction.
Conclusion: mt-tRNAGlu variants are important risk factors for NSHL.
Plain Language Summary: The main aim of our study was to explore the association between the mt-tRNAGlu variants and hearing loss. We found that m.T14709C, m.A14683G, m.A14692G and m.A14693G variants were associated with hearing impairments, these variants localized at extremely conserved nucleotides of mt-tRNAGlu and may result a failure in tRNA metabolism, furthermore, patients with mt-tRNAGlu variants exhibited much lower levels of mtDNA copy number, ATP as compared with controls, whereas ROS increased. As a result, mt-tRNAGlu variants may serve as biomarkers for mitochondrial deafness, and screening for tRNAGlu variants is recommended for early detection and diagnosis of mitochondrial deafness.
Keywords: deafness, mitochondrial tRNAGlu variants, pediatrics, tRNA metabolism
Introduction
Deafness is one of the most common human health problems, affecting one in 1000 newborns.1 It is anticipated that the number of deaf people will be more than 28 million by the end of 2030 in the US.2 To date, the etiology of hearing loss is not well understood, but increasing evidence has suggested that deafness can be caused by genetic and environmental factors.3 In fact, the genetic impact has been found more than 50% patients with hearing loss. To date, around 124 genes, as well as 1000 variants have been identified to be related to hearing loss (https://hereditaryhearingloss.org/).4 In addition to nuclear genes, mitochondrion is very important organelle whose primary role is to generate ATP via oxidative phosphorylation (OXPHOS). Moreover, mitochondria have their own genetic codes, named mtDNA, which is 16,569 bp in length.5 Variants in mtDNA are important causes of aminoglycoside-induced and non-syndromic hearing loss (AINSHL) in many families worldwide.6–8 In addition to the well-known m.A1555G or m.C1494T mutation, mitochondrial tRNA (mt-tRNA) gene is another hot spot for pathogenic variants associated with deafness.9 Variants in mt-tRNA genes may alter tRNA structure and functions, including the processing of RNA precursors, modification of specific nucleotides, and maintenance of secondary and tertiary structures. Failure of mt-tRNA metabolism and protein synthesis caused by these variants may lead to mitochondrial dysfunction, which is involved in deafness.10,11 More recently, several mt-tRNA variants had been reported to be related to hearing loss, including mt-tRNAIle A4317G;12 mt-tRNACys C5783T;13 mt-tRNASer(UCN) T7505C,14 however, the pathophysiology of these variants remained poorly elucidated.
In the present study, we screened for the frequencies of mt-tRNAGlu variants in a cohort of 180 children with NSHL and 100 controls using PCR and direct sequencing analyses. Phylogenetic conservation and mtDNA haplogroup analyses were performed to assess the pathogenicity of mt-tRNA variants. We also performed clinical, genetic, molecular, and biochemical assessments of a Chinese pedigree carrying the putative pathogenic mt-tRNAGlu variant.
Materials and Methods
Patients
From January 2020 to January 2023, 180 deaf children including 100 males and 80 females, aged from 1 to 5 years, with an average age of 3.5 years were recruited from Hangzhou First People’s Hospital and Quzhou People’s Hospital. Furthermore, 100 healthy subjects including 60 males and 40 females, aged 3–8 years, with an average age of 6 years, were enrolled as controls. This study was reviewed by the ethics committees of Hangzhou First People’s Hospital (Approval No: 2020–285-01) and Quzhou People’s Hospital (Approval No: 2021–028). Informed consent was obtained by each individual (parents) enrolled in the study. All the procedures were performed with the principles of the Declaration of Helsinki.
Audiological Examinations
Age-appropriate audiological and neurotological examinations of hearing loss were performed as detailed previously, including pure-tone audiometry (PTA), auditory brainstem response, acoustic immittance measurement and distortion product otoacoustic emission.15 The PTA was performed in a sound-controlled room at frequencies ranging from 250 to 8000 Hz, as suggested in a recent study.16 The levels of hearing loss were divided into five grades according to a previous study: normal: <20 decibels (dB); mild: 20–40 dB; moderate: 41–70 dB; severe: 71–95 dB; and profound >95 dB.17
Mutational Analysis of Mt-tRNAGlu Gene
To screen for mt-tRNAGlu variants, we first performed PCR amplification of mt-tRNAGlu in all participants enrolled in this study. The primer sequence for amplification of mt-tRNAGlu was: forward-5’-GCA TAA TTA AAC TTT ACT TC-3’, reversed-5’-AGA ATA TTG AGG CGC CAT TG-3’. After amplification, the PCR products were purified and analyzed by direct Sanger sequencing. The sequence data were then compared with the revised Cambridge reference sequences (rCRS) to detect the mt-tRNAGlu variants (GenBank accession number: NC_012920.1).18
Data Analysis
A total of 14 species were selected for conservation analysis, and the conservation index (CI) was calculated by comparing the human mtDNA with that of the other 13 vertebrates. A CI of ≥ 75% was considered functional significance.19
Molecular Characterization of One Chinese Family with Mt-tRNAGlu A14692G Variant
We ascertained a Chinese pedigree (Figure 1) via Hangzhou First People’s Hospital. The entire mitochondrial genomes of matrilineal relatives (II-1, II-6, III-6, and IV-3) was PCR-amplified using 24 primers and sequenced. The mtDNA variants were detected by comparison with the rCRS (GenBank accession number: NC_012920.1).18 Furthermore, Phylotree (http://www.phylotree.org/) and East Asian phylogeny were used to determine the mtDNA haplogroup of this pedigree.20,21
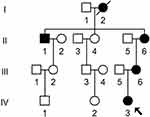 |
Figure 1 One Han Chinese family with NSHL, arrow indicated the proband. |
Mutational Analysis of Common Deafness-Related Genes
To further explore the roles of nuclear genes variants to deafness expression, we conducted mutational screening of common deafness associated genes (GJB2, GJB3, GJB6, TRMU and SLC26A4) in matrilineal relatives (II-1, II-6, III-6 and IV-3), together with other healthy subjects from this family. The primers for GJB2 amplification were: forward-5’-TAT GAC ACT CCC CAG CAC AG-3’, and reverse-5’-GGG CAA TGC TTA AAC TGG C-3’.22 The primers for GJB3 were: forward-5’- GTC ACC TAT TCA TTC ATA CGA TGG-3’ and reverse-5’-TCA CTC AGC CCC TGT AGG AC-3’.23 The primer sequences for amplification of the GJB6 were: forward-5’-CCT TAA AAT AAA GTT GGC TTC AG-3’, reverse-5’-GGA ACT TTC AGG TTG GTA TTG-3’.24 The primer sequences for TRMU were: forward-5’- ACA GCG CAG AAG AAG AGC AGT-3’, reverse-5’- ACA ACG CCA CGA CGG ACG-3’.25 The five primer sequences for SLC26A4 were as follows: forward-5’-CGT GTA GCA GCA GGA AGT AT-3’, and reverse-5’-TTA AAT AAA AAA GAC TGA CT-3’; forward-5’-TGG GGA AAA AGG ATG GTG GT-3’, and reverse-5’-CCA ACC CCT TCT TTA GCT GA-3’; forward-5’-GCA GGA TAG CTC AAG GAA TT-3’, and reverse-5’-TCA TCA GGG AAA GGA AAT AA-3’; forward-5’-TCT CCT TGA TGT CTT GCT TA-3’, and reverse-5’-CCC ATG TAT TTG CCC TGT TG-3’; and forward-5’-CTG GGC AAT AGA ATG AGA CT-3’, and reverse-5’-ATC TGT AGA AAG GTT GAA TA-3’.26 After PCR amplification and direct Sanger sequencing, the data were compared with the wild-type sequences of GJB2, GJB3, GJB6, TRMU and SLC26A4 (GenBank accessible numbers: M86849, AF052692, NG_008323, AF448221 and NM_000441.1, respectively) to detect variants.
Qualification of mtDNA Copy Number
mtDNA copy number was assessed using real-time PCR in four patients with NSHL (II-1, II-6, III-6 and IV-3) carrying the m. A14692G and m.G7444A variants, as well as in four control subjects (II-2, II-5, III-1 and III-5). The primers used for amplification of mt-ND1 were as follows: forward: 5’-AAC ATA CCC ATG GCC AAC CT-3’, revised: 5’-AGC GAA GGG TTG TAG TAG CCC-3’. The primers for the amplification of β-globin were as follows: forward:5’-GAA GAG CCA AGG ACA GGT AC-3’; reversed: 5’-CAA CTT CAT CCA CGT TCA CC-3’. Real-time PCR was performed using a LightCycler® 480II system (Roche Diagnostics GmbH, Mannheim Germany) and each measurement was repeated in triplicate.
Isolation of Polymononuclear Leukocytes (PMNs)
The PMNs from four subjects with NSHL (II-1, II-6, III-6 and IV-3) carrying the m. A14692G and m.G7444A variants, as well as in four control subjects (II-2, II-5, III-1 and III-5) without these variants were isolated according to our previous study.27 PMNs were counted in a Neubauer chamber and cultured in Hanks’ buffered salt solution.
ATP Analysis
The cellular ATP levels in eight cells were measured by using the Cell Titer-Glo® Luminescent Cell Viability Assay kit (Promega), according to the protocols provided by the manufacturer.28
Analysis of ROS Production in Cells
The fluorometry was used to determine the ROS level. Approximately 2×106 cells were first incubated with the fluorescent probe 2,7-dichlorodihydrofluorescein (DCFH) for about 30 min, subsequently, the cells were analyzed by fluorescence plate reader, as mentioned in our previous study.29
Statistical Analysis
Statistical significance was evaluated by an independent Student’s t-test using SPSS software (version 22.0; SPSS, Inc., Chicago, IL, USA). p<0.05 was considered statistically significant.
Results
Screening for Deafness-Associated Mt-tRNAGlu Variants
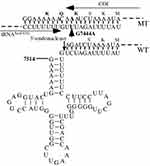
To determine the frequencies of the deafness-related mt-tRNAGlu variants, we first performed PCR amplification of mt-tRNAGlu gene in all participants (Figure 2), which indicated that the targeted PCR product was 1050-bp. Sanger-sequencing suggested that four variants were identified in the current study: m.T14709C, m.A14683G, m.A14692G, and m.A14693G (Table 1). Among these, the m.T14709C variant was found in two subjects with hearing loss (1.11%), the m.A14692G and m.A14683G variants were identified in one individual with deafness (0.56%), and the m.A14693G variant occurred in four patients with hearing loss (2.22%). These variants were not detected in the control groups.
 |
Table 1 Molecular Characterization of Deafness-Associated Mt-tRNAGlu Mutations |
 |
Figure 2 PCR amplification of mt-tRNAGlu gene in subjects with NSHL, arrow indicated the PCR product, which was 1050-bp. |
Molecular Features of Deafness-Associated Mt-tRNAGlu Variants
Next, we performed phylogenetic conservation analysis of the variants identified in this study. For this purpose, a total of 14 species of mtDNA sequences were selected. As shown in Table 1 and Figure 3, we found that all variants were highly conserved (CI=100% for all).
Structurally, as shown in Figure 4, the m.T14709C variant was located at the anticodon stem of mt-tRNAGlu gene (position 37), whereas the m.A14683G variant was localized at the TψC loop of mt-tRNAGlu gene (position 64), which created a novel base-pairing (64A-50G) and may change the secondary structure of this mt-tRNA. Moreover, both m.A14692G and m.A14693G variants occurred in the TψC loop (positions 55 and 54). Nucleotide at positions 55 and 54 are often chemically modified and thus contribute to the structural and functional importance of mt-tRNA.30,31 Therefore, it can be anticipated that the alteration of mt-tRNA structure by these variants may lead to the failure of mt-tRNAGlu metabolism.
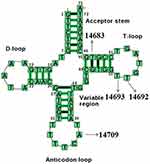 |
Figure 4 Cloverleaf structure of mt-tRNAGlu gene, arrows indicated the positions of m.T14709C, m.A14693G, m.A14692G and m.A14683G variants. |
Clinical and Molecular Analysis of One Pedigree with NSHL
We ascertained a Chinese pedigree with maternally inherited NSHL at Hangzhou First People’s Hospital. Of the seven matrilineal relatives, four had suffered varying degrees of hearing loss. The proband (IV-3), aged 2 years, suffered from profound bilateral hearing loss (105 dB for the right ear and 116 dB for the left ear). As shown in Figure 5 and Table 2, subjects (II-1, II-6 and III-6) were also deaf. Intriguingly, none of the patients had a history of aminoglycoside use.
 |
Table 2 Summary of Clinical and Molecular Data for Several Members of the Chinese Family |
 |
Figure 5 Audiological examination of matrilineal relatives of one pedigree with NSHL, (X) left ear; (O) right ear. |
Screening for mtDNA Variants
Because of maternal transmission, which suggested that mtDNA dysfunction may be involved in the pathogenesis of NSHL. We conducted mutational screening of the entire mitochondrial genome of four matrilineal relatives (II-1, II-6, III-6 and IV-3). As shown in Table 3, PCR-Sanger sequencing revealed a set of genetic polymorphisms belonging to mitochondrial haplogroup B4c1.21 Of these, there were eight variants in the D-loop, two variants in 12S rRNA, three variants in 16S rRNA, and two variants in mt-tRNA genes (m.G7444A and m.A14692G), while the rest were mainly located in protein-coding genes. In addition, seven missense variants were identified: ND2 C5178A (Leu to Met), CO1/tRNASer(UCN) G7444A (Ter to Lys), A6 A8701G (Thr to Ala) and A8860G (Thr to Ala), ND3 A10398G (Thr to Ala), Cytb C14766T (Thr to Ile) and A15326G (Thr to Ala). These missense variants and tRNA variants were further assessed by phylogenetic analysis, especially in bovine,32 mouse33 and Xenopus laevis.34 We found that except for the m.G7444A and m.A14692G variants (Figures 6 and 7), others were not well conserved, suggesting that these variants may be involved in deafness expression.
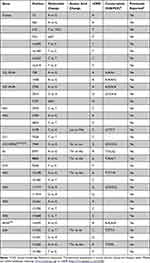 |
Table 3 mtDNA Variants in One Chinese Family with Hearing Impairment |
 |
Figure 6 Identification of m.A14692G and m.G7444A variants by direct sequencing analysis. |
Mutational Analysis of Nuclear Genes
Variants in GJB2, GJB3, GJB6, TRMU and SLC26A4 played active roles in NSHL.35,36 To examine the contributions of these gene variants to NSHL, we analyzed the exons of these genes in matrilineal relatives of this pedigree (II-1, II-6, III-6, and IV-3), as well as in controls. However, we did not identify any variants of these genes, suggesting that nuclear modified genes may not play a role in hearing impairment.
Analysis of mtDNA Content
To test whether the m.A14692G and m.G7444A variants affected mitochondrial function, we examined the mtDNA content in four individuals with hearing loss and four control subjects using Real-time PCR. As can be seen in Figure 8A, deaf patients exhibited much lower mtDNA copy numbers than controls (p=0.0010).
 |
Figure 8 Analysis of mtDNA copy number in matrilineal relatives of the pedigree with NSHL. (A) mtDNA copy number; (B) ATP content; (C) ROS qualifications. |
ATP Decreased in Mutant Cells
We next measured the ATP levels in mutant and control cell lines, as shown in Figure 8B, cells with m.A14692G and m.G7444A variants showed markedly decreased in ATP levels as compared with controls (p<0.0001).
ROS Levels Increased in Cells with m.A14692G and m.G7444A Variants
We also examined the levels of ROS production in cell lines with and without m.A14692G and m.G7444A variants (Figure 8C), as compared with controls, we noticed that mutant cells exhibited much higher levels of ROS production (p=0.0001).
Discussion
In the present study, we examined the frequency of mt-tRNAGlu variants in a cohort of 180 children with NSHL. Four potential pathogenic variants: m.T14709C, m.A14692G, m.A14693G and m.A14683G were identified using Sanger-Sequencing. These variants were further evaluated using the following criteria: (1) present in <1% of controls; (2) evolutionary conservation; and (3) potential structural and functional alterations. Of these, the m.T14709C variant was first reported in a patient with maternally inherited diabetes and deafness (MIDD).37 Biochemical analysis revealed that the m.T14709C variant affected the Complexes I and IV activities,38 furthermore, this variant decreased the steady-state level of tRNAGlu and subsequently influenced the protein synthesis.39 Thus, the m.T14709C was definitely pathogenic for NSHL.
Moreover, the m.A14693G variant occurred at the extremely conserved position of mt-tRNAGlu (position 55), a position that was critical for tRNA chemical modification.3 Previously study indicated that the m.A14693G variant may modulate the clinical expression of deafness-associated m.A1555G mutation.40 Besides, the m.A14683G variant created a new Watson-Crick base-paring (64A-50G).41 Intriguingly, m.T15965C variant which occurred at the same position as the TψC stem of mt-tRNAPro gene has been associated with Parkinson diseases.42 Therefore, the m.A14683G variant, which was similar to the m.T15965C variant, may also cause mitochondrial dysfunction that was responsible for NSHL.
Among the cases harboring the mt-tRNAGlu variants, only one child with the m.A14692G variant had an obvious family history of NSHL. Interestingly, none of the members in this pedigree had a history of aminoglycoside use, notably, the age at onset of NSHL varied from 1 to 69 years, with an average of 38 years. Sequence analysis of the entire mitochondrial genome from matrilineal relatives suggested the presence of two interesting variants: mt-tRNAGlu A14692G and CO1/tRNASer(UCN) G7444A. At the molecular level, the m.A14692G variant affected a highly conserved uridine at position 55 in the TΨC loop of mt-tRNAGlu. The uridine was modified to pseudouridine (Ψ55), which plays an important role in the structure and function of this mt-tRNA.43,44 The destabilization of base-pairing (18A-Ψ55) caused by the m.A14692G variant perturbed the conformation and stability of mt-tRNAGlu. An approximately ~65% decrease in the steady-state level of mt-tRNAGlu was observed in the cells carrying this variant. Failure in mt-tRNAGlu metabolism impaired mitochondrial translation, especially for polypeptides with a high proportion of glutamic acid codons such as MT-ND1, MT-ND6 and MT-CO2 in mutant cells.45 Thus, the m.A14692G variant was definitely pathogenic as it caused mitochondrial dysfunction.
In addition, the m.G7444A variant resulted in a read-through of the stop codon AGA of the CO1 message, thereby adding three amino acids (Lys-Gln-Lys) to the C-terminal of the polypeptide.46 Thus, the variant polypeptide may retain a partial function (Figure 7). Alternatively, the m.G7444A variant was adjacent to the site of 3’ end endonucleolytic processing of the L-strand RNA precursor, spanning mt-tRNASer(UCN) and ND6 mRNA.47 Furthermore, the homoplasmic m.A7445G variant was reported to reduce tRNASer(UCN) levels by ~70% and to cause a 45% reduction in mitochondrial protein synthesis in cybrid cells containing this variant.47 Therefore, the m.G7444A variant, which was similar to the m.A7445G variant, may have an impact on mitochondrial function. Our results indicated that patients with both the m.A14692G and m.G7444A variants had lower mtDNA copy numbers, ATP production than controls, whereas the levels of ROS increased significantly. In fact, mtDNA copy number was a relative measure of the cellular number or mass of mitochondria.48 Recent experimental study suggested that alterations in mtDNA copy number played a fundamental role in the increase in ROS, maintenance of mtDNA copy number was essential for the preservation of mitochondrial function and cell growth.49 Moreover, the increased ROS would affect the quality of gametes, lead to the cochlear cell death and apoptosis,50,51 and in turn, impaired OXPHOS and decreased the ATP production. Therefore, the m.A14692G and m.G7444A variants led to mitochondrial dysfunctions that were involved in NSHL. However, the absent of any functional variants in GJB2, GJB3, GJB6, TRMU and SLC26A4 suggested that nuclear genes may not play important roles in deafness expression. Hence, the combination of m.A14692G and m.G7444A variants may account for high penetrance and expressivity of NSHL in this pedigree.
In conclusion, our study indicated that mt-tRNAGlu variants were important risk factors for NSHL, m.T14709C, m.A14692G, m.A14693G and m.A14683G were associated with NSHL. Screening for mt-tRNAGlu variants was recommended for early diagnosis and prevention of NSHL. However, the homoplasmic forms of mtDNA variants suggested that the mutation itself was not sufficient to produce the clinical phenotypes, hence, other modified factors such as environmental factors, epigenetic modifications and personal lifestyles contributed to deafness expression.
Limitations
The main limitations of the current study were the relatively small sample size, and further studies including more deaf patients and controls are needed to verify these conclusions.
Conclusions
In summary, our data indicated that variants in the mt-tRNAGlu gene were important contributors to NSHL, and screening for variants in this gene is recommended for the early diagnosis and detection of NSHL.
Abbreviations
mtDNA, mitochondrial DNA; NSHL, non-syndromic hearing loss; ROS, reactive oxygen species; OXPHOS, oxidative phosphorylation; AINSHL, aminoglycoside-induced and non-syndromic hearing loss; mt-tRNA, mitochondrial tRNA; PTA, pure-tone audiometry; dB, decibels; rCRS, revised Cambridge reference sequences; CI, conservation index; DCFH, 2,7-dichlorodihydrofluorescein; MIDD, maternally inherited diabetes and deafness.
Data Sharing Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request (Yu Ding: [email protected]).
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committees of Hangzhou First People’s Hospital (Approval No: 2020-285-01) and Quzhou People’s Hospital (Approval No: 2021-028). Prior to the commencement of the research, our team obtained written informed consent from each patient.
Consent for Publication
Each participant provided their consent for publication.
Acknowledgment
We thank the members of the Department of Clinical Laboratory, Quzhou People’s Hospital for their discussion.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the grants from Ministry of Public Health of Zhejiang Province (No: 2021RC022) and Quzhou Bureau of Science and Technology (No: 2022K51).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Jafarlou F, Najafi B, Sameni SJ. Is newborn hearing screening cost effective? Economic consideration for policy makers. Int J Prev Med. 2021;12:155. doi:10.4103/ijpvm.IJPVM_270_20
2. Marinelli JP, Lohse CM, Fussell WL, et al. Association between hearing loss and development of dementia using formal behavioural audiometric testing within the Mayo Clinic Study of Aging (MCSA): a prospective population-based study. Lancet Healthy Longev. 2022;3(12):e817–e824. doi:10.1016/S2666-7568(22)00241-0
3. Ding Y, Leng J, Fan F, et al. The role of mitochondrial DNA mutations in hearing loss. Biochem Genet. 2013;51(7–8):588–602. doi:10.1007/s10528-013-9589-6
4. Bitner-Glindzicz M. Hereditary deafness and phenotyping in humans. Br Med Bull. 2002;63(1):73–94. doi:10.1093/bmb/63.1.73
5. Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. doi:10.1038/nrg1606
6. Moassass F, Al-Halabi B, Nweder MS, et al. Investigation of the mtDNA mutations in Syrian families with non-syndromic sensorineural hearing loss. Int J Pediatr Otorhinolaryngol. 2018;113:110–114. doi:10.1016/j.ijporl.2018.07.028
7. Ibrahim I, Dominguez-Valentin M, Segal B, et al. Mitochondrial mutations associated with hearing and balance disorders. Mutat Res. 2018;810:39–44. doi:10.1016/j.mrfmmm.2018.03.003
8. Mutai H, Watabe T, Kosaki K, et al. Mitochondrial mutations in maternally inherited hearing loss. BMC Med Genet. 2017;18(1):32. doi:10.1186/s12881-017-0389-4
9. Zheng J, Ji Y, Guan MX. Mitochondrial tRNA mutations associated with deafness. Mitochondrion. 2012;12(3):406–413. doi:10.1016/j.mito.2012.04.001
10. Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45(1):299–329. doi:10.1146/annurev-genet-110410-132531
11. Saoji M, Sen A, Cox RT. Loss of individual mitochondrial ribonuclease P complex proteins differentially affects mitochondrial tRNA processing in vivo. Int J Mol Sci. 2021;22(11):6066. doi:10.3390/ijms22116066
12. Cui Y, He DJ. Mitochondrial tRNAIle A4317G mutation may be associated with hearing impairment in a Han Chinese family. Mol Med Rep. 2018;18(6):5159–5165. doi:10.3892/mmr.2018.9519
13. Meng F, Jia Z, Zheng J, et al. A deafness-associated mitochondrial DNA mutation caused pleiotropic effects on DNA replication and tRNA metabolism. Nucleic Acids Res. 2022;50(16):9453–9469. doi:10.1093/nar/gkac720
14. Xue L, Chen Y, Tang X, et al. A deafness-associated mitochondrial DNA mutation altered the tRNASer(UCN) metabolism and mitochondrial function. Mitochondrion. 2019;46:370–379. doi:10.1016/j.mito.2018.10.001
15. Zheng J, Bai X, Xiao Y, et al. Mitochondrial tRNA mutations in 887 Chinese subjects with hearing loss. Mitochondrion. 2020;52:163–172. doi:10.1016/j.mito.2020.03.005
16. Yu X, Li S, Ding Y. Maternally transmitted nonsyndromic hearing impairment may be associated with mitochondrial tRNAAla 5601C>T and tRNALeu(CUN) 12311T>C mutations. J Clin Lab Anal. 2022;36(4):e24298. doi:10.1002/jcla.24298
17. Ding Y, Teng YS, Zhuo GC, et al. The mitochondrial tRNAHis G12192A mutation may modulate the clinical expression of deafness-associated tRNAThr G15927A mutation in a Chinese pedigree. Curr Mol Med. 2019;19(2):136–146. doi:10.2174/1566524019666190308121552
18. Andrews RM, Kubacka I, Chinnery PF, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. doi:10.1038/13779
19. Levin L, Zhidkov I, Gurman Y, et al. Functional recurrent mutations in the human mitochondrial phylogeny: dual roles in evolution and disease. Genome Biol Evol. 2013;5(5):876–890. doi:10.1093/gbe/evt058
20. van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30(2):E386–94. doi:10.1002/humu.20921
21. Kong QP, Bandelt HJ, Sun C, et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15(13):2076–2086. doi:10.1093/hmg/ddl130
22. Rayess HM, Weng C, Murray GS, et al. Predictive factors and outcomes of cochlear implantation in patients with connexin 26 mutation: a comparative study. Am J Otolaryngol. 2015;36(1):7–12. doi:10.1016/j.amjoto.2014.08.010
23. Gao Y, Zhang Q, Zhang S, et al. A Connexin Gene (GJB3) mutation in a Chinese family with erythrokeratodermia variabilis, ichthyosis and nonsyndromic hearing loss: case report and mutations update. Front Genet. 2022;13:797124. doi:10.3389/fgene.2022.797124
24. Ebrahimkhani S, Asaadi Tehrani G. Evaluation of the GJB2 and GJB6 polymorphisms with autosomal recessive nonsyndromic hearing loss in Iranian population. Iran J Otorhinolaryngol. 2021;33(115):79–86. doi:10.22038/ijorl.2020.45196.2483
25. Gaignard P, Gonzales E, Ackermann O, et al. Mitochondrial infantile liver disease due to TRMU gene mutations: three new cases. JIMD Rep. 2013;11:117–123. doi:10.1007/8904_2013_230
26. Zhou Y, Li C, Li M, et al. Mutation analysis of common deafness genes among 1201 patients with non-syndromic hearing loss in Shanxi province. Mol Genet Genomic Med. 2019;7(3):e537. doi:10.1002/mgg3.537
27. Ding Y, Xia BH, Zhang CJ, et al. Mitochondrial tRNALeu(UUR) C3275T, tRNAGln T4363C and tRNALys A8343G mutations may be associated with PCOS and metabolic syndrome. Gene. 2018;642:299–306. doi:10.1016/j.gene.2017.11.049
28. Ding Y, Xia BH, Zhuo GC, et al. Premature ovarian insufficiency may be associated with the mutations in mitochondrial tRNA genes. Endocr J. 2019;66(1):81–88. doi:10.1507/endocrj.EJ18-0308
29. Ding Y, Xia BH, Zhang CJ, et al. Mutations in mitochondrial tRNA genes may be related to insulin resistance in women with polycystic ovary syndrome. Am J Transl Res. 2017;9(6):2984–2996.
30. Ding Y, Gao B, Huang J. Mitochondrial cardiomyopathy: the roles of mt-tRNA mutations. J Clin Med. 2022;11(21):6431. doi:10.3390/jcm11216431
31. Jia Z, Meng F, Chen H, et al. Human TRUB1 is a highly conserved pseudouridine synthase responsible for the formation of Ψ55 in mitochondrial tRNAAsn, tRNAGln, tRNAGlu and tRNAPro. Nucleic Acids Res. 2022;50(16):9368–9381. doi:10.1093/nar/gkac698
32. Gadaleta G, Pepe G, De Candia G, et al. The complete nucleotide sequence of the rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28(6):497–516. doi:10.1007/BF02602930
33. Bibb MJ, Van Etten RA, Wright CT, et al. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26(2 Pt 2):167–180. doi:10.1016/0092-8674(81)90300-7
34. Roe BA, Ma DP, Wilson RK, et al. The complete nucleotide sequence of the xenopus laevis mitochondrial genome. J Biol Chem. 1985;260(17):9759–9774. doi:10.1016/S0021-9258(17)39303-1
35. Luo LF, Hou CC, Yang WX. Nuclear factors: roles related to mitochondrial deafness. Gene. 2013;520(2):79–89. doi:10.1016/j.gene.2013.03.041
36. Finsterer J, Fellinger J. Nuclear and mitochondrial genes mutated in nonsyndromic impaired hearing. Int J Pediatr Otorhinolaryngol. 2005;69(5):621–647. doi:10.1016/j.ijporl.2004.12.002
37. Perucca-Lostanlen D, Taylor RW, Narbonne H, et al. Molecular and functional effects of the T14709C point mutation in the mitochondrial DNA of a patient with maternally inherited diabetes and deafness. Biochim Biophys Acta. 2002;1588(3):210–216. doi:10.1016/s0925-4439(02)00167-9
38. Meulemans A, Seneca S, Smet J, et al. A new family with the mitochondrial tRNAGLU gene mutation m.14709T>C presenting with hydrops fetalis. Eur J Paediatr Neurol. 2007;11(1):17–20. doi:10.1016/j.ejpn.2006.10.004
39. McFarland R, Schaefer AM, Gardner JL, et al. Familial myopathy: new insights into the T14709C mitochondrial tRNA mutation. Ann Neurol. 2004;55(4):478–484. doi:10.1002/ana.20004
40. Ding Y, Li Y, You J, et al. Mitochondrial tRNA(Glu) A14693G variant may modulate the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in a Han Chinese family. J Genet Genomics. 2009;36(4):241–250. doi:10.1016/S1673-8527(08)60111-3
41. An S, Qi D, Chen Y, et al. Mitochondrial tRNA Glu A14683G may be a novel mutation associated with inherited hypertension. Int J Clin Exp Med. 2018;11:269–274.
42. Grasbon-Frodl EM, Kösel S, Sprinzl M, et al. Two novel point mutations of mitochondrial tRNA genes in histologically confirmed Parkinson disease. Neurogenetics. 1999;2(2):121–127. doi:10.1007/s100480050063
43. Allnér O, Nilsson L. Nucleotide modifications and tRNA anticodon-mRNA codon interactions on the ribosome. RNA. 2011;17(12):2177–2188. doi:10.1261/rna.029231.111
44. Yasukawa T, Ueda T, Ueda T, et al. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J Biol Chem. 2000;275(6):4251–4257. doi:10.1074/jbc.275.6.4251
45. Wang M, Liu H, Zheng J, et al. A deafness- and diabetes-associated tRNA mutation causes deficient pseudouridinylation at position 55 in tRNAGlu and mitochondrial dysfunction. J Biol Chem. 2016;291(40):21029–21041. doi:10.1074/jbc.M116.739482
46. Liu Q, Liu P, Ding Y, et al. Mitochondrial COI/tRNASer(UCN) G7444A mutation may be associated with aminoglycoside-induced and non-syndromic hearing impairment. Mol Med Rep. 2015;12(6):8176–8178. doi:10.3892/mmr.2015.4484
47. Guan MX, Enriquez JA, Fischel-Ghodsian N, et al. The deafness-associated mitochondrial DNA mutation at position 7445, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase subunit ND6 gene expression. Mol Cell Biol. 1998;18(10):5868–5879. doi:10.1128/MCB.18.10.5868
48. Memon AA, Vats S, Sundquist J, et al. Mitochondrial DNA copy number: linking diabetes and cancer. Antioxid Redox Signal. 2022;37(16–18):1168–1190. doi:10.1089/ars.2022.0100
49. Jeng JY, Yeh TS, Lee JW, et al. Maintenance of mitochondrial DNA copy number and expression are essential for preservation of mitochondrial function and cell growth. J Cell Biochem. 2008;103(2):347–357. doi:10.1002/jcb.21625
50. Hawk MA, Schafer ZT. Mechanisms of redox metabolism and cancer cell survival during extracellular matrix detachment. J Biol Chem. 2018;293(20):7531–7537. doi:10.1074/jbc.TM117.000260
51. Ding Y, Teng Y, Guo Q, et al. Mitochondrial tRNAGln 4394C>T mutation may contribute to the clinical expression of 1555A>G-induced deafness. Genes. 2022;13(10):1794. doi:10.3390/genes13101794
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.