Back to Journals » Infection and Drug Resistance » Volume 8
Histopathological changes induced in an animal model by potentially pathogenic Enterococcus faecalis strains recovered from ready-to-eat food outlets in Osun State, Nigeria
Authors Olawale A, David O, Oluyege A, Osuntoyinbo R, Laleye SA, Famurewa O
Received 26 January 2014
Accepted for publication 15 May 2014
Published 26 June 2015 Volume 2015:8 Pages 181—187
DOI https://doi.org/10.2147/IDR.S61381
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Adetunji Kola Olawale,1,2 Oluwole Moses David,2,3 Adekemi Olubukunola Oluyege,2 Richard Temitope Osuntoyinbo,4 Solomon Anjuwon Laleye,5 Oladiran Famurewa,2
1Department of Applied Sciences, Osun State Polytechnic, Iree, 2Department of Microbiology, University of Ado-Ekiti, Ado-Ekiti, Nigeria; 3Phytomedicine Research Centre, Department of Botany, University of Fort Hare, Alice, South Africa; 4Department of Microbiology, Waterford Regional Hospital, Waterford, Republic of Ireland; 5Department of Microbiology, Adekunle Ajasin University, Akungba-Akoko, Nigeria
Abstract: Enterococci have been implicated as an emerging important cause of several diseases and multiple antibiotic resistance. However, there is little information about the prevalence of pathogenic and/or antibiotic-resistant Enterococcus faecalis in ready-to-eat foods in Nigeria. Here we report the pathogenic potential of three selected antibiotic-resistant E. faecalis strains isolated from food canteens and food outlets with different virulence determinant genes, including EFC 12 (with gel+, esp+, cylA+, and asa1+), EFT 148 (with gel+, ace+, and asa1+), and EFS 18 (with esp+ and cylA+) in an animal model. Enterococcemia, hematological parameters, and histopathological changes in organ tissues were examined in experimental animals. The results showed differences in enterococcemia and hematological parameters between the control group and experimental animal group. Enterococcemia was observed for 7 days, and the animal group infected with EFC 12 showed the highest growth rate, followed by EFT 148, with the lowest growth rate seen in the EFS 18-infected group. White blood cell count, packed cell volume, and platelets were significantly reduced (P<0.05) in the experimental animals compared with the controls. White blood cells decreased drastically during the study period in rats challenged with EFC 12 (from 7,800 to 6,120 per mm3) but levels remained higher in the control group (from 9,228 to 9,306 per mm3). Histopathological changes included areas of pronounced hemorrhage, necrosis, and distortion in liver tissues, which were more marked in rats infected with EFC 12, followed by EFT 148, then EFS 18. The results of this study suggest the presence of potentially pathogenic E. faecalis strains in food canteens and food outlets; hence, there is a need for strict adherence to good hygiene practices in the study area owing to the epidemiological significance of foods.
Keywords: pathogenic potentials, antibiotic resistance, Enterococcus faecalis, food canteen
Introduction
Enterococci are important bacteria in foods essentially due to their involvement in spoilage, fermentation, and use as probiotics in humans and slaughter animals.1 Certain specific enterococcal strains have been used as probiotic adjunct cultures in the production of cheddar cheese owing to their ability to improve microbial balance in the intestine.2,3 However, reports show that the safety of food samples containing enterococci remains a challenge that requires careful consideration and attention.2,4 Enterococci are important pathogens that cause various infections in humans and other vertebrate animals.5–7 There are also reports of infections in livestock and poultry in veterinary practice.8,9
Some Enterococcus spp. are resistant to antibiotics and possess virulence factors such as adhesins, invasins, pili, and hemolysin,8 and several of these virulence factors have been described.3 Eaton and Gasson10 showed that gelatinase (gel) genes may be silent and the phenotype may be negative, even though a gel gene is present. Antibiotic-resistant enterococci, especially vancomycin-resistant enterococci, are also increasingly reported.11,12 Within the genus Enterococcus, E. faecalis is an important part of the normal intestinal flora in humans and animals. It is the second regular bacterium, next to Escherichia coli as an ecological agent.13,14 E. faecalis is ranked second or third in frequency among bacteria isolated from hospitalized patients.15
The resistance of enterococci to pasteurization temperatures and their ability to adapt to different substrates and environmental conditions indicate that the organisms can be found in food products manufactured from raw materials (milk or meat) and in heat-treated food products. This means essentially that this bacterium could withstand the usual conditions of food production. In addition, it could contaminate finished products during food processing. Enterococci can become an important part of the fermented food microbiota, especially in fermented cheeses and meats. The findings of Hayakawa et al16 suggesting the presence of a “non-hospital” pool of vancomycin-resistant E. faecalis are very worrying.
Antibiotic-resistant enterococci are widespread in food, including meat, dairy products, and ready-to-eat foods, as well as within enterococcal strains used as probiotics.17,18 Acquired resistance traits for a number of antibiotics in enterococci from food origin have been reported throughout Europe,17 while the role of antibiotic-resistant enterococci, especially vancomycin-resistant enterococci, has been emphasized, implicating food as a possible natural reservoir in the dissemination of antibiotic resistance traits in the environment.18
The role of enterococci in disease has consistently raised suspicion with regard to the safety of these organisms for use in foods and as probiotics. There has been a degree of suspicion regarding a link between the use of some antibiotics in livestock and humans becoming colonized by antibiotic-resistant enterococci via the food chain.19,20 However, information about the prevalence of pathogenic E. faecalis from food origin, especially ready-to-eat foods, remains scanty, particularly in the study area. Therefore, the present study examined the pathogenic potential of selected antibiotic-resistant E. faecalis with certain virulence factors isolated from food canteens and outlets in Osun State, Nigeria.
Materials and methods
Bacterial isolates
Three E. faecalis strains recovered from canteen samples (food, plates, and palms of food handlers) previously identified by a biochemical protocol and confirmed by polymerase chain reaction using 16S primers21,22 were obtained from the Department of Microbiology, Ekiti State University, Ado-Ekiti, Nigeria. Susceptibility testing of the isolates as well as their virulence potential (Table 1) had been determined and are reported elsewhere.23
Sources of experimental animals
Male albino rats of the Wistar strain and aged 28 days were sourced from the animal house, University of Ibadan, Nigeria, and divided into four (one control and three experimental) study groups, with five rats in each group.
Preparation of inoculum for enterococcal challenge test
Overnight culture of each of the isolates grown in peptone water was harvested by centrifugation at 14,636 × g for 15 minutes at 26°C. The bacterial pellet was suspended in 2 mL of sterile normal saline, adjusted to a final concentration of 109 cfu/mL, diluted to 5×105/mL per 100 μL, and fed to the animals orally with a 28 gauge, 0.5 inch intubator. This bacterial suspension has been reported to be optimal for inducing a reproducible, longer-term, non-acute infection with morbidity and mortality in lower-order mammals.24 The population of bacterial cells fed into each rat was confirmed by the viable plate count technique. The animals were observed daily for morbidity and mortality, and any rat exhibiting noticeable changes, such as a scruffy coat or lethargy, was sacrificed. Infection experiment was repeated to confirm results.
Determination of enterococcemia and hematological changes
Three groups of albino rats were fed orally with enterococcal strains (EFC 12, EFS 18, and EFT 148) that possess different virulence determinant genes through an intubator as described by Singh et al25 while the control group was fed with the same volume of sterile normal saline. After challenge, enterococcemia was determined in blood samples.
Blood was collected from each animal’s tail to investigate enterococcemia as follows. Two drops of blood were placed in a test tube containing 1 mL of sterile peptone water and allowed to stand for 20 minutes. The sample was then plated on Bile Aesculin Agar (Oxoid, Basingstoke, UK) and incubated at 37°C for 24 hours. The blood samples were also smeared and stained on grease-free glass slides and viewed under a microscope (AmScope, Irvine, CA, USA) for the presence of bacteria with a cocci shape typical of enterococci.
Hematological changes were examined after the 1st, 2nd, and 3rd week of infection. Packed cell volume, red blood cells, white blood cells (WBCs), platelets, and hemoglobin were determined using the methods described by Cheesbrough.26
Histopathological changes were examined in the experimental animals following sacrifice 3 weeks after challenge. The methods of Gentry-Weeks et al27 were used to collect, process, and examine the samples. Pathological changes in the samples were observed by microscopy (AmScope) with a camera attachment.
Statistical analyses
The hematological data were subjected to analysis of variance using Statistical Package for Social Sciences version 17 software (SPSS Inc., Chicago, IL, USA).
Results and discussion
Growth of the test organisms was monitored in the experimental animals to investigate enterococcemia. The three strains of E. faecalis used were EFC 12 with four virulence genes (gel+, esp+, cylA+, and asa1+), EFT 148 with three virulence genes (gel+, ace+, and asa1+) and EFS 18 with two virulence genes (esp+ and cylA+). With time, there was a reduction in the enterococcal load in all experimental groups, with a sharp decrease from the onset of the experiment to day 7. EFC 148 had the smallest and EFC 12 the highest enterococcal load by day 7 (Figure 1).
The hematological findings indicated a significant (P<0.05) reduction in WBC count, packed cell volume, and platelets in the experimental animals when compared with the controls. Over the period of the study, WBC levels decreased markedly in rats challenged with EFC 12 (from 7,800 to 6,120 per mm3) but remained higher in control groups (from 9,228 to 9,306 per mm3). There was a reduction in platelet count in the experimental group compared with the control group. The WBC count, which was markedly lower in animals challenged with gel+ E. faecalis may be due to the cytotoxic, tissue-destructive, bone marrow depressive, or inhibitory effects of the gelatinase.3,24,28 Further, in the EFT 148-infected and EFS 18-infected groups, WBC counts were lower (7,110–7,012 and 7,610–7,060 per mm3, respectively) but this was not up to that obtained in the gel+ EFC 12 group (Table 2). Increased WBC counts have been associated with microbial infections, leukemia, and tissue necrosis.26 Animals in the group infected with EFC 12 had the lowest packed cell volume, hemoglobin concentration, and red blood cell counts (34.01%, 10.03 g/dL, and 3.12×106/mm3, respectively). Hemolysins can inhibit leukocyte function29 and cause lysis of erythrocytes that may provide iron and nutrients in tissue while low packed cell volume value has been attributed to anemia.30 A decreased red blood cell count indicates destruction of circulating erythrocytes or impairment of blood-forming (erythropoietic) centers in rats.31
Gelatinase has been associated with virulence determinants in animal models, and its ability to hydrolyze gelatin, collagen, and certain bioactive peptides suggests that it participates in the initiation and propagation of the inflammatory process in infections involving gelatinase-producing organisms such as E. faecalis.9,32,33 In our study, the gelatinase gene was expressed in two (EFC 12 and EFT 148) of the three E. faecalis strains examined. These two strains caused significant enterococcemia in the experimental animals. The gelatin-hydrolyzing activity has been suggested to be different from that of caseinase activity.34,35 Sedgley et al36 reported that the gelatinase gene (gelE) was detected in all endodontic isolates of E. faecalis and that two thirds of the isolates expressed gelatinase activity. David et al33 reported similar findings when investigating clinical isolates. All these studies concluded that evidence of potential virulence determinants was identified in Enterococcus spp., specifically production of gelatinase in combination with one or more other virulence determinants. Studies by Hubble et al37 and Sedgley et al38 indicated that expression of gelE contributed to increased dissemination of E. faecalis and was associated with increased in vitro adhesion of E. faecalis.
In addition, gelatinase is required for efficient biofilm formation and is a major contributor to the pathogenesis of enterococcal endocarditis.39 Therefore, it is evident that bacterial infection accounts for the WBC reduction seen in the experimental animal groups in our study. Extracellular proteases, gelE and sprE, are two known virulence factors that contribute immensely to the pathogenesis of E. faecalis.33,39,40
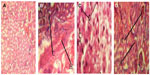
Histopathological investigation showed areas of pronounced hemorrhage, necrosis, and distortion in the liver tissues (Figure 2A–D), which were more marked in rat tissues infected with EFC 12 than in rat tissues infected with EFS 18 and EFT 148. Other tissues examined did not reveal any significant difference compared with control tissue. The combination of cytolysin and Esp seems to confer more virulence on EFS 18 than EFT 148. An association between hemolysin production and virulence is well known for bacteria such as streptococci, pneumococci, listeria, clostridia, and some strains or serotypes of E. coli.41–43 Previous research suggests that hemolysin contributes to virulence in experimental and human enterococcal infections. In a rabbit model of endophthalmitis,44 infections caused by cytolytic E. faecalis resulted in 99% loss of retinal function at postoperative day 3, with near total destruction of retinal architecture, compared with loss of 74% in infections caused by non-cytolytic strains, which produced few changes.
A study from Japan reported hemolytic strains in 60% of isolates from adults with systemic infection due to E. faecalis in contrast with only 17% of fecal isolates from healthy adults colonized with E. faecalis.24 Enterococcus spp. have intrinsic low-level resistance to gentamicin, and some strains can acquire high-level resistance to this drug.45,46 The finding of Chow et al24 suggest that enterococcal hemolysin contributes to virulence when associated with aggregation substance in experimental animal endocarditis.
Biofilm formation in E. faecalis, although not investigated in the present study, but has been strongly associated with the presence of esp, which is a major contributor to pathogenesis.39,43,47–49 It has been suggested that the presence of esp rather than the phenotype (adherence or biofilm formation) is a good marker for identification of strains that are highly adherent to abiotic surfaces. The presence of a pathogenicity island was also not investigated in this study, but it is usually found in E. faecalis, and has been reported to contain several virulence determinant genes, which play different roles in the course of infectious disease.50–52 In the present study, the mortality rate was higher (80%) in the EFC 12-infected animals, while those infected with EFS 18 and EFT 148 had a mortality rate of 60%. This is basically consistent with the findings of Zhou et al.8 The virulence of the organisms may therefore be attributed to the possible presence of a pathogenicity island as well.
Conclusion
The hematological changes and severe tissue damage observed in this study established pathogenicity in the enterococcal-infected experimental animals. Therefore, we conclude that food canteens and food outlets in the study area are a reservoir for potentially pathogenic antibiotic-resistant E. faecalis. Hence, there is a need for improved environmental and personal hygiene in order to ensure good manufacturing practice by food handlers in the area. Also, food canteens and food outlets should be subjected to periodic inspection by the appropriate authorities. Surveillance programs should be put in place to ensure strict compliance. Our study agrees with previous reports on the hazards of ready-to-eat foods vended to school children.53 Therefore, it is mandatory that food handlers/vendors be subject to food safety and food hygiene training and that effective national evaluation criteria be developed.54
Disclosure
The authors report no conflicts of interest in this work.
References
Franz CM, Huch M, Abriouel H, Holzapfel W, Gálvez A. Enterococci as probiotics and their implications in food safety. Int J Food Microbiol. 2011;151:125–140. | |
Giraffa G. Functionality of enterococci in dairy products. Int J Food Microbiol. 2003;88:215–222. | |
Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749–1757. | |
Ogier JC, Serror P. The safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol. 2008;126:291–301. | |
Singh KV, Murray BE. Efficacy of ceftobiprole medocaril against Enterococcus faecalis in a murine urinary tract infection model. Antimicrob Agents Chemother. 2012;56:3457–3460. | |
Vesić D, Kristich CJ. MurAA is required for intrinsic cephalosporin resistance of Enterococcus faecalis. Antimicrob Agents Chemother. 2012;56:2443–2451. | |
Prajsnar TK, Renshaw SA, Ogryzko NV, Foster SJ, Serror P, Mesnage S. Zebrafish as a novel vertebrate model to dissect enterococcal pathogenesis. Infect Immun. 2013;81:4271–4279. | |
Zhou X, Wang D, Wang X-L. Characterization of pathogenic or non-pathogenic Enterococcus faecalis isolated from lambs from Xinjiang, a remote North-west province of China. Afr J Microbiol Res. 2011;5:2827–2833. | |
Zhou X, Wang X, Guo B, Wang X. Isolation and identification of Enterococcus faecalis and detection of its virulence factor genes in lambs presenting with encephalitis in Xinjiang Province, China. Afr J Microbiol Res. 2013;7:2238–2244. | |
Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67:1628–1635. | |
Hall AD, Steed ME, Arias CA, Murray BE, Rybak MJ. Evaluation of standard- and high-dose daptomycin versus linezolid against vancomycin-resistant Enterococcus isolates in an in vitro pharmacokinetic/pharmacodynamic model with simulated endocardial vegetations. Antimicrob Agents Chemother. 2012;56:3174–3180. | |
Tsai HY, Liao CH, Chen YH, et al. Trends in susceptibility of vancomycin-resistant Enterococcus faecium to tigecycline, daptomycin, and linezolid and molecular epidemiology of the isolates: results from the tigecycline in vitro surveillance in Taiwan (TIST) study, 2006 to 2010. Antimicrob Agents Chemother. 2012;56:3402–3405. | |
Drahovska H, Slobodnikova L, Kocincova D, et al. Antibiotic resistance and virulence factors among clinical and food enterococcus isolated in Slovakia. Folia Microbiol. 2004;49(6):763–768. | |
Drahovská H, Slobodníková L, Kocíncová D, et al. Antibiotic resistance and virulence factors among clinical and food Enterococcus isolated in Slovakia. Folia Microbiol (Praha). 2004;49:763–768. | |
Kayser FH. Safety aspects of Enterococcus from the medical point of view. Int J Food Microbiol. 2003;88:255–263. | |
Hayakawa K, Marchaim D, Martin ET, et al. Comparison of the clinical characteristics and outcomes associated with vancomycin-resistant Enterococcus faecalis and vancomycin-resistant E. faecium bacteremia. Antimicrob Agents Chemother. 2012;56:2452–2458. | |
Franz CM, Muscholl-Silberhorn AB, Yousif NM, Vancanneyt M, Swings J, Holzapfel WH. Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl Environ Microbiol. 2001;67:4385–4389. | |
Giraffa G. Enterococci from foods. FEMS Microbiol Rev. 2002;26:163–171. | |
Giridhara Upadhyaya PM, Ravikumar KL, Umapathy BL. Review of virulence factors of Enterococcus: an emerging nosocomial pathogen. Indian J Med Microbiol. 2009;70:301–306. | |
Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155 Pt 6:1749–1757. | |
Desai PJ, Pandit D, Mathur M, Gogate A. Prevalence, identification and distribution of various species of enterococci isolated from clinical specimens with special reference to urinary tract infection in catheterized patients. Indian J Med Microbiol. 2001;19:132–137. | |
Creti R, Imperi L, Bertuccini F, et al. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J Med Microbiol. 2004;53:13–20. | |
Olawale AK, Salako RJ, Olawale AO, Famurewa O. Antibiotic-Resistant Enterococcus faecalis isolated from food canteens in Osun States, Nigeria. Bri Microbiol Res J. 2015;6(4):196–206. | |
Chow JW, Thal LA, Perri MB, et al. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37:2474–2477. | |
Singh KV, Qin X, Weinstock GM, Murray BE. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J Infect Dis. 1998;178:1416–1420. | |
Cheesbrough M. Medical Laboratory Manual for Tropical Countries. London, UK: Butterworth and Co, Ltd; 2003. | |
Gentry-Weeks C, Estay M, Loui C, Baker D. Intravenous mouse infection model for studying the pathology of Enterococcus faecalis infections. Infect Immun. 2003;71:1434–1441. | |
Diarra MS, Rempel H, Champagne J, Masson L, Pritchard J, Topp E. Distribution of antimicrobial resistance and virulence genes in Enterococcus spp. and characterization of isolates from broiler chickens. Appl Environ Microbiol. 2010;76:8033–8043. | |
Skals M, Jorgensen NR, Leipziger J, Praetorius HA. α-Hemolysin from Escherichia coli uses endogenous amplification through P2X receptor activation to induce hemolysis. Proc Natl Acad Sci U S A. 2009;105:4030–4035. | |
Adanlawo IG. Toxicological studies on Saponins extract from the fruit of Solanum anguiri. PhD Thesis. Ilorin, Nigeria: Department of Biochemistry, University of Ilorin; 2003. | |
Ogueke CC, Ogbulie JN, Njoku HO. Antimicrobial properties and preliminary phytochemical analysis of ethanolic extract of Alstonia bonnei. Niger J Microbiol. 2006;20:896–899. | |
Waters CM, Antiporta MH, Murray BE, Dunny GM. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J Bacteriol. 2003;185:3613–3623. | |
David OM, Oluduro OA, Shittu OA, Famurewa O. Identification of changes during infection with gelatinase-producing and gelatinase-defective strains of Enterococcus faecalis using live-animal model. Vet Res. 2011;4:126–132. | |
Furumura MT, Figueiredo PMS, Carbonell GV, da Costa Darini AL, Yano T. Virulence-associated characteristics of Enterococcus faecalis strains isolated from clinical sources. Braz J Microbiol, 2006;37:230–236. | |
Patidar RK, Gupta MK, Singh V. Phenotypic detection of virulence traits and antibiotic susceptibility of endodontic Enterococcus faecalis isolates. Am J Microbiol Res. 2013;1:4–9. | |
Sedgley CM, Nagel AC, Shelburne CE, Clewell DB, Appelbe O, Molander A. Quantitative real-time PCR detection of oral Enterococcus faecalis in humans. Arch Oral Biol. 2005;50:575–583. | |
Hubble TS, Hatton JF, Nallapareddy SR, Murray BE, Gillespie MJ. Influence of Enterococcus faecalis proteases and the collagen-binding proten Ace, on adhesion of dentin. Oral Microbiol Immunol. 2003;18:121–126. | |
Sedgley CM, Lennan SL, Clewell DB. Prevalence, phenotype and genotype of oral enterococci. Oral Microbiol Immunol. 2004;19:95–101. | |
Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, Hancock LE. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immun. 2010;78:4936–4943. | |
Zeng J, Teng F, Murray BE. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect Immun. 2005;73:1606–1612. | |
Lam MM, Seemann T, Bulach DM, et al. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol. 2012;194:2334–2341. | |
Muñoz-Atienza E, Gómez-Sala B, Araújo C, et al. Antimicrobial activity, antibiotic susceptibility and virulence factors of lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 2013;13:15. | |
Frank KL, Guiton PS, Barnes AM, et al. AhrC and Eep are biofilm infection-associated virulence factors in Enterococcus faecalis. Infect Immun. 2013;81:1696–1708. | |
Stevens SX, Jensen HG, Jett BD, Gilmore MS. A hemolysin-encoding plasmid contributes to bacterial virulence in experimental Enterococcus faecalis endophthalmitis. Invest Ophthalmol Vis Sci. 1992;33:1650–1656. | |
Huycke MM, Spiegel CA, Gilmore MS. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1626–1634. | |
Leendertse M, Heikens E, Wijnands LM, et al. Enterococcal surface protein transiently aggravates Enterococcus faecium-induced urinary tract infection in mice. J Infect Dis. 2009;200:1162–1165. | |
Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. | |
Zoletti GO, Pereira EM, Schuenck RP, Teixeira LM, Siqueira JF Jr, dos Santos KR. Characterization of virulence factors and clonal diversity of Enterococcus faecalis isolates from treated dental root canals. Res Microbiol. 2011;162:151–158. | |
Suzuki N, Yoneda M, Hirofuji T. Mixed red-complex bacterial infection in periodontitis. Int J Dent. 2013;2013:587279. | |
Heikens E, Bonten MJ, Willems RJ. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J Bacteriol. 2007;189:8233–8241. | |
Heikens E, van Schaik W, Leavis HL, Bonten MJ, Willems RJ. Identification of a novel genomic island specific to hospital-acquired clonal complex 17 Enterococcus faecium isolates. Appl Environ Microbiol. 2008;74:7094–7097. | |
Dupont H, Vael C, Muller-Serieys C, et al. Prospective evaluation of virulence factors of Enterococcus isolated from patients with peritonitis: impact on outcome. Diagn Microbiol Infect Dis. 2008;60:247–255. | |
Kim MJ, Kim SA, Kang YS, Hwang IG, Rhee MS. Microbial diversity and prevalence of foodborne pathogens in cheap and junk foods consumed by primary schoolchildren. Lett Appl Microbiol. 2013;57:47–53. | |
Egan MB, Raats MM, Grubb SM, et al. A review of food safety and food hygiene training studies in the commercial sector. Food Control. 2007;18:1180–1190. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




 ) EFC 12 with gel+, esp+, cylA+, and asa1+ genes, (
) EFC 12 with gel+, esp+, cylA+, and asa1+ genes, ( ) EFT 148 with gel+, ace+, and asa1+ genes and (
) EFT 148 with gel+, ace+, and asa1+ genes and ( ) EFS 18 with gel−, esp+, and cylA+ genes.
) EFS 18 with gel−, esp+, and cylA+ genes.