Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 17
circPVT1 Inhibits the Proliferation and Aids in Prediction of the Prognosis of Bladder Cancer
Authors Zhou H, Cui X, Zhu L, Xu Z, Wang Z , Shao J
Received 2 September 2023
Accepted for publication 21 December 2023
Published 5 January 2024 Volume 2024:17 Pages 1—11
DOI https://doi.org/10.2147/PGPM.S427147
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Hongyi Zhou,1,* Xueping Cui,2,* Leilei Zhu,1 Zhuoqun Xu,1 Zhuo Wang,3 Jianfeng Shao4
1Department of Urology, The Affiliated Wuxi People’s Hospital of Nanjing Medical University, Wuxi, Jiangsu Province, 214023, People’s Republic of China; 2Department of Pharmacy, The Sixth Affiliated Hospital of Wenzhou Medical University, The People’s Hospital of Lishui, Lishui, Zhejiang Province, 323000, People’s Republic of China; 3Department of Geriatrics, The Affiliated Wuxi People’s Hospital of Nanjing Medical University, Wuxi, Jiangsu Province, 214023, People’s Republic of China; 4Department of Urology, Wuxi No. 2 People’s Hospital (Jiangnan University Medical Center), Wuxi, Jiangsu Province, 214023, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianfeng Shao; Zhuo Wang, Email [email protected]; [email protected]
Background: Circular RNA PVT1 (circPVT1) is aberrantly expressed in several cancers, but its functional role and clinical relevance in bladder urothelial carcinoma (BLCA) remain unknown. This study aimed to identify the expression level of circPVT1 in BLCA and investigated its functional relevance with BLCA progression both in vitro and in vivo.
Methods: GEPIA, UALCAN, and OncoLnc were referred to presented data. Quantitative real-time PCR (qPCR) was used for the measurement of transnational expression of genes in BLCA specimens and cell lines. Immunohistochemistry (IHC) and fluorescence in situ hybridization analysis (FISH) assays were performed to detect HER2 amplification, Pearson’s correlation analysis to analyze the correlation between circPVT1 expression and clinical characteristics, Cox regression and K-M survival analyses to analyze prognostic factors. A nomogram was constructed for predicting prognosis. The proliferation of cells was measured by CCK-8 and colony formation assay, and the proliferation in vivo was evaluated using nude mouse models. qPCR was used to detect the expression of proliferation-related genes.
Results: circPVT1 was but mRNA PVT1 was not significantly overexpressed in BLCA. A high circPVT1 expression was associated with a better survival and negative HER2, but not with age, gender, and T stage. circPVT1 was an independent prognostic factor for the overall survival of BLCA patients. Knocking down circPVT1 promoted BLCA proliferation in vitro and in vivo. Knocking down circPVT1 upregulated ERBB2, MKI67, and PCNA expression and downregulated TP53 expression, but exerted no influence on CCND1 and CCNB1 expression.
Conclusion: circPVT1 is a tumor suppressor and novel prognostic biomarker for BLCA.
Keywords: circPVT1, bladder cancer, biomarker, proliferation, HER2 amplification, BLCA
Introduction
Bladder urothelial carcinoma (BLCA) has high morbidity and mortality in China.1,2 Despite great advances in the research on BLCA, its mechanism is still not fully clear. Recent studies have shown that circRNAs play an important role in their occurrence and have become a research hotspot.3–5 circRNA plasmacytoma variant translocation 1 (circPVT1), transcribed from the long noncoding RNA region with PVT1 locus on chromosome 8q24, is upregulated to drive the development of various cancers.6,7 PVT1 from online data analysis was abnormally expressed in BLCA. However, the role of circPVT1 in BLCA is still unknown.
It is necessary to study clinical value and molecular function of circPVT1 in BLCA. To address these issues, we need to explore whether differential expression of circPVT1 is possible in clinical and cell lines. Combined with statistical analysis, we investigate whether its expression has clinical prognostic value. In addition, we further confirmed in vitro and in vivo whether it has the function of promoting proliferation. In summary, we will reveal its clinical significance and functional role at the clinical, cellular, and animal levels.
Materials and Methods
Clinical Samples
Twenty pairs of BLCA and its normal resection margin tissues without muscle invasive bladder cancer (MIBC) and in T1G3 high risk non-muscle invasive bladder cancer (NMIBC) were obtained from The Affiliated Wuxi People’s Hospital of Nanjing Medical University, and frozen at −80°C for RT-qPCR detection. The 162 BLCA samples were fixed in formalin and measured by IHC. The study was approved by the ethics committee of The Affiliated Wuxi People’s Hospital of Nanjing Medical University (approval number: KYLLH2018019) and consistent with the Declaration of Helsinki. All patients involved in this study had signed an informed consent form, with a follow-up period ranging from 3 to 97 mouths. The tumor was staged by two pathologists blinded to patient data, according to the guidelines of the American Joint Committee on Cancer (AJCC). The patient’s pathological characteristics are shown in Table 1.
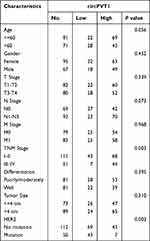 |
Table 1 Correlation Between circPVT1 Expression and Clinicopathological Characteristics in Bladder Cancer |
Bioinformatics Analysis
The PVT1 mRNA levels in BLCA and normal bladder epithelium tissues were determined by integrated analysis from Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia2.cancer-pku.cn/) and The University of Alabama at Birmingham Cancer data analysis Portal (UALCAN, http://ualcan.path.uab.edu/). Survival analysis of PVT1 mRNA expression in BLCA was performed using Kaplan–Meier curve from GEPIA (UALCAN and OncoLnc (http://www.oncolnc.org/).
RNA Isolation and RT-qPCR Assay
Total RNA was extracted from frozen tissue samples or cell lines using Trizol reagent (Invitrogen, Carlsbad, CA, United States). Vazyme reverse transcription kit (Nanjing, China, Vazyme) was used to reverse transcribe RNA to cDNA. RT-qPCR was carried out on an ABI 7500 RT-PCR System (Applied Biosystems, United States) using SYBR Green Master Mix (Takara, Japan). All experiments are performed at least in triplicate. The primers used in this study were synthesized and verified by GenePharma (Shanghai, China) and are shown in Supplementary Table 1. The 2-∆∆Ct method was used to quantify the relative expression levels.
HER2 Amplification Assay
Immunohistochemistry (IHC) assay and fluorescence in situ hybridization (FISH) assay were performed for the staining of HER2. All cases were scored for HER2 status by two pathologists, according to the results of IHC. According to the American Society of Clinical Oncology (ASCO) guidelines, a HER2-positive status was defined as an IHC score of 3+, and an HER2-negative status as an IHC score of 0 or 1+. An IHC score of 2+ was tested by FISH.8 For any sample with a score of less than 3+, its status was further validated by FISH. A ratio below 1.8 indicated negative and that larger than 2.2 indicated positive for ERBB2 gene amplification. A ratio between 1.8 and 2.2 was considered as equivocal.
Cell Lines and Cell Culture
Seven human urinary bladder cancer cell lines (5637, T24, TCCSUP, JMSU1, HT136, RT112, and UMUC3) and normal human urothelial cell line SV-HUC-1 were purchased from the American Type Culture Collection (ATCC; Beijing, China) and cultured according to ATCC suggestions. All medium was supplemented with 10% fetal bovine serum (FBS, Gibco) and 100 IU Penicillin/100 µg/mL Streptomycin and cultured in 37°C/5% CO2 incubator.
Cell Transfection
The lentivirus-mediated short hairpin RNA against circPVT1 (si-circPVT1) and its control (si-Control) were obtained from Genepharma (Shanghai, China). The above plasmids or oligonucleotides were transfected into T24 and UMUC3 cells by Lipofectamine 3000 Reagent (Invitrogen, Carlsbad, CA, USA). Transfection efficiency was detected using qRT-PCR.
RNase R Digestion
Total RNA isolated from T24 and UMUC3 cells was treated with or without RNase R (Solarbio, Beijing, China), with MOCK serving as control. After incubation for 15 min at 37°C, the samples were used for qRT-PCR assay.
Proliferation Assay
The growth rates of T24 and UMUC3 cells were tested by the proliferation assay. Living cells were first seeded in 96-well plates (100 µL/well) at a density of 1× 104 cells/mL, followed by incubations for 1, 2, 3, 4, 5, and 6 days, respectively. Afterward, every 10 µL of CCK-8 solution was added to each well, and the cells were incubated for 2 h at 37°C. Further, 100 µL of dimethyl sulfoxide (DMSO) was added to each well to solubilize the formazan product. The absorbance was then recorded at 450 nm using a microplate reader (Bio-Rad, CA, USA).
Clone Formation Assay
Every 800 cells were plated in six-well plates, cultured for 2 days, fixed in 4% paraformaldehyde solution for 10 min, and then stained with 0.5% crystal violet (Servicebio, Wuhan, China) for 10 min. The colonies were counted for statistical analysis.
Tumor Nude Models
Male BALB/C nude mice (6 weeks of age) were purchased from Shanghai Laboratory Animal Research Center (Shanghai, China) and maintained in static micro-isolator cages. Every 1×107 T24 cells transfected with si-circPVT1 or si-Control were suspended in 110μL of phosphate-buffered saline and then injected subcutaneously into the groin of mice. Four weeks later, the xenograft tumor was stripped off, and its size and weight were calculated. The animal study and the experimental protocol were approved by the Jiangnan University Experimental Animal Care Commission (approval number: JN.No20211015b0480131) and conducted following Chinese guidelines for animal welfare (GB/T35892–2018).
Statistical Methods and Software
The difference between BLCA and normal tissues was analyzed by the paired t-test using R (version R4.0.3, https://www.r-project.org/). Nomogram was constructed using the “rms” package. The Kaplan–Meier method was used to estimate the proportion of patients having survived over a period, and the Log rank test with a two-sided P value to compare Kaplan–Meier curves. The correlation between the clinical factors in the two groups was evaluated using the chi-square test or Fisher's exact test. Multiple logistic regression analysis was conducted to screen the significant variables for nomogram constructions. P<0.05 was considered statistically significant. The pictures were edited using Adobe Photoshop CS6 software.
Results
circPVT1 is Highly, but mRNA PVT1 is Not Aberrantly Expressed in BLCA
Online data revealed that the transcription level of PVT1 in BLCA was significantly higher than that in normal tissues (P<0.05, Supplementary Figure 1A and B), while other data showed that this difference was not significant (P>0.05, Supplementary Figure 1A and C). The PVT1 transcriptional expression levels were not significantly different between BLCA stages (P>0.05, Supplementary Figure 1D). However, the promoter methylation level of PVT1 in BLCA was significantly lower than that in the normal tissues (P<0.05, Supplementary Figure 1E). Those online data could not support whether the transcriptional expression of PVT1 was abnormal or not in BLCA. Then, RT-PCR data showed no significant difference in PVT1 mRNA expression between tumor and normal tissues (P>0.05, Figure 1A). This finding was consistent in BLCA cell lines (563, T24, TCCSUP, JMSU1, HT1376, and RT112, those P>0.05, Figure 1D), but not in UMUC3 (P < 0.05, Figure 1D). Because circPVT1 originates from the circularization of the exon 2 of PVT1 gene, we detected whether circPVT1 was abnormally expressed or not. Interestingly, circPVT1 was highly expressed in BLCA, according to that in normal tissues (P<0.001, Figure 1B). These results are also consistent within BLCA cell lines (all P < 0.05, Figure 1E). We found that the whole expression of cicrPVT1 and mRNA PVT1 was significantly higher in BLCA (P<0.001, Figure 1C). The association between cicrPVT1 and PVT1 mRNA was obvious in BLCA tissues (R2=0.6, P<0.001, Figure 1G), but not in normal tissues (P>0.05, Figure 1F). These data also showed that circPVT1 expression might be a better diagnostic factor (AUC=0.93 ±0.037) than mRNA PVT1 (AUC=0.71±0.081, Figure 1H). In T24 and UMUC3 cell lines, RNase R treatment demonstrated that circPVT1 was more stable than PVT1 mRNA (Figure 1I and J). All these data indicate that circPVT1 is up-regulated and stable in BLCA and might serve as a potential diagnostic biomarker.
High circPVT1 Expression is Associated with Better Survival and HER2-Negative
A high PVT1 mRNA expression was significantly correlated with a poor overall survival (P<0.05, supplementary Figure 2A) in the GEPIA database, but not in the Oncolnc database (P=0.12, supplementary Figure 2B). The PVT1 mRNA expression was not associated with gender (P=0.095, supplementary Figure 2C), race (P=0.076, supplementary Figure 2D), smoking habit (P=0.2, supplementary Figure 2E), body weight (P=0.75, supplementary Figure 2F), suggesting that mRNA PVT1 expression was not associated with overall survival.
Therefore, we investigated the clinical value of circPVT1 expression in BLCA. A total of 162 BLCA sample tissues were collected and detected by qRT-PCR method. Based on a cutoff value 38.3, circPVT1 was determined to be highly expressed circPVT1 in 112 samples and lowly expressed in 50 samples. The statics results from Table 1 reveal that circPVT1 expression was significantly correlated with TNM stage and HER2 amplification status (P < 0.05), not with age, gender, T stage, and N stage (P < 0.05), indicating that circPVT1 was a potential BLCA suppressor.
circPVT1 is an Independent Prognostic Factor for Overall Survival (OS) in BLCA
The univariate COX analysis showed that circPVT1 expression, TNM stage, and HER2 status were significantly associated with OS (Table 2). The multivariate analysis revealed that only circPVT1 was significantly associated with OS (Table 2). Besides, we also found that HER2 status was a prognostic factor. Representative images of HER2 staining by IHC and FISH are shown in Figure 2A–D. The HER2 negative amplification (The average HER2/CEP17 ratio of is 0.96, Figure 2A) and positive amplification (The average HER2/CEP17 ratio of is 5.86, Figure 2B) were detected by FISH at the DNA level. The HER2 negative amplification (Figure 2C) and positive amplification (Figure 2D) were detected by IHC at the protein level. Kaplan–Meier curve showed that HER2 positive amplification was significantly associated with a shorter survival time and a lower survival rate (Figure 2E) and that circPVT1 high expression was associated with a longer survival time and a higher survival rate (Figure 2F). Taking circPVT1 expression and HER2 amplification for consideration, three pairwise comparisons were considerably different (Figure 2G). A nomogram was constructed based on circPVT1 expression, HER2 amplification, and clinical characteristics (Figure 3A). This model was efficient to predict the 3-year (Figure 3B) or 5-year OS (Figure 3C). These findings indicate that circPVT1 might be an independent prognostic factor for OS.
 |
Table 2 Univariate and Multivariate Cox Regression Analyses of Prognostic Factors in Bladder Cancer |
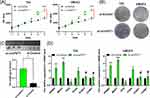
Knocking Down circPVT1 Promotes BLCA Proliferation in vitro and in vivo
To unravel biological function of circPVT1 in BLCA, we knocked down circPVT1 expression in T24 and UMUC3 cells by small interfering RNA. The CCK-8 assays showed that the viability of both T24 and UMUC3 cell lines was significantly higher in the si-circPVT1 group than in the si-Control group (Figure 4A). Colony formation assays showed that more colonies of both T24 and UMUC3 cell lines were formed in the si-circPVT1 group than in the si-Control group (Figure 4B). The mouse tumor model also demonstrated that the weight in the si-circPVT1 group was significantly larger than that in the si-Control group (Figure 4C). These data indicate that knocking down circPVT1 promotes BLCA proliferation in vitro and in vivo. qRT-PCR revealed that the ERBB2, KI67, and PCNA expression levels were significantly higher, and the TP53 expression in the si-circPVT1 group was significantly lower than those in the si-Control group, while those in CCND1 and CCNB1 showed no between-group differences (Figure 4D). These data indicated that knocking down circPVT1 promoted ERBB2, KI67, and PCNA expression and inhibited TP53 expression.
Discussion
Cancer-related circRNAs, such as circRNA-ST6GALNAC6, circRNA hsa_circ_0014130, and CircRNA 001418, have slipped into the research hotspot.9–11 Our study revealed that circPVT1 was significantly up-regulated in BLCA, while the expression of mRNA PVT1 did not change. Moreover, circPVT1 was, but mRNA PVT1 was not significantly correlated with OS. This indicated a higher clinical value of circPVT1 than mRNA PVT1.
circPVT1 is more easily detectable in tissues than mRNA PVT1, because of RNase R digestion. Our data revealed that circPVT1 was negatively correlated with HER2 amplification and TNM stage. HER2 is not a specific marker for BLCA. What is more, circPVT1 was an independent prognostic biomarker. Experimental data further validated that circPVT1 was a cancer suppressor. These results are different from some previous reports showing that a high circPVT1expression is correlated with unfavorable prognosis in cancer patients. In vivo and in vitro experiments uncovered that knocking down circPVT1 promoted the proliferation of BLCA by upregulating ERBB2, KI67, and PCNA and downregulating TP53. ERBB2 plays a vital role in promoting cancer proliferation.12,13 KI67 and PCNA can mark the proliferation of cancer cells.14 TP53 is well known to inhibit cancer cell proliferation.15 Our data demonstrate that circPVT1 can inhibit the proliferation of BLCA. However, some studies have reported that circPVT1 promotes the proliferation of other cancers, including lung cancer,16–18 thyroid cancer,19,20 and gallbladder cancer.21 These suggest the dual role of circPVT1 in cancer development, either as a driver or a suppressor. Many miRNAs can be targeted by circPVT1, such as miR-30e,22 miR-21-5p,23 miR‑429.18 As we all know, co-expression of circRNAs and miRNAs is spatiotemporal and dynamic. Therefore, we speculate that the function of circPVT1 may change with its combination with various targets.
The discovery of prognostic factors plays a vital role in the treatment of bladder cancer. In addition to the traditional pathological staging to evaluate prognosis, new prognosis factors are gradually explored and discovered. For example, the factor of the patient’s nutritional status,24 the factor of the systemic immune inflammation index,25 the factor of neutral granulocytes to lymphocytes in the urinary tract cancer in the bladder patients,26 and so on. Here, we find that circPVT1 acts as a novel biomarker and reveal its molecular function, which enriches the content of the new molecular biomarker and the perception of its molecules. However, circPVT1 translated into clinical daily practice still go faraway, recent studies revealed that neoadjuvant and adjuvant following radical treatments and27 adjuvant immunotherapy28 have played a vital role in clinical studies so far. Those factors could be considered for further validating. Besides, we should consider other prognostic markers interfering with effects such as HGT129 and variant histologies.30
There are still several limitations that ought to be considered. First, the samples were limited, and Ta vs T1 vs MIBC should be classified. Second, an over-expression circPVT1 assay, more phenotype studies, HE sections and the IHC for targeted proteins should be performed. Third, the targets of circPVT1 were not evaluated. Therefore, our findings should be validated in a larger patient cohort. In addition, RNA pulldown, luciferase reporter assay, and rescue experiments are needed to clarify the molecular mechanism of circPVT1.
Conclusion
circPVT1 acts as a tumor suppressor and may be employed to predict the prognosis of BLCA.
Acknowledgments
The authors would like to thank the members at the Department of Pathology, Wuxi People’s Hospital Affiliated to Nanjing Medical University and Cancer Drug Resistance Research Laboratory, Wuxi Medical College, Jiangnan University.Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by Top Talent Support Program for young and middle-aged people of Wuxi Health Committee (HB2023004) and Wuxi Health Committee (Z202325).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lobo N, Afferi L, Moschini M, et al. Epidemiology, screening, and prevention of bladder cancer. Eur Urol Oncol. 2022;5(6):628–639. doi:10.1016/j.euo.2022.10.003
2. Al Saidi I, Mohamedabugroon A, Sawalha A, et al. Epidemiology of bladder cancer in the Arab World: 2019 global burden of disease data. Asian Pac J Cancer Prev. 2022;23(9):2907–2919. doi:10.31557/APJCP.2022.23.9.2907
3. Xie J, Jiang H, Zhao Y, et al. Prognostic and diagnostic value of circRNA expression in prostate cancer: a systematic review and meta-analysis. Front Oncol. 2022;12:945143. doi:10.3389/fonc.2022.945143
4. Zhu G, Chang X, Kang Y, et al. CircRNA: a novel potential strategy to treat thyroid cancer (Review). Int J Mol Med. 2021;48(5). doi:10.3892/ijmm.2021.5034
5. Liang Y, Liu N, Yang L, et al. A brief review of circRNA biogenesis, detection, and function. Curr Genomics. 2021;22(7):485–495. doi:10.2174/1389202922666210331130722
6. Lin Z, Tang X, Wang L, et al. Prognostic and clinicopathological value of circPVT1 in human cancers: a meta-analysis. Cancer Rep. 2021;4(5):e1385. doi:10.1002/cnr2.1385
7. Ghafouri-Fard S, Khoshbakht T, Taheri M, et al. A concise review on the role of CircPVT1 in tumorigenesis, drug sensitivity, and cancer prognosis. Front Oncol. 2021;11:762960. doi:10.3389/fonc.2021.762960
8. Zhang H, Ren G, Wang X, et al. HER-2 gene amplification by fluorescence in situ hybridization (FISH) compared with immunohistochemistry (IHC) in breast cancer: a study of 528 equivocal cases. Breast Cancer Res Treat. 2012;134(2):743–749. doi:10.1007/s10549-012-2101-x
9. Wang L, Wu S, He H, et al. CircRNA-ST6GALNAC6 increases the sensitivity of bladder cancer cells to erastin-induced ferroptosis by regulating the HSPB1/P38 axis. Lab Invest. 2022;102(12):1323–1334. doi:10.1038/s41374-022-00826-3
10. Li G, Guo BY, Wang HD, et al. CircRNA hsa_circ_0014130 function as a miR-132-3p sponge for playing oncogenic roles in bladder cancer via upregulating KCNJ12 expression. Cell Biol Toxicol. 2022;38(6):1079–1096. doi:10.1007/s10565-021-09668-z
11. Peng G, Meng H, Pan H, et al. CircRNA 001418 promoted cell growth and metastasis of bladder carcinoma via EphA2 by miR-1297. Curr Mol Pharmacol. 2021;14(1):68–78. doi:10.2174/1874467213666200505093815
12. Grote I, Bartels S, Christgen H, et al. ERBB2 mutation is associated with sustained tumor cell proliferation after short-term preoperative endocrine therapy in early lobular breast cancer. Mod Pathol. 2022;35(12):1804–1811. doi:10.1038/s41379-022-01130-7
13. Friedrich RE, Nornberg LKN, Hagel C. ERBB2 and ERBB3 growth factor receptors, neuregulin-1, CD44 and Ki-67 proliferation index in neurofibromatosis type 1-associated peripheral nerve sheath tumors. Anticancer Res. 2022;42(5):2327–2340. doi:10.21873/anticanres.15712
14. Jayaraman S, Pazhani J, Priyaveeraraghavan V, et al. PCNA and Ki67: prognostic proliferation markers for oral cancer. Oral Oncol. 2022;130:105943. doi:10.1016/j.oraloncology.2022.105943
15. Velez MG, Kosiorek HE, Egan JB, et al. Differential impact of tumor suppressor gene (TP53, PTEN, RB1) alterations and treatment outcomes in metastatic, hormone-sensitive prostate cancer. Prostate Cancer Prostatic Dis. 2022;25(3):479–483. doi:10.1038/s41391-021-00430-4
16. Shi J, Lv X, Zeng L, et al. CircPVT1 promotes proliferation of lung squamous cell carcinoma by binding to miR-30d/e. J Exp Clin Cancer Res. 2021;40(1):193. doi:10.1186/s13046-021-01976-w
17. Huang M, Li T, Wang Q, et al. Silencing circPVT1 enhances radiosensitivity in non-small cell lung cancer by sponging microRNA-1208. Cancer Biomark. 2021;31(3):263–279. doi:10.3233/CBM-203252
18. Cao L, Zhou X, Ding X, et al. Knockdown of circ‑PVT1 inhibits the progression of lung adenocarcinoma and enhances the sensitivity to cisplatin via the miR‑429/FOXK1 signaling axis. Mol Med Rep. 2021;24(4). doi:10.3892/mmr.2021.12323
19. Zheng X, Rui S, Wang XF, et al. circPVT1 regulates medullary thyroid cancer growth and metastasis by targeting miR-455-5p to activate CXCL12/CXCR4 signaling. J Exp Clin Cancer Res. 2021;40(1):157. doi:10.1186/s13046-021-01964-0
20. Zeng L, Yuan S, Zhou P, et al. Circular RNA Pvt1 oncogene (CircPVT1) promotes the progression of papillary thyroid carcinoma by activating the Wnt/beta-catenin signaling pathway and modulating the ratio of microRNA-195 (miR-195) to vascular endothelial growth factor A (VEGFA) expression. Bioengineered. 2021;12(2):11795–11810. doi:10.1080/21655979.2021.2008639
21. Wang S, Su TT, Tong H, et al. CircPVT1 promotes gallbladder cancer growth by sponging miR-339-3p and regulates MCL-1 expression. Cell Death Discov. 2021;7(1):191. doi:10.1038/s41420-021-00577-y
22. Jia Y, Gu W. Up-regulation of circPVT1 in T cell acute lymphoblastic leukemia promoted cell proliferation via miR-30e/DLL4 induced activating NOTCH signaling. Pathol Res Pract. 2021;224:153536. doi:10.1016/j.prp.2021.153536
23. Hao Y, Lu C, Zhang B, et al. CircPVT1 up-regulation attenuates steroid-induced osteonecrosis of the femoral head through regulating miR-21-5p-mediated Smad7/TGFbeta signalling pathway. J Cell Mol Med. 2021;25(10):4608–4622. doi:10.1111/jcmm.16294
24. Claps F, Mir MC, Van Rhijn BWG, et al. Impact of the controlling nutritional status (CONUT) score on perioperative morbidity and oncological outcomes in patients with bladder cancer treated with radical cystectomy. Urol Oncol. 2023;41(1):49 e13–49 e22. doi:10.1016/j.urolonc.2022.09.023
25. Grossmann NC, Schuettfort VM, Pradere B, et al. Impact of preoperative systemic immune-inflammation Index on oncologic outcomes in bladder cancer patients treated with radical cystectomy. Urol Oncol. 2022;40(3):106 e11–e19. doi:10.1016/j.urolonc.2021.10.006
26. Von Deimling M, Schuettfort VM, D’Andrea D, et al. Predictive and prognostic role of the neutrophil-to-lymphocyte ratio in muscle invasive bladder cancer treated with neoadjuvant chemotherapy and radical cystectomy. Clin Genitourin Cancer. 2023;21(4):430–441. doi:10.1016/j.clgc.2023.01.008
27. Claps F, Mir MC, Zargar H. Molecular markers of systemic therapy response in urothelial carcinoma. Asian J Urol. 2021;8(4):376–390. doi:10.1016/j.ajur.2021.05.001
28. Mir MC, Campi R, Loriot Y, et al. Adjuvant systemic therapy for high-risk muscle-invasive bladder cancer after radical cystectomy: current options and future opportunities. Eur Urol Oncol. 2022;5(6):726–731. doi:10.1016/j.euo.2021.04.004
29. Lopez-Beltran A, Blanca A, Cimadamore A, et al. T1 bladder carcinoma with variant histology: pathological features and clinical significance. Virchows Arch. 2022;480(5):989–998. doi:10.1007/s00428-021-03264-6
30. Claps F, Van De Kamp MW, Mayr R, et al. Prognostic impact of variant histologies in urothelial bladder cancer treated with radical cystectomy. BJU Int. 2023;132(2):170–180. doi:10.1111/bju.15984
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.