Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 19
Analysis of Key Genes and miRNA-mRNA Networks Associated with Glucocorticoids Treatment in Chronic Obstructive Pulmonary Disease
Authors Wu JJ , Zhang PA , Chen MZ, Zhang Y, Du WS , Li XN, Ji GC, Jiang LD, Jiao Y, Li X
Received 23 September 2023
Accepted for publication 21 February 2024
Published 28 February 2024 Volume 2024:19 Pages 589—605
DOI https://doi.org/10.2147/COPD.S441716
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Jian-Jun Wu,1 Ping-An Zhang,1 Ming-Zhe Chen,2 Yi Zhang,1 Wei-Sha Du,1 Xiao-Ning Li,1 Guo-Chao Ji,1 Liang-Duo Jiang,3 Yang Jiao,4,* Xin Li5,*
1Respiratory Department, The Third Affiliated Hospital, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 2Infectious Disease Department, Henan Provincial Hospital of Traditional Chinese Medicine, Zhengzhou, Henan, People’s Republic of China; 3Respiratory Department, Dongzhimen Hospital, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 4Respiratory Department, Dongfang Hospital, Beijing University of Chinese Medicine, Beijing, People’s Republic of China; 5Glaucoma Department, Eye Hospital, China Academy of Chinese Medical Sciences, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xin Li, Glaucoma Department, Eye Hospital, China academy of Chinese Medical Sciences, Beijing, People’s Republic of China, Email [email protected]
Background: Some patients with chronic obstructive pulmonary disease (COPD) benefit from glucocorticoid (GC) treatment, but its mechanism is unclear.
Objective: With the help of the Gene Expression Omnibus (GEO) database, the key genes and miRNA-mRNA related to the treatment of COPD by GCs were discussed, and the potential mechanism was explained.
Methods: The miRNA microarray dataset (GSE76774) and mRNA microarray dataset (GSE36221) were downloaded, and differential expression analysis were performed. Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed on the differentially expressed genes (DEGs). The protein interaction network of the DEGs in the regulatory network was constructed with the STRING database, and the key genes were screened through Cytoscape. Potential downstream target genes regulated by differentially expressed miRNAs (DEMs) were predicted by the miRWalk3.0 database, and miRNA-mRNA regulatory networks were constructed. Finally, some research results were validated.
Results: ① Four DEMs and 83 DEGs were screened; ② GO and KEGG enrichment analysis mainly focused on the PI3K/Akt signalling pathway, ECM receptor interaction, etc.; ③ CD2, SLAMF7, etc. may be the key targets of GC in the treatment of COPD; ④ 18 intersection genes were predicted by the mirwalk 3.0 database, and 9 pairs of miRNA-mRNA regulatory networks were identified; ⑤ The expression of miR-320d-2 and TFCP2L1 were upregulated by dexamethasone in the COPD cell model, while the expression of miR-181a-2-3p and SLAMF7 were downregulated.
Conclusion: In COPD, GC may mediate the expression of the PI3K/Akt signalling pathway through miR-181a-2-3p, miR-320d-2, miR-650, and miR-155-5p, targeting its downstream signal factors. The research results provide new ideas for RNA therapy strategies of COPD, and also lay a foundation for further research.
Keywords: COPD, GC, GEO database, miRNA-mRNA, DEMs, DEGs
Introduction
Chronic obstructive pulmonary disease (COPD) is a chronic inflammatory disease of the airway that is characterized by chronic inflammation of the airway, lung parenchyma, and pulmonary vessels. Alveolar macrophages, T lymphocytes (especially CD8+), and neutrophils are increased in different parts of the lung, and eosinophils are increased in some patients.1
Among the drugs for the treatment of chronic airway inflammatory diseases, glucocorticoids (GCs) are recognized as the most rapid and potent anti-inflammatory drugs. They have achieved good clinical efficacy in the treatment of bronchial asthma2,3 and COPD.4–11 In patients with moderate to severe acute exacerbation of COPD (AECOPD), systemic use of GCs improves forced expiratory volume in one second (FEV1) and oxygenation status and shortens rehabilitation and hospitalization time.12 Compared with systemic GCs, inhaled corticosteroids (ICSs) have fewer adverse reactions and can replace or partially replace systemic GCs.13–15
However, some studies have shown that patients with COPD do not benefit from inhaled corticosteroid (ICS) therapy alone. Specifically, ICS treatment cannot effectively delay the long-term decline in FEV1 or reduce the mortality of patients with COPD.16,17 GCs resistance caused by HDAC2 activity decreases, dysfunction of corticosteroid glucocorticoid receptor-α and GR-β increases in epithelial cells, etc., may be a reason why COPD patients cannot benefit from GC treatment.18–20 Recent studies have shown that microRNAs are associated with GCs resistance in COPD.21
MicroRNAs (miRNAs) are a kind of noncoding single-stranded RNA that are conserved and widely distributed in eukaryotes. The size is approximately 19~25 nucleotides, which mainly plays a posttranscriptional regulatory role by inhibiting the expression or translation of target genes.22,23 Studies have shown that multiple miRNAs are involved in the occurrence and development of COPD.24–30 In patients with moderate to severe COPD, short-term and long-term treatment with ICS can affect the expression of miRNAs.31 However, there are few reports on the molecular mechanism of miRNA-mediated COPD, especially in the treatment of COPD by GCs. The regulatory mechanism and molecular network of GC-mediated miRNAs regulating mRNA expression and inhibiting the COPD inflammatory response are not clear. This study intended to mine key genes and construct a miRNA-mRNA regulatory network by analysing miRNA and mRNA expression microarray datasets related to GC treatment of COPD in the GEO database. To explore its mechanism of action and provide an important theoretical reference and scientific basis for GC treatment of COPD.
Materials and Methods
miRNA and mRNA Microarray
The microarray data of the miRNA and mRNA expression profiles related to GC treatment of COPD were retrieved from the GEO database of NCBI. The screening criteria were as follows: ① patients with COPD; ② the experimental group was treated with GCs (ICS or oral GCs), and the control group was treated with placebo. Finally, the miRNA microarray dataset GSE76774 and mRNA microarray dataset GSE36221 that met the requirements were downloaded. Both studies were derived from the GLUCOLD study in the Netherlands. Based on the GPL8786 platform (Affymetrix multispecies miRNA-1 array), GSE76774 included 69 patients with moderate and severe COPD (smokers or ex-smokers). Bronchopulmonary biopsy was performed before treatment (22 cases in the treatment group and 23 cases in the control group), 6 months (17 cases in the treatment group and 24 cases in the control group), and 30 months (17 cases in the treatment group and 20 cases in the control group) after treatment. Based on the GPL6244 platform (Affymetrix Human Gene 1.0 ST Array), GSE36221 included 90 patients with moderate and severe COPD (smokers or ex-smokers). Bronchopulmonary biopsy was performed before treatment (19 cases in the treatment group and 21 cases in the control group), 6 months (23 cases in the treatment group and 17 cases in the control group), and 30 months (19 cases in the treatment group and 17 cases in the control group) after treatment. The database or software involved in the research is shown in Table 1.
 |
Table 1 Database or Software Involved in the Research Design |
Methods
Data Processing and Differential Expression Analysis
The microarray data of miRNA and mRNA expression profiles before treatment and 6 months and 30 months after treatment were analysed by the limma package of R software. The screening criteria before treatment were set as p > 0.05; the screening criteria after 6 months and 30 months of treatment were set as p < 0.05 and | log2-fold change (FC) | > 0.3. The differentially expressed data were visualized by a volcano map and cluster map.
Functional Enrichment Analysis of DEGs
Using the clusterProfiler package in R software, the DEGs analysed by GSE36221 were analysed by Gene Ontology (GO) and pathway enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG). Among them, GO analysis included biological process (BP), cellular component (CC), and molecular function (MF), with P < 0.05.
Construction of the DEG Protein Interaction Network
To further identify the relationship between different genes, the STRING (search tool for the retrieval of interacting genes) database was used for protein–protein interactions (PPIs), and the confidence score was set to > 0.4. The PPI network was visualized by Cytoscape software. The key genes were identified through the “cytohubba” module. The data were imported from the network result file obtained from the string database, and the degree algorithm was selected to obtain the top 25 key genes.
Prediction of miRNA Target Genes and Construction of the miRNA-mRNA Regulatory Network
The miRWalk Database was used to predict the target genes of differentially expressed miRNAs (DEMs), and the intersection between the predicted target genes and DEGs analysed by GSE36221 was obtained by Venny 2.1. According to the regulatory relationship between miRNAs and mRNAs, the relationship between miRNAs and mRNAs was clarified. The miRNA-mRNA regulatory network was visualized by Cytoscape software.
Cell Experiment
To verify the experimental results, miR-320d-2, miR-181a-2-3p, TFCP2L1, and SLAMF7 were randomly selected for validation using a COPD cell model.
Materials
BEAS-2B human bronchial epithelial cells were purchased from Beina Biotechnology Co., Ltd. (BNCC359274), Daqianmen cigarettes were purchased from Shanghai Tobacco Co., Ltd., TFCP2L1 antibody, SLAMF7 antibody were purchased from Proteintech Co., Ltd.
Method
Cell culture After the cell concentration was adjusted to 5×104/mL by complete medium, the cells were uniformly inoculated on 96-well plates at 100ul/ well, and cultured at 37°C and 5%CO2 for 24h.
Cigarette Smoke Extract (CSE) Preparation
One unfiltered burning cigarette (consisting of 12 mg of tar, 0.9 mg of nicotine, 14 mg of carbon monoxide) was connected to a 50mL syringe through a rubber hose. 300mL of smoke was collected through continuous negative pressure suction. The smoke was then injected into an airtight container containing 25mL DMEM of high-sugar medium and thoroughly mixed. Then 1mol/L of NaOH was used to adjust the PH of the mixture to 7.4, and finally 0.22um filter was used to remove bacteria. The mixture at this point was defined as 100% CSE solution. The optical density value of CSE solution was detected with A UV spectrophotometer at the wavelength of 320nm, so that the optical density value of different batches was about 0.2.
CSE Concentration Screening
BEAS-2B cells were incubated with 3%, 5%, 8%, and 10% CSE solutions for 12, 24, and 48 hours, respectively. 0% CSE was used as the control group. Each group has 6 replicates. CCK8 solution was added with a dose of 10 μL/well and incubated for 1–3 hours. Finally, the OD value was measured at 450nm on the enzyme marker. According to the results, 5% CSE was selected for the next experiment.
Dexamethasone Concentration Screening
BEAS-2B cells were incubated with 10−10mol/L, 10−9mol/L, 10−8mol/L, 10−7mol/L and 10−6mol/L dexamethasone solutions for 12, 24, and 48 hours, respectively. 0% dexamethasone was used as the control group. The remaining experimental methods were the same as before. According to the results, 10−8mol/L dexamethasone was selected for the next experiment.
Cell Grouping and Medication
Cell was divided into control group (Control), model group (Model) and dexamethasone group (DXM). The control group was cultured in complete medium (90% DMEM+10% FBS+1% dual antibody) for 24 hours. The model group was cultured with 5%CSE for 24h. The dexamethasone group was cultured with 5%CSE for 12h, and then cultured with 10−8mol/L dexamethasone for 12h.
Western Blot
The protein was extracted by the Protein Extraction Kit, and then quantified by the BCA method. The expression levels of TFCP2L1 and SLAMF7 in cells were detected by the WB method. The internal reference was GAPDH. The experiment was repeated three times.
RT-PCR
Total RNA of each group was extracted by Trizol reagent, then reverse transcribed into cDNA and amplified by PCR. Amplification program: 95 °C for 5 minutes, followed by 95 °C for 10 seconds, continued at 60 °C for 30 seconds, and finally 72 °C for 30 seconds, a total of 35 cycles. The expression levels of miR-320d-2, miR-181a-2-3p, TFCP2L1, and SLAMF7 were calculated by 2 −ΔΔCT method. The primer sequences are shown below: miR320d-2 Forward: AAAAGCTGGGTTGAGAGGA; miR181a-2-3P Forward: ACCACCGACCGTTGACTGTACC; U6-F Forward: CTCGCTTCGGCAGCACA, U6-R Reverse: AACGCTTCACGAATTTGCGT; SLAMF7 Forward: CAAGGTGTTCAAGCCGAAGG, SLAMF7 Reverse: GAGCACTCTGTGAGGATGGT; TFCP2L1 Forward: CTTGTCACCATACAGCCAGAAG; TFCP2L1 Reverse: ACATGCAGCACGTACTCCT; GAPDH Forward: CCTCTGACTTCAACAGCGACAC; GAPDH Reverse: TGGTCCAGGGGTCTTACTCC.
Statistical Analysis
The data was statistically analyzed by SPSS 25.0 and visualized by GraphPad Prism 8.0. P<0.05 indicated a statistically significant difference.
Results
DEMs
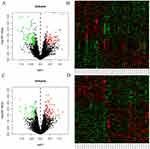
Before treatment, there were 816 miRNAs with no difference between the two groups. After six months of treatment, compared with the control group, there were 31 DEMs in the treatment group (10 upregulated and 21 downregulated). See Figure 1A and B for a volcano diagram and cluster diagram. After 30 months of treatment, compared with the control group, there were 23 DEMs in the treatment group (13 upregulated and 8 downregulated). See Figure 1C and D for volcano diagram and cluster diagram. Venny 2.1 was used to obtain DEMs with sustained, co directional,and differential expression. Four DEMs were ultimately obtained, including one upregulated expression (miR-320d) and three downregulated expression (miR-650, miR-155-5p, miR-181a-3p), as shown in Table 2.
 |
Table 2 Continuously Express miRNAs in the Same Direction |
DEGs
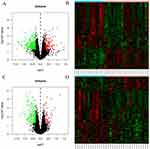
Before treatment, there were 22,666 mRNAs with no difference between the two groups. After six months of treatment, compared with the control group, there were 387 DEGs in the treatment group (132 upregulated and 255 downregulated). See Figure 2A and B for a volcano diagram and cluster diagram. After 30 months of treatment, compared with the control group, there were 279 DEGs in the treatment group (55 upregulated and 224 downregulated). See Figure 2C and D for volcano diagram and cluster diagram. Venny 2.1 was used to obtain DEGs with sustained, co directional, and differential expression. 83 DEGs (20 were upregulated and 63 were downregulated) were ultimately obtained. The main DEGs are shown in Table 3.
 |
Table 3 Main DEGs |
DEGs Function Analysis Results
The GO and KEGG pathway enrichment analyses of 83 DEGs were carried out by the clusterProfiler package of R software. A total of 218 GO enrichment entries were obtained, including 66 BP entries, 5 CC entries, and 11 MF entries. BP was mainly enriched in the production of molecular mediator of immune response; positive regulation of cell activation; immunoglobulin production; positive regulation of leukocyte activation; B-cell activation; lymphocyte differentiation; immune response-regulating cell surface receptor signalling pathway; mononuclear cell differentiation, etc. CC was mainly enriched in immunoglobulin complex; external side of plasma membrane; immunoglobulin complex, circulating; blood microparticle, etc. MF was mainly enriched in antigen binding; immunoglobulin receptor binding; immunoglobulin binding; glycosaminoglycan binding; integrin binding, etc. See Figure 3 for the first five BP, CC, and MF entries. KEGG pathway enrichment analysis identified four signalling pathways: PI3K/Akt signalling pathway; ECM receptor interaction; primary immunodeficiency; and adhesive spots (Figure 4).
 |
Figure 3 Bar chart of GO enrichment analysis of DEGs. |
 |
Figure 4 Bar chart of KEGG enrichment analysis of DEGs. |
Construction of the DEG PPI Network and Screening of Key Genes
The string platform was used to construct PPI networks for 83 DEGs, as shown in Figure 5. The results show that 58 node proteins (nodes) play a direct or indirect role in the treatment of COPD with GC, involving a total of 59 interactions (edges). Cytoscape was used for network topology analysis. According to the screening conditions, key genes such as CD2, KIT, FCER1A, SLAMF1, SLAMF 7, GPX3, and PRELP were obtained, as shown in Figure 6.
 |
Figure 5 DEGs PPI network. |
 |
Figure 6 Key genes. |
MiRNA Target Gene Prediction and Regulatory Network Construction
The miRWalk database was used to predict the downstream targets of mRNA, and a total of 6378 genes were obtained. A total of 18 overlapping genes were obtained from the intersection with the above 83 DEGs, as shown in Table 4. According to the negative regulatory relationship between miRNA and mRNA, nine pairs of miRNA-mRNA pairs were finally selected. The miRNA-mRNA regulation network was constructed and visualized by Cytoscape software, as shown in Figure 7. The flow chart of the whole research method and results are shown in Figure 8.
 |
Table 4 Intersection Genes of miRNA Target Genes and DEGs |
 |
Figure 7 miRNA-mRNA regulatory networks. |
 |
Figure 8 Flow chart. |
Cell Experiment
We determined the expression of SLAMF7 and TFCP2L1 using WB, and the expression of SLAMF7, TFCP2L1, miR-320d-2 and miR-181a-2-3p using PCR The study showed that the expression of miR-320d-2 and TFCP2L1 were low in the COPD group, and the expression of miR-181a-2-3p and SLAMF7 were high in the COPD group. After dexamethasone treatment, the expression of miR-320d-2 and TFCP2L1 were upregulated, while the expression of miR-181a-2-3p and SLAMF7 were downregulated (Figures 9 and 10). The results of the cell experiment were consistent with previous studies.
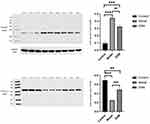 |
Figure 9 WB of Partial miRNA-mRNA. **, ##P value<0.01, ***, ###P value<0.001. |
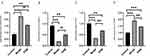 |
Figure 10 PCR of Partial miRNA-mRNA (A) miR-181a-2-3p (B) miR-320d-2 (C) SLAMF7 (D) TFCP2L1. **, ##P value<0.01, ***, ###P value<0.001. |
Discussion
In this study, 83 DEGs (such as CD2, KIT, FCER1A, SLAMF1, SLAMF7, and NMI), 4 DEMs (miR-320d, miR-155-5p, miR-650, and miR-181a-3p) and 9 miRNA-mRNA regulatory pairs (such as miR-320d-SLAMF1, miR-320d-SLAMF7, miR-181a-2-3p-TFCP2L1) related to the treatment of COPD by GCs were screened, and a related biological pathway, the PI3K/Akt signalling pathway, was obtained (Figure 11).
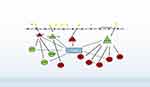 |
Figure 11 Key genes and miRNA-mRNA networks associated with glucocorticoids treatment in COPD. |
This study found that miRNAs such as miR-320d, miR-155-5p, miR-650, and miR-181a-2-3p changed continuously during the treatment of COPD by GCs, in which the expression of miR-650, miR-155, and miR-181a-3p were downregulated and the expression of miR-320d was upregulated. MiR-155 and miR-320d are associated with the inflammatory response24,25 and oxidative stress32 in COPD. Clinical studies have shown that compared with nonsmokers, the expression of miR-155 in lung tissue25 and induced sputum24 of patients with COPD. The expression of miR-155 increased in lung tissue,25 alveolar macrophages,25 and bronchoalveolar lavage fluid24 of mice exposed to cigarette smoke (CS). However, the inflammatory cells, cytokines, and chemokines in the bronchoalveolar lavage (BAL) of miR-155 knockout mice exposed to CS decreased, and lung inflammation induced by CS decreased significantly. Moreover, this inflammation was alleviated by a nasal drip of a specific miR-155 inhibitor.25 miR-155 is also associated with oxidative stress in COPD.33 In model rats of COPD, the effective components of the Bufei Yishen recipe can inhibit oxidative stress induced by PM2.5 through the miR-155/FOXO3a pathway.33 Clinical studies have also confirmed that miR-320d is related to the inflammatory response in COPD. Compared with nonsmokers, mir-320d is upregulated in plasma-derived exosomal RNA in patients with COPD.32 MiR-320d is associated with the inflammatory response of the airway epithelium in COPD.31 It has been demonstrated that GCs play an anti-inflammatory role in COPD31 and asthma34 through miR-155 and miR-320d. MiR-320d is a novel ICS mediator that regulates the proinflammatory response of the COPD airway epithelium.31 MiR-155 is the target of GCs in the treatment of allergic asthma. The anti-inflammatory effect of GCs can be reversed by inhibiting the expression of miR-155.34 However, whether miR-155 has the same effect in theanti-inflammatory treatment of COPD remains to be confirmed. GCs may affect the inflammatory response and oxidative stress of COPD by regulating expression of multiple miRNAs, such as miR-155 and miR-320d, but its specific intervention mechanism is unknown.
This study found that DIO2, TFCP2L1, DCLK1, GALNT15, PER1, SLAMF7, PRELP, SLAMF1, TSC22D3, NMI, METTL7A, MS4A2, FAM107A, IL33, CD96, GPX3, TIMP4, APOD, and other key genes were continuously changed during COPD treatment by GCs, in which the expression of DIO2, DCLK1, SLAMF7, SLAMF1, NMI, MS4A2, IL33, and CD96 were downregulated, and the expression of TFCP2L1, GALNT15, PER1, PRELP, TSC22D3, METTL7A, FAM107A, GPX3, TIMP4, and APOD were upregulated. Studies have shown that GPX3,35,36 TSC22D337, DIO2,38 Per1,39,40 IL-33,41 and other genes are related to the inflammatory response and oxidative stress of COPD. The expression of GPX3 decreased in HBE cells of COPD,35 lung tissue of patients with severe emphysema,36 and lung tissue of patients with advanced COPD.35 In the treatment of COPD, PPARγ agonists may play an antioxidant role by blocking the downregulation of GPX3. Glucocorticoid-induced leucine zipper (TSC22D3) is a regulator of GC anti-inflammatory effects.37 Beclomethasone-17-monopropionate (17-BMP) can upregulate the expression of TSC22D3 in alveolar macrophages of COPD patients and smokers.42 TSC22D3 can interact with multiple pathways, including nuclear factor-κB (NF-κB), activator protein 1 (AP-1), Raf-1, and Ras.37 GC-induced leucine zipper (TSC22D3) can inhibit the transcriptional response of NF-κB and AP-1,43 and its mechanism is related to the increase in TSC22D3 expression induced by GCs through transactivation and inhibition of NF-κB signalling, resulting in proinflammatory cytokine release from airway epithelial cells.44 Per1 is an important part of the biological molecular oscillation system. Compared with nonsmokers, the level of Per1 in lung tissue and PBMCs in patients with COPD increased. The oscillation and dysregulated expression of biological clock genes such as BMAL1, Per1, and Per2 are related to the aetiology, chronic inflammation, and imbalance of autophagy levels in COPD.39,40 IL-33 may become a new target for COPD therapy.41 Elevated levels of IL-33 in lung biopsy samples, epithelial and endothelial cells, serum or plasma, and sputum in patients with COPD are associated with decreased lung function.41 Nonhaematopoietic lung cells are not only an important cell source ofIL-33 but also express ST2 and respond to IL-33 stimulation (regardless of source), thus participating in lung inflammation and tissue remodelling.45 The risk of COPD associated with the loss of function of IL-33 (rs146597587) is reduced. In contrast, the increased function of IL-33 and IL1RL1 variants increases the risk of COPD.46 Compared with placebo, itepekimab targeting IL-33 can provide COPD patients with previous smoking potential benefits in reducing the frequency of exacerbation and improving lung function.41
In this study, GO and KEGG analyses were carried out for the screened DEGs. The results showed that the effect of GCs on COPD was mainly related to the production of molecular mediators of the immune response, positive regulation of cell activation, the PI3K/Akt signalling pathway, and so on. The therapeutic effect of GCs on COPD may be related to the intervention of the PI3K/Akt pathway and regulation of its downstream target genes through miR-155 and miR-320d. Although this hypothesis has not been confirmed in COPD, miRNAs have been widely confirmed to target and regulate their downstream genes by intervening in the PI3K/Akt pathway. Multiple miRNAs, such as miR-155,30 miR-320d,47 miR-650,48 and miR-181,49,50 can be used as upstream factors to regulate the PI3K/Akt pathway. The PI3K/Akt pathway is related to SLAMF7, NMI, TIMP-4, and other genes.48–50 MiR-155 plays a role through the PI3K/Akt pathway in an LPS intraperitoneal injection-induced sepsis mouse model.30 Curcumin inhibits the inflammatory response of macrophages in an LPS-induced sepsis mouse model, and its mechanism is related to curcumin interfering with the PI3K/Akt pathway through miR-155.30 The PI3K/Akt signalling pathway is related to SLAMF7, NMI, TIMP-4, and ETC.48–50 Studies have shown that SLAMF7 is expressed in LPS-stimulated monocytes via PI3K and NF-κB-dependent pathways and inhibits the production of proinflammatory cytokines, TNF-α and IL-12p70.51 SLAMF7 regulates the activation of NK cells by recruiting EAT-2 and activating the PI3K and phospholipase Cgamma signalling pathways in human NK cells.52,53
This study found that there were nine pairs of miRNA-mRNA related to the treatment of COPD by GCs: miR-320d-SLAMF1, miR-320d-SLAMF7, miR-320d-NMI, miR-181a-2-3p-TFCP2L1, miR-650-FAM107A, miR-650-GPX3, miR-650-PRELP, miR-650-TIMP4, and miR-650-APOD. The in vitro experiments also confirmed the above experimental results. These DEMs and DEGs are the key nodes of GCs in the treatment of COPD.
Although this study constructed a potential miRNA-mRNA regulatory network and key genes for GC treatment of COPD based on bioinformatics, there are certain limitations: ①The reports on miR-650 and miR-181a-2-3p mainly focus on tumours, but there are no reports in COPD, which needs further research. ②Patients with stable COPD mostly use ICS/LABA, and some patients can benefit from it. The effect of ICS/LABA on miRNA and mRNA needs to be further studied.
Conclusion
GCs may intervene the expression of PI3K/Akt signal pathway through mir-320d, miR-320d and mir-155-5p, and target its downstream signal factors, to play an anti-inflammatory role in the treatment of COPD.
Abbreviations
COPD, chronic obstructive pulmonary disease; GC, glucocorticoid; GO, Gene ontology; KEGG, Kyoto encyclopedia of genes and genomes.
Data Sharing Statement
The data supporting the findings of this study are available within the article.
Ethics Approval and Informed Consent
This research was reviewed and permitted by the Scientific research ethics committee of the Third Affiliated Hospital of Beijing University of Chinese Medicine.
Author Contributions
ALL authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The work was supported by National Natural Science Foundation of China (grant numbers 82274462), Beijing Municipal Natural Science Foundation (grant numbers 7232284) and The Fifth Batch of National Excellent Clinical Talents of Traditional Chinese Medicine Research Project (National Administration of Traditional Chinese Medicine Renjiaoshi [2022] No. 1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Brightling C, Greening N. Airway Inflammation in COPD: progress to precision medicine. Eur Respir J. 2019;54(2):1900651. doi:10.1183/13993003.00651-2019
2. McGeachie MJ, Sordillo JE, Dahlin A, et al. Expression of SMARCD1 interacts with age in association with asthma control on inhaled corticosteroid therapy. Respir Res. 2020;21(1):31. doi:10.1186/s12931-020-1295-4
3. Navanandan N, Moran E, Smith H, Hoch H, Mistry RD. Primary care provider preferences for glucocorticoid management of acute asthma exacerbations in children. J Asthma. 2021;58(4):547–553. doi:10.1080/02770903.2019.1709869
4. Zheng J-P, Zhang J, Ma L-J, et al. Clinical outcomes of using nebulized budesonide as the initial treatment for acute exacerbations of chronic obstructive pulmonary disease: a Post-Hoc analysis. Int J Chron Obstruct Pulmon Dis. 2019;14:2725–2731. doi:10.2147/COPD.S196615
5. Zhang J, Zheng J, Huang K, Chen Y, Yang J, Yao W. Use of glucocorticoids in patients with COPD exacerbations in China: a retrospective observational study. Ther Adv Respir Dis. 2018;12:1753466618769514. doi:10.1177/1753466618769514
6. Chronic Obstructive Pulmonary Disease Group of Chinese Thoracic Society; Chronic Obstructive Pulmonary Disease Committee of Chinese Association of Chest Physician. 中华医学会呼吸病学分会慢性阻塞性肺疾病学组, 中国医师协会呼吸医师分会慢性阻塞性肺疾病工作委员会. 慢性阻塞性肺疾病诊治指南(2021年修订版) [Guidelines for the diagnosis and management of chronic obstructive pulmonary disease (revised version 2021)]. Zhonghua Jie He He Hu Xi Za Zhi. 2021;44(3):170–205. Chinese. doi:10.3760/cma.j.cn112147-20210109-00031
7. Nannini LJ, Poole P, Milan SJ, Holmes R, Normansell R. Combined corticosteroid and long-acting beta₂-agonist in one inhaler versus placebo for chronic obstructive pulmonary disease. Cochrane Database Syst Rev. 2013;2013(11):CD003794. doi:10.1002/14651858.CD003794.pub4
8. Vestbo J, Leather D, Diar Bakerly N, et al. Effectiveness of fluticasone furoate-vilanterol for COPD in clinical practice. N Engl J Med. 2016;375(13):1253–1260. doi:10.1056/NEJMoa1608033
9. Lipson DA, Barnhart F, Brealey N, et al. Once-daily single-inhaler triple versus dual therapy in patients with COPD. N Engl J Med. 2018;378(18):1671–1680. doi:10.1056/NEJMoa1713901
10. Sonnex K, Alleemudder H, Knaggs R. Impact of smoking status on the efficacy of inhaled corticosteroids in chronic obstructive pulmonary disease: a systematic review. BMJ Open. 2020;10(4):e037509. doi:10.1136/bmjopen-2020-037509
11. Calverley PMA, Anderson JA, Brook RD, et al. Fluticasone furoate, vilanterol, and lung function decline in patients with moderate chronic obstructive pulmonary disease and heightened cardiovascular risk. Am J Respir Crit Care Med. 2018;197(1):47–55. doi:10.1164/rccm.201610-2086OC
12. 2023 GOLD Report. Available from: https://goldcopd.org/2023-gold-report-2/.
13. Ding Z, Li X, Lu Y, et al. A randomized, controlled multicentric study of inhaled budesonide and intravenous methylprednisolone in the treatment on acute exacerbation of chronic obstructive pulmonary disease. Respir Med. 2016;121:39–47. doi:10.1016/j.rmed.2016.10.013
14. Sun X, He Z, Zhang J, et al. Compare the efficacy of inhaled budesonide and systemic methylprednisolone on systemic inflammation of AECOPD. Pulm Pharmacol Ther. 2015;31:111–116. doi:10.1016/j.pupt.2014.09.004
15. Pleasants RA, Wang T, Xu X, et al. Nebulized corticosteroids in the treatment of COPD exacerbations: systematic review, meta-analysis, and clinical perspective. Respir Care. 2018;63(10):1302–1310. doi:10.4187/respcare.06384
16. Reddy AT, Lakshmi SP, Banno A, Reddy RC. Glucocorticoid receptor α mediates Roflumilast’s ability to restore dexamethasone sensitivity in COPD. Int J Chron Obstruct Pulmon Dis. 2020;15:125–134. doi:10.2147/COPD.S230188
17. Ko FWS, Sin DD. Twenty-five years of respirology: advances in COPD. Respirology. 2020;25(1):17–19. doi:10.1111/resp.13734
18. Hodge G, Jersmann H, Tran HB, Holmes M, Reynolds PN, Hodge S. Lymphocyte senescence in COPD is associated with loss of glucocorticoid receptor expression by pro-inflammatory/cytotoxic lymphocytes. Respir Res. 2015;16(1):2. doi:10.1186/s12931-014-0161-7
19. Wu J, Li X, Qin Y, et al. Jinwei Tang modulates HDAC2 expression in a rat model of COPD. Exp Ther Med. 2018;15(3):2604–2610. doi:10.3892/etm.2018.5707
20. Stolz D, Matera MG, Rogliani P, et al. Current and future developments in the pharmacology of asthma and COPD: ERS Seminar, Naples 2022. Breathe. 2023;19(2):220267. doi:10.1183/20734735.0267-2022
21. Mei D, Tan WSD, Wong WSF. Pharmacological strategies to regain steroid sensitivity in severe asthma and COPD. Curr Opin Pharmacol. 2019;46:73–81. doi:10.1016/j.coph.2019.04.010
22. Tang H, Mao J, Ye X, et al. SHIP-1, a target of miR-155, regulates endothelial cell responses in lung fibrosis. FASEB J. 2020;34(2):2011–2023. doi:10.1096/fj.201902063R
23. Hobbs BD, Tantisira KG. MicroRNAs in COPD: small molecules with big potential. Eur Respir J. 2019;53(4):1900515. doi:10.1183/13993003.00515-2019
24. Conickx G, Avila Cobos F, van den Berge M, et al. microRNA profiling in lung tissue and bronchoalveolar lavage of cigarette smoke-exposed mice and in COPD patients: a translational approach. Sci Rep. 2017;7(1):12871. doi:10.1038/s41598-017-13265-8
25. De Smet EG, Van Eeckhoutte HP, Avila Cobos F, et al. The role of miR-155 in cigarette smoke-induced pulmonary inflammation and COPD. Mucosal Immunol. 2020;13(3):423–436. doi:10.1038/s41385-019-0241-6
26. Zhuang Y, Hobbs BD, Hersh CP, Kechris K. Identifying miRNA-mRNA networks associated with COPD phenotypes. Front Genet. 2021;12:748356. doi:10.3389/fgene.2021.748356
27. Kaur G, Maremanda KP, Campos M, et al. Distinct exosomal miRNA profiles from BALF and lung tissue of COPD and IPF patients. Int J Mol Sci. 2021;22(21):11830. doi:10.3390/ijms222111830
28. Rojas-Quintero J, Polverino F. Tweaking lung inflammation in COPD: the “Mirky” ways of miRNAs. Am J Physiol Lung Cell Mol Physiol. 2021;321(6):L1089–L1090. doi:10.1152/ajplung.00435.2021
29. Climent M, Viggiani G, Chen Y-W, Coulis G, Castaldi A. MicroRNA and ROS crosstalk in cardiac and pulmonary diseases. Int J Mol Sci. 2020;21(12):4370. doi:10.3390/ijms21124370
30. Ma F, Liu F, Ding L, et al. Anti-Inflammatory effects of curcumin are associated with down regulating microRNA-155 in LPS-treated macrophages and mice. Pharm Biol. 2017;55(1):1263–1273. doi:10.1080/13880209.2017.1297838
31. Faiz A, Steiling K, Roffel MP, et al. Effect of long-term corticosteroid treatment on microRNA and gene-expression profiles in COPD. Eur Respir J. 2019;53(4):1801202. doi:10.1183/13993003.01202-2018
32. Sundar IK, Li D, Rahman I. Small RNA-sequence analysis of plasma-derived extracellular vesicle miRNAs in smokers and patients with chronic obstructive pulmonary disease as circulating biomarkers. J Extracell Vesicles. 2019;8(1):1684816. doi:10.1080/20013078.2019.1684816
33. Li J, Wang J, Li Y, et al. Effective-component compatibility of bufei yishen formula protects COPD rats against PM2.5-induced oxidative stress via miR-155/FOXO3a pathway. Ecotoxicol Environ Saf. 2021;228:112918. doi:10.1016/j.ecoenv.2021.112918
34. Zhou H, Li J, Gao P, Wang Q, Zhang J. miR-155: a novel target in allergic asthma. Int J Mol Sci. 2016;17(10):1773. doi:10.3390/ijms17101773
35. Reddy AT, Lakshmi SP, Banno A, Reddy RC. Role of GPx3 in PPARγ-induced protection against COPD-associated oxidative stress. Free Radic Biol Med. 2018;126:350–357. doi:10.1016/j.freeradbiomed.2018.08.014
36. Golpon HA, Coldren CD, Zamora MR, et al. Emphysema lung tissue gene expression profiling. Am J Respir Cell Mol Biol. 2004;31(6):595–600. doi:10.1165/rcmb.2004-0008OC
37. Ayroldi E, Riccardi C. Glucocorticoid-Induced Leucine Zipper (GILZ): a new important mediator of glucocorticoid action. FASEB J. 2009;23(11):3649–3658. doi:10.1096/fj.09-134684
38. Gałecka E, Kumor-Kisielewska A, Górski P. Association of serum deiodinase type 2 level with chronic obstructive pulmonary disease in the Polish Population. Acta Biochim Pol. 2019;66(2). doi:10.18388/abp.2018_2761
39. Hu Y, He T, Zhu J, et al. The link between circadian clock genes and autophagy in chronic obstructive pulmonary disease. Mediators Inflamm. 2021;2021:2689600. doi:10.1155/2021/2689600
40. Yao H, Sundar IK, Huang Y, et al. Disruption of Sirtuin 1-mediated control of circadian molecular clock and inflammation in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2015;53(6):782–792. doi:10.1165/rcmb.2014-0474OC
41. Kim RY, Oliver BG, Wark PAB, Hansbro PM, Donovan C. COPD Exacerbations: targeting IL-33 as a New Therapy. Lancet Respir Med. 2021;9(11):1213–1214. doi:10.1016/S2213-2600(21)00182-X
42. Plumb J, Robinson L, Lea S, et al. Evaluation of glucocorticoid receptor function in COPD lung macrophages using beclomethasone-17-monopropionate. PLoS One. 2013;8(5):e64257. doi:10.1371/journal.pone.0064257
43. Newton R. Anti-Inflammatory glucocorticoids: changing concepts. Eur J Pharmacol. 2014;724:231–236. doi:10.1016/j.ejphar.2013.05.035
44. Eddleston J, Herschbach J, Wagelie-Steffen AL, Christiansen SC, Zuraw BL. The anti-inflammatory effect of glucocorticoids is mediated by glucocorticoid-induced leucine zipper in epithelial cells. J Allergy Clin Immunol. 2007;119(1):115–122. doi:10.1016/j.jaci.2006.08.027
45. Drake LY, Prakash YS. Contributions of IL-33 in non-hematopoietic lung cells to obstructive lung disease. Front Immunol. 2020;11:1798. doi:10.3389/fimmu.2020.01798
46. Cazzola M, Ora J, Cavalli F, Rogliani P, Matera MG. An overview of the safety and efficacy of monoclonal antibodies for the chronic obstructive pulmonary disease. Biologics. 2021;15:363–374. doi:10.2147/BTT.S295409
47. Zhang Z, Zhang J, Li J, et al. miR-320/ELF3 axis inhibits the progression of breast cancer via the PI3K/AKT pathway. Oncol Lett. 2020;19(4):3239–3248. doi:10.3892/ol.2020.11440
48. Wang K, Chen Y, Zhao Z, Feng M, Zhang S. Identification of potential core genes and miRNAs in testicular seminoma via bioinformatics analysis. Mol Med Rep. 2019;20(5):4013–4022. doi:10.3892/mmr.2019.10684
49. Liu J, Xing Y, Rong L. miR-181 regulates cisplatin-resistant non-small cell lung cancer via downregulation of autophagy through the PTEN/PI3K/AKT pathway. Oncol Rep. 2018;39(4):1631–1639. doi:10.3892/or.2018.6268
50. Rezaei T, Amini M, Hashemi ZS, et al. microRNA-181 serves as a dual-role regulator in the development of human cancers. Free Radic Biol Med. 2020;152:432–454. doi:10.1016/j.freeradbiomed.2019.12.043
51. Kim JR, Horton NC, Mathew SO, Mathew PA. CS1 (SLAMF7) inhibits production of proinflammatory cytokines by activated monocytes. Inflamm Res. 2013;62(8):765–772. doi:10.1007/s00011-013-0632-1
52. Gutierrez-Guerrero A, Mancilla-Herrera I, Maravillas-Montero JL, Martinez-Duncker I, Veillette A, Cruz-Munoz ME. SLAMF7 selectively favors degranulation to promote cytotoxicity in human NK cells. Eur J Immunol. 2022;52(1):62–74. doi:10.1002/eji.202149406
53. Tassi I, Colonna M. The cytotoxicity receptor CRACC (CS-1) recruits EAT-2 and activates the PI3K and phospholipase cgamma signaling pathways in human NK cells. J Immunol. 2005;175(12):7996–8002. doi:10.4049/jimmunol.175.12.7996
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.