Back to Journals » Journal of Inflammation Research » Volume 17
Analysis and Validation of Critical Signatures and Immune Cell Infiltration Characteristics in Doxorubicin-Induced Cardiotoxicity by Integrating Bioinformatics and Machine Learning
Authors Huang C, Pei J, Li D, Liu T, Li Z, Zhang G, Chen R, Xu X, Li B, Lian Z , Chu XM
Received 13 October 2023
Accepted for publication 23 January 2024
Published 2 February 2024 Volume 2024:17 Pages 669—685
DOI https://doi.org/10.2147/JIR.S444600
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Chao Huang,1,* Jixiang Pei,1,* Daisong Li,1 Tao Liu,2 Zhaoqing Li,1 Guoliang Zhang,1 Ruolan Chen,1 Xiaojian Xu,1 Bing Li,3,4 Zhexun Lian,1 Xian-Ming Chu1,5
1Department of Cardiology, the Affiliated Hospital of Qingdao University, Qingdao, Shandong, 266100, People’s Republic of China; 2The Affiliated Qingdao Central Hospital of Qingdao University, The Second Affiliated Hospital of Medical College of Qingdao University, Qingdao, Shandong, 266042, People’s Republic of China; 3Department of Genetics and Cell Biology, Basic Medical College, Qingdao University, Qingdao, 266000, People’s Republic of China; 4Department of Dermatology, The Affiliated Haici Hospital of Qingdao University, Qingdao, 266033, People’s Republic of China; 5The Affiliated Cardiovascular Hospital of Qingdao University, Qingdao, 266071, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhexun Lian; Xian-Ming Chu, Tel +86-532-82913257, Email [email protected]; [email protected]
Purpose: Doxorubicin-induced cardiotoxicity (DIC) is a severe side reaction in cancer chemotherapy that greatly impacts the well-being of cancer patients. Currently, there is still an insufficiency of effective and reliable biomarkers in the field of clinical practice for the early detection of DIC. This study aimed to determine and validate the potential diagnostic and predictive values of critical signatures in DIC.
Methods: We obtained high-throughput sequencing data from the GEO database and performed data analysis and visualization using R software, GO, KEGG and Cytoscape. Machine learning methods and weighted gene coexpression network (WGCNA) were used to identify key genes for diagnostic model construction. Receiver operating characteristic (ROC) analysis and a nomogram were used to assess their diagnostic values. A multiregulatory network was built to reveal the possible regulatory relationships of critical signatures. Cell-type identification by estimating relative subsets of RNA transcript (CIBERSORT) analysis was used to investigate differential immune cell infiltration. Additionally, a cell and animal model were constructed to investigate the relationship between the identified genes and DIC.
Results: Among the 3713 differentially expressed genes, three key genes (CSGALNACT1, ZNF296 and FANCB) were identified. A nomogram and ROC curves based on three key genes showed excellent diagnostic predictive performance. The regulatory network analysis showed that the TFs CREB1, EP300, FLI1, FOXA1, MAX, and MAZ modulated three key genes. An analysis of immune cell infiltration indicated that many immune cells (activated NK cells, M0 macrophages, activated dendritic cells and neutrophils) might be related to the progression of DIC. Furthermore, there may be various degrees of correlation between the three critical signatures and immune cells. RT‒qPCR demonstrated that the mRNA expression of CSGALNACT1 and ZNF296 was significantly upregulated, while FANCB was significantly downregulated in DOX-treated cardiomyocytes in vitro and in vivo.
Conclusion: Our study suggested that the differential expression of CSGALNACT1, ZNF296 and FANCB is associated with cardiotoxicity and is also involved in immune cell infiltration in DIC. They might be potential biomarkers for the early occurrence of DIC.
Keywords: doxorubicin, cardiotoxicity, biomarker, machine learning, immune infiltration
Introduction
With the significant advancement in early detection and treatment, the number of cancer survivors has increased,1 but there have also been multiple adverse outcomes, such as chemotherapy-induced cardiotoxicity.2 Doxorubicin (DOX), a member of the anthracycline antibiotic family, is extensively used in the chemotherapy of various cancers, such as hematological malignancies and other tumors.3 Unfortunately, approximately a quarter of cancer survivors experience DOX-induced cardiotoxicity (DIC), which significantly limits its clinical application.4 Although DIC has attracted the attention of cardiologists, there are limited methods available for early detection and treatment in clinical practice.
The pathophysiological mechanisms of DIC are complex and not yet fully elucidated, including oxidative stress, mitochondrial dysfunction, inflammatory response, metabolic disorders, and cell death.5–8 An increasing amount of evidence has indicated that immune cells have crucial functions in cardiac homeostasis and disease.9,10 Further investigation is required to explore the precise function and molecular mechanism of immune cells in the development of DIC. The rapid developments of multiomics and high-throughput sequencing provide novel tools for identifying early biomarkers and exploring therapeutic targets.11
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) has the ability of unlimited proliferation and differentiation into cardiomyocytes, so it is widely used in the study and treatment of cardiac disease.12,13 Recently, Burridge et al indicated that the viability of using hiPSC-CMs as a platform for determining and validating the genetic foundation and molecular mechanisms underlying DIC.14 In addition, many studies have also used hiPSC-CMs as the cell source of DIC.15–17 On these bases, we downloaded high-throughput gene expression data from NCBI GEO database.
In the current work, we explored key genes of DIC using comprehensive bioinformatics approaches and machine-learning strategies. Furthermore, we constructed a multiregulatory network to uncover the possible regulatory connections of key genes. Similarly, machine learning was carried out to construct a diagnostic nomogram model for DIC prediction on the basis of the three key genes (ZNF296, FANCB and CSGALNACT1) that were discovered in DIC pathogenic genes. Therefore, the study of its related molecular mechanism may be helpful for its further clinical application in DIC. We attempted to explore the connection between key genes and immunity. Finally, our study validated the differential expression of the key genes by constructing the model treated with doxorubicin in vitro and in vivo. This study collectively identified a stable and achievable biomarker in DIC, which could potentially serve as a target for diagnosing and preventing cancer survivors who have undergone DOX treatment.
Materials and Methods
Data Acquisition and Homogenization
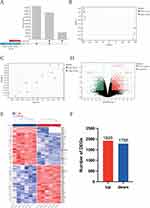
Our information was gathered by accessing the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), which is a public repository of high-throughput gene expression data, chips, and microarrays.18 High-throughput gene expression data were downloaded from the NCBI GEO database, including GSE76314, GSE157282 and GSE181517. GPL11154 served as the complementary high-throughput detection platform for GSE76314; GSE157282 was based on platform GPL24676. GSE181517 was based on platform GPL18573. The GSE76314 dataset,14 which describes the differences in cell-derived cardiomyocytes (hiPSC-CMs), was used as the training set (3 doxorubicin samples vs 3 control samples). The GSE157282 dataset19 [doxorubicin samples vs 3 controls samples], which describes the differences in the hiPSC-CMs, was also used as the training set. GSE18151720 was used as the validation set to explore the dynamic changes in the critical signature, which included 3 doxorubicin samples and 3 control samples. The scale function in R version 4.2.0 software was used to combine GSE76314 and GSE157282 into one dataset and calibrate the batch using the SVA package.21 For uniform manifold approximation and projection (UMAP), after the removal of the batch effect, the samples among various datasets are clustered and intertwined with each other, indicating that the batch effect is better removed. Principal component analysis (PCA) was used to verify the reproducibility of the data, and the R package ggord was used to construct the PCA plots.22
Analysis of Differentially Expressed Genes
In this study, we performed differential expression analysis using the R Bioconductor package “Limma” (V3.46.0) to identify differentially expressed genes.23 Differentially expressed genes (DEGs) were defined as those with adjusted P values and genes with adjusted P values < 0.05 and |Log2-fold-change (log2FC)|≥ 1 after adjusting the P values using Benjamini‒Hochberg’s false discovery rate (FDR). The R software ggplot2 V3.3.5 package was used to create volcano plots, and the R software Heatmap V1.0.12 package was utilized to generate heatmaps for the top 100 DEGs from each dataset.
Functional Enrichment Analysis of DEGs
A functional enrichment analysis of DEGs was performed to clarify their role using the R software clusterProfiler and Goplot package. These analyses were discerningly marked by significance, indicated by the adjusted P value < 0.05. A comprehensive enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology (GO) was conducted and visualized, including biological process (BP), cellular component (CC), and molecular function (MF) for all DEGs.
Screening and Validation of Critical Gene Signatures
In this study, random forests (RF),24 least absolute shrinkage and selection operator (LASSO) logistic regression,25 and weighted gene coexpression network analysis (WGCNA)26 were utilized to screen novel and pivotal signatures associated with DIC. The RF model, LASSO analysis, and WGCNA were executed using the R software random Forest package, R software glmnet package and R software WGCNA package, respectively. Following the individual algorithmic analyses, the points of intersection among the three classification models were identified, setting the stage for the next step of the analysis. To assess the diagnostic efficacy of these three diagnostic signatures in doxorubicin-induced cardiotoxicity, the area under the curve (AUC) and the ROC curves were established using R software’s pROC package. In adherence to statistical standards, a threshold of two-sided P < 0.05 was deemed to be statistically significant. To verify the expression levels of key markers, their differences between the doxorubicin and control groups were assessed in the training and validation sets.
Multifactor Regulatory Network Analysis of Key Genes
Guardians of gene expression include long noncoding RNAs (lncRNAs), microRNAs (miRNAs), and transcription factors (TFs) in transcription and posttranscription prototypes. To demonstrate the regulatory networks of key signatures, lncRNAs, mRNAs, TFs, and miRNAs were predicted by RNAInter,27 hTFtarget,28 and mirDIP.29 Then, we used Cytoscape software30 to visualize the multifactor regulatory network.
Analysis of Immune Cell Infiltration on Critical Gene Signatures
Prior to CIBERSORT analysis, principal component analysis (PCA) of the gene expression profile was plotted and visualized by the R software factoextra package. Analysis of 22 different types of immune cells using CIBERSORT demonstrated the differential expression of cells between the doxorubicin group and the control group using a box diagram (P value < 0.05 was deemed to be statistically significant). Afterwards, Spearman correlation analysis between immune cells and immune cells, as well as between key signatures and immune cells, was demonstrated by a heatmap through the R software corrplot V0.92 package.
Cell Line Culture and Treatment
AC16 immortalized human cardiomyocyte line was procured from MeisenCTCC (Zhejiang, China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) 1% and penicillin/streptomycin and grown in a CO2 incubator maintained at atmospheric oxygen levels and 5% CO2. When the AC16 cell density reached 80–90%, the cells were exposed to 1 µM doxorubicin (Sigma‒Aldrich) or vehicle (PBS). Whole cells were collected after 24 h of treatment.
Animal Experiments
The Establishment of Animal Model
Twenty male wild-type C57BL/6 mice aged 8 weeks (18–22g) were purchased from the SiPeiFu (Beijing) Biotechnology Co., Ltd., Beijing City, and they were randomly divided into two groups: normal control (NC) group and DOX treatment (DOX) group. In NC group, mice were i.p.injected with the equivalent volume of saline two times (on days 1 and 8, respectively); in DOX group, mice were intraperitoneally injected with DOX on day 1 (10 mg/kg) and day 8 (10 mg/kg), and acute DIC model was established in vivo (cumulative dose of 20 mg/kg).31 At the end of the experiment, each group of mice was anesthetized and killed, and their hearts were taken for subsequent experiments. The experimental procedures were implemented in accordance with the China Animal Welfare Legislation, reviewed and approved by the Qingdao University Committee on Ethics in the Care and Use of Laboratory.
Cardiac Functions Examination
At the end of the treatment, the mice heart function was examined through echocardiography Imaging System (VINNO 6 LAB, VINNO). Cardiac ventricular structure and systolic function were examined by two-dimensional imaging motion mode to record parasternal long-axis fields. Parameters included the left ventricular (LV) anterior wall thickness in the end-diastolic/end- systolic stages, the LV posterior wall thickness, the LV internal diameter; LV ejection fraction, and LV fractional shortening.
The Hematoxylin & Eosin Staining of Heart Tissues
Hearts of mice were fixed in 10% formalin, and then treated with dehydration, embedding and sectioning. Hematoxylin & eosin staining was performed to examine cardiac histological morphology.
RNA Extraction and Quantitative Real-Time PCR for Cell and Animal Verification Experiments
The TRIzol method was used to extract total RNA from AC16 cells and heart tissue samples, and spectrophotometry was used to measure the concentration and purity. According to the manufacturer’s instructions, equivalent amounts (1 μg) of purified RNA were used as a template to synthesize cDNA using ABScrlpt III RT Master Mix for qPCR with a gDNA Remover Kit (Cat# RK20429, ABclonal, Wuhan, China). The manufacturer’s instructions were followed to perform quantitative PCR using a QuantStudio 3 (Bio-Rad) and 2X Universal SYBR Green Fast qPCR Mix (Cat#RK21203, ABclonal, Wuhan, China). The relative expression levels were determined by the 2–ΔΔCT method and are presented relative to GAPDH mRNA. Primer sequences are provided in Table S1.
Statistical Analysis
GraphPad Prism 9.0 was utilized for conducting the statistical analysis. The unpaired Student’s t-test was used to compare the DOX group (DOX) and control group (NC). The assays were conducted at least three times or more. A two-tailed p value <0.05 was considered statistically significant.
Results
Identification of DEGs
Figure 1 clarifies the depicted flow chart of the present work. Both the GSE76314 and GSE157282 datasets were merged (Figure 2A) for the following analysis. The utilization of UMAP was performed to visually represent the clustering of read counts and identify batch effects (Figure 2B). Next, we merged these two datasets into one dataset and rectified the batch using the SVA software package. Figure 2C shows that the interbatch variations are effectively removed after data normalization. Finally, a normalized gene expression matrix file containing 12 samples (6 doxorubicin samples and 6 control samples) was obtained. These DEGs are distributed in the volcano map (Figure 2D). The first 100 temperatures are shown in the heatmap (Figure 2E). In the results of differential expression analysis, a total of 3713 genes were identified as DEGs, including 1925 upregulated genes and 1788 downregulated genes (Figure 2F).
 |
Figure 1 Flow chart of this study. |
GO and KEGG Pathway Analysis of DEGs
Subsequently, we performed enrichment analyses for overlapping DEGs in KEGG and GO. Among the KEGG pathways, the pathways in cancer, erpes simplex virus 1 infection, rap1 signaling pathway, alcoholism, and cGMP-PKG signaling pathway were considered the pathways with the highest degree of enriched pathways (Figure 3A). In significantly enriched BP, these included the positive regulation of nucleobase containing compound metabolic process, positive regulation of biosynthetic process, cell population proliferation, cell cycle, and apoptotic process (Figure 3B). Subsequently, chromosome, chromatin, microtubule cytoskeleton, transcription regulator complex, and chromosomal region were the most enriched terms in CC (Figure 3C). For MF, the top 5 enriched items were transcription regulator activity, sequence-specific DNA binding, ribonucleotide binding, DNA binding transcription factor activity, and adenyl nucleotide binding (Figure 3D).
Screening of Critical Signatures
Here, three key genes were screened using the RF, LASSO, and WGCNA machine learning algorithms. Using the WGCNA algorithm (Figure S1), we confirmed 11 extraordinary coexpression modules (Figure 4A). Figure 4B clarifies that there is a correlation between multiple modules in DIC. Given that the turquoise and dark red modules are most significantly associated with DIC, we screened genes in the turquoise and dark red modules for follow-up exploration and successfully identified 3370 genes (Figure 4C–E, Table S2). Moreover, the results showed that 15 genes were identified by the LASSO algorithm (Figure 4F and G, Table S2), and 90 genes were determined with the RF algorithm (Figure 4H–J, Table S2). Subsequently, a Venn diagram shows that FANCB, ZNF296 and CSGALNACT1 obtained by the three algorithms are overlapping genes (Figure 4K).
Verification of FANCB, ZNF296 and CSGALNACT1 in DIC
To evaluate the possible significance of the key genes in DIC, we created ROC curves. First, we verified the expression of FANCB, ZNF296 and CSGALNACT1 in DIC. The results showed that compared with the control group, the expression of ZNF296 and CSGALNACT1 in the training and validation sets was significantly upregulated, and the expression of FANCB was significantly downregulated (all P < 0.01, Figure 5A and B). Second, the AUCs of the three key genes in the training set were 1 (Figure 5C), indicating that they were accurate and reliable in terms of predictive value. In the validation set of GSE181517, the AUCs for FANCB, ZNF296 and CSGALNACT1 were also all 1 (Figure 5D). Finally, we created nomogram models to diagnose DIC using the hallmark genes FANCB, ZNF296 and CSGALNACT1 (Figure 5E).
Construction of a Multifactor Regulatory Network Based on Key Genes
Next, we explored the potential regulatory mechanisms by studying the regulatory networks involving lncRNAs, miRNAs and TFs and conducted a comprehensive visual analysis (Figure 6). Here, to construct a multifactor regulatory network, interaction pairs involving miRNAs, lncRNAs and TFs with three key genes were extracted. In the multifactor regulatory network, FANCB was managed by 5 miRNAs and 1 lncRNA, including hsa-miR-4695-5p, hsa-miR-3612, hsa-miR-2113, hsa-miR-7109-3p, hsa-miR-135a-2-3p, and CISTR, and the possible TFs for FANCB were ATF3, CEBPA, CEBPB, CREB1, E2F1, EP300, ETS1, FLI1, FOXA1, GABPA, HDAC1, HDAC2, KLF5, MAX, and MAZ. Additionally, the ZNF296 network included 2 lncRNAs, 7 miRNAs, and 66 TFs, and the CSGALNACT1 network contained 167 miRNAs and 25 TFs. Obviously, the results of our work suggest that the TFs CREB1, EP300, FLI1, FOXA1, MAX, and MAZ modulate all 3 marker genes.
Analysis of Immune Cell Infiltration and Correlation Analysis Between Key Genes
Recent evidence has shown that immune cells play a critical role in the pathophysiological process of DIC.6,32,33 Therefore, we investigated the link between key features in DIC and immune infiltration. The swatches of different groups are well divided in the training set in PCA (Figure 7A). To study 22 types of immune cell phenotypes, we utilized the CIBERSORT algorithm to analyze the training set. Compared with the control samples, DIC samples exhibited a greater percentage of activated NK cells, M0 macrophages, activated dendritic cells and neutrophils (all P < 0.05). However, the proportions of memory B cells, CD8 T cells, resting memory CD4 T cells, and resting NK cells in DIC were relatively lower than those in the control (all P < 0.05) (Figure 7B). Twenty immune cell types were included for further investigation after two immune cell types with undetectable levels were excluded. It can be seen from the related heatmaps (Figure 7C) that naive B cells and CD8 T cells, naive B cells and resting memory CD4 T cells, CD8 T cells and neutrophils, resting NK cells and activated M0 macrophages displayed the most significant negative correlations (P < 0.0001). Naive B cells and neutrophils, activated mast cells and monocytes, and M0 macrophages and neutrophils exhibited the most significant positive correlations (P < 0.0001). Our aim was to detect the link between key genes and 20 types of immune cell phenotypes in doxorubicin cardiotoxicity. According to the correlation analysis results, FANCB displayed the most positive correlations with resting NK cells and CD8 T cells [r = 0.70, -log10 (p value) = 1.94] while demonstrating negative correlations with activated dendritic cells [r = −0.76, -log10 (p value) = 2.35]. ZNF296 displayed the most positive correlations with activated dendritic cells [r = 0.76, -log10 (p value) = 2.40] and revealed negative correlations with resting NK cells [r = −0.80, -log10 (p value) =2.72]. CSGALNACT1 displayed the most positive correlations with M0 macrophages [r = 0.92, -log10(p value) = 4.60] and revealed negative correlations with resting NK cells [r = −0.79, -log10(p value) =2.65] (Figure 7D).
Verification of FANCB, ZNF296 and CSGALNACT1 in AC16 Human Cardiomyocyte-Like Cells
To verify whether the mRNA expression of FANCB, ZNF296 and CSGALNACT1 was consistent with the results of the database analysis, we constructed a doxorubicin-induced injury model in vitro.34 The results showed that FANCB was expressed at a low level (Figure 8A), while ZNF296 and CSGALNACT1 were expressed at a high level in the DOX group (Figure 8B and C).
 |
Figure 8 The mRNA expression of FANCB (A), ZNF296 (B) and CSGALNACT1 (C) in AC16 human cardiomyocyte cells by RT‒qPCR. (**P < 0.01, ***P < 0.001). |
The Verification Experiments in vivo
Firstly, mouse model of DIC is illustrated in Figure 9A, and the heart weight to tibia length ratio (HW/TL) was significantly reduced in the DOX group (Figure 9B). The echocardiography results demonstrate a downward trend of cardiac systolic and diastolic function in the DOX group (Figure 9C), as evidenced by significantly decreased ejection fraction (Figure 9D), decreased fractional shortening (Figure 9E). HE staining illustrated that DOX treatment exacerbated cardiomyocyte tissue disorders compared with control group (Figure 9F). These results indicate that we successfully established a DIC model. Finally, the mRNA levels of FANCB was decreased in the hearts of DOX treated group relative to the control group (Figure 9G), while ZNF296 and CSGALNACT1 is contrary to the FANCB (Figure 9H and I).
Discussion
Doxorubicin prolongs the survival time of tumor patients, but it also leads to severe side effects, especially cardiotoxicity, which significantly affects the effectiveness of anticancer treatment.35,36 Hence, it is crucial to recognize secure and effective biomarkers to detect DIC. Currently, with the rapid development of high-throughput sequencing and multiomics technologies,37,38 some new methods have been used to detect and diagnose DIC.11
In this study, 3713 DEGs were discovered using a comprehensive bioinformatics analysis between DIC and the controls. The results indicated that the significantly enriched terms included positive regulation of nucleobase-containing compound metabolic process in BP, chromosome in CC, and transcription regulator activity in MF. Of note, multiple algorithms were applied to identify key genes in DIC, including WGCNA, RF and LASSO. Ultimately, CSGALNACT1, ZNF296 and FANCB were determined to be key genes. Additionally, through ROC curves and nomogram analysis, it was shown that the expression levels of CSGALNACT1, ZNF296 and FANCB could accurately distinguish DIC from controls. Similarly, we validated the stability of CSGALNACT1, ZNF296 and FANCB expression levels through distinct validation datasets. From the results, CSGALNACT1, ZNF296 and FANCB may be potential factors for DIC, which suggests that they have vital value for clinical application in DIC. Additionally, our regulatory network showed that the TFs CREB1, EP300, FLI1, FOXA1, MAX, and MAZ modulated all 3 marker genes. However, the transcriptional regulation of CSGALNACT1, ZNF296 and FANCB by CREB1, EP300, FLI1, FOXA1, MAX and MAZ needs further study.
Zinc finger protein 296 (ZNF296 or Zfp296) is a reprogramming factor composed of 445 amino acids that belongs to the C2H2 zinc finger protein family and is involved in cell proliferation, survival, differentiation, and carcinogenesis.39 ZNF296 is expressed in mouse testes, bone marrow, and embryos.40 Previous evidence suggests that ZNF296 is an effective biomarker for breast cancer detection.41 Furthermore, recent studies have found that ZNF296 is a regulator of H3K9me3 and is closely associated with the pluripotency of embryonic stem cells.42 Another study indicated that ZNF296 was associated with the formation of induced pluripotent stem (iPS) cells.43 Interestingly, a microarray analysis based on transcriptomic signatures showed that ZNF296 was upregulated in tuberculosis and could be used as a diagnostic biomarker.44 In the present study, we also found that ZNF296 was significantly upregulated in the DOX-treated group, which was validated through platforms and across cell experiments. Although the role of ZNF296 in DIC has not been clearly discussed, it may be a potential diagnostic biomarker.
Furthermore, CSGALNCT-1, also named chondroitin sulfate N-acetylgalactosaminyltransferase 1, plays a vital role as a glycosyltransferase in the biosynthesis of chondroitin sulfate. It is also involved in important biological processes, such as cartilage development and polysaccharide metabolism.45 Previous microarray studies have indicated that CSGALNACT1 is a prognostic biomarker in multiple myeloma.46 According to a recent study, CSGALNACT1 was found to have a notable upregulation in lung-derived mesenchymal stem cells (LMSCs) from emphysema patients, which can regulate extracellular matrix (ECM), but the specific function remains unclear.47 Significantly, chondroitin sulfate (CS) is a natural macromolecule catalyzed by CSGALNACT1 and a major component of the extracellular matrix.48 Another study published recently showed that abnormal accumulation of chondroitin sulfate glycosaminoglycans led to cardiac fibrosis.49 Interestingly, our study showed that CSGALNACT1 was expressively upregulated in cardiomyocytes in the doxorubicin-treated group. Therefore, we speculate that CSGALNACT1 may be involved in DIC, although the specific mechanisms require further investigation. In conclusion, CSGALNACT1 is considered a suitable biomarker for DIC.
FANCB, fully defined as Fanconi anemia complementation group B and known as FAAP95, is an essential constituent of the Fanconi anemia (FA) core complex. It has a vital function in hematopoiesis and the development of germ cells.50,51 Some studies have indicated that the FA pathway serves as an indispensable mechanism for the DNA damage response.52 Further study has shown that FANCB is known as a DNA repair gene.53 Previous studies have concluded that DNA damage is one of the molecular mechanisms in DIC. Interestingly, our study found that the expression of FANCB in cardiomyocytes treated with doxorubicin was significantly reduced, which was consistent with the verification of the cell experiment. Therefore, we speculate that FANCB is directly or indirectly involved in DIC via DNA damage/repair. Although the exact mechanism remains unclear, this study provides a novel perspective for understanding doxorubicin-induced cardiomyocyte injury.
Noncoding RNAs (ncRNAs) widely participate in cell physiological and pathological processes by regulating the epigenetic modification of gene expression, which has attracted great attention.54,55 Increasing evidence has reported the significant role of ncRNA in the DIC.56 Therefore, we further predicted upstream ncRNAs targeting three key genes, ZNF296, FANCB, and CSGALNACT1, and ultimately constructed a gene regulatory network including microRNAs and lncRNAs. Additionally, we also investigated the regulatory network between transcription factors and three core genes. Notably, the transcription factors CREB1, EP300, FLI1, FOXA1, MAX, and MAZ were simultaneously regulated by three key genes.
In recent years, a growing body of research has highlighted the potential impact of immune cells on DIC, including neutrophils,57,58 macrophages,59,60 and NK cells.61 In our study, significant differences were observed between the DOX-treated group and the control group in terms of immune cell infiltration. Specifically, the peaks of M0 macrophages, activated NK cells, activated dendritic cells, and neutrophils were higher, while the proportions of memory B cells, CD8 T cells, resting CD4 T cells, and resting NK cells were lower. Previous studies have shown that 24 hours after doxorubicin treatment, neutrophils significantly infiltrate the heart, accompanied by a decrease in cardiac function, vascular structure damage, and increased deposition of vascular collagen, leading to fibrosis,58 which is consistent with our study findings. Notably, macrophages, a type of innate immune system cell, were also found to be upregulated in our study. Some studies found that in an acute doxorubicin-induced cardiotoxicity mouse model, doxorubicin increased the proportion of M1 macrophages while suppressing M2 macrophages.62 A macrophage profile tracking study based on CX3CR1 found that M1 macrophages may be the major group during the early stage of cardiac injury.63 Although studies have suggested that immune cells play an important role in DIC, the specific regulatory mechanisms remain unclear.
Therefore, we attempted to reveal the potential function of immune cells by establishing a connection between key genes and immune cells. In our research, CSGALNACT1, ZNF296 and FANCB showed different degrees of correlation with immune cells, including resting NK cells and activated dendritic cells. In a prior study, it was demonstrated that CSGALNACT1 was linked to the activation of lymphocytes and the production of inflammatory cytokines (IL-6 and IFN-r cytokine) in experimental autoimmune encephalomyelitis (EAE).64 In our data, CSGALNACT1 displayed the most positive correlations with M0 macrophages and showed negative correlations with resting NK cells. Our findings provide novel insights into the immune mechanisms of CSGALNACT1, ZNF296, and FANCB during DIC.
There are some limitations in our study. First, although we have integrated multiple datasets, the sample size was still relatively small. Second, due to the lack of relevant clinical data, we did not perform prognostic analysis. Third, we validated the mRNA expression of critical key genes in DOX-treated cardiomyocytes, but further validation, such as Western blotting and immunofluorescence staining, is necessary. To improve these deficiencies, we will conduct further clinical studies to confirm the findings with more detailed clinical information and a wide range of samples.
Conclusion
In summary, we concluded that (1) three key genes, CSGALNACT1, ZNF296, and FANCB, were identified that might be probable diagnostic biomarkers in DIC and may contribute to immunity regulation simultaneously; (2) immune cell infiltration was closely related to DIC; and (3) the inflammatory response signaling pathway, biosynthetic process, and apoptotic process may be the key mechanisms in DIC. This study provides a new strategy for screening and evaluating biomarkers in clinical practice while also laying the foundation for the underlying role of immune cell responses in DIC.
Abbreviations
DIC, Doxorubicin induced cardiotoxicity; mRNA, Message RNA; TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; AUC, Area under the curve; DEGs, differentially expressed genes; lncRNA, Long noncoding RNA; miRNA, microRNA; TF, Transcription factor; ROC, Receiver operating characteristic; RF, Random forests; LASSO, Least absolute shrinkage and selection operator; WGCNA, Weighted gene coexpression network analysis; FANCB, FA complementation group B; ZNF296, Zinc finger protein 296; CSGALNACT1, Chondroitin sulfate N-acetylgalactosaminyltransferase 1; DMEM, Dulbecco’s modified Eagle’s medium.
Data Sharing Statement
The following accession numbers were included in this study that were downloaded and analyzed in the public datasets and could be accessed in the GEO data repository (https://www.ncbi.nlm.nih.gov/geo/): GSE76314, GSE157282, and GSE181517. Further enquiries can be directed to the corresponding author.
Ethics Committee Approval
The animal experiments were approved by the Ethics Committee of Qingdao University.
Acknowledgments
Thank you for the help of the members from the Department of Cardiology at the Affiliated Hospital of Qingdao University.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by The National Natural Science Foundation of China (no. 82172574, no. 81871231), The Natural Science Foundation of Shandong Province (no. ZR2020MH016, no. ZR202209280042), Research Planning Project of Shandong Higher Medical Education Research Center (no. YJKT202171), and Shandong Taishan Scholars Young Experts Program (No. tsqn202103056).
Disclosure
All the authors declared that they had no competing interests for this work.
References
1. Miller KD, Nogueira L, Devasia T, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72(5):409–436. doi:10.3322/caac.21731
2. Chang HM, Moudgil R, Scarabelli T, Okwuosa TM, Yeh ETH. Cardiovascular complications of cancer therapy: best practices in diagnosis, prevention, and management: part 1. J Am Coll Cardiol. 2017;70(20):2536–2551. doi:10.1016/j.jacc.2017.09.1096
3. Young RC, Ozols RF, Myers CE. The anthracycline antineoplastic drugs. N Engl J Med. 1981;305(3):139–153. doi:10.1056/nejm198107163050305
4. Zamorano JL, Lancellotti P, Rodriguez Munoz D, et al. 2016 ESC Position Paper on cancer treatments and cardiovascular toxicity developed under the auspices of the ESC Committee for Practice Guidelines: the Task Force for cancer treatments and cardiovascular toxicity of the European Society of Cardiology (ESC). Eur Heart J. 2016;37(36):2768–2801. doi:10.1093/eurheartj/ehw211
5. Wallace KB, Sardao VA, Oliveira PJ. Mitochondrial determinants of doxorubicin-induced cardiomyopathy. Circ Res. 2020;126(7):926–941. doi:10.1161/CIRCRESAHA.119.314681
6. Ni C, Ma P, Wang R, et al. Doxorubicin-induced cardiotoxicity involves IFNgamma-mediated metabolic reprogramming in cardiomyocytes. J Pathol. 2019;247(3):320–332. doi:10.1002/path.5192
7. Brandao SR, Carvalho F, Amado F, Ferreira R, Costa VM. Insights on the molecular targets of cardiotoxicity induced by anticancer drugs: a systematic review based on proteomic findings. Metabolism. 2022;134:155250. doi:10.1016/j.metabol.2022.155250
8. Kong CY, Guo Z, Song P, et al. Underlying the mechanisms of doxorubicin-induced acute cardiotoxicity: oxidative stress and cell death. Int J Biol Sci. 2022;18(2):760–770. doi:10.7150/ijbs.65258
9. Swirski FK, Nahrendorf M. Cardioimmunology: the immune system in cardiac homeostasis and disease. Nat Rev Immunol. 2018;18(12):733–744. doi:10.1038/s41577-018-0065-8
10. Sun K, Li YY, Jin J. A double-edged sword of immuno-microenvironment in cardiac homeostasis and injury repair. Signal Transduct Target Ther. 2021;6(1):79. doi:10.1038/s41392-020-00455-6
11. Sawicki KT, Sala V, Prever L, et al. Preventing and treating anthracycline cardiotoxicity: new insights. Annu Rev Pharmacol Toxicol. 2021;61:309–332. doi:10.1146/annurev-pharmtox-030620-104842
12. Kawamura M, Miyagawa S, Fukushima S, et al. Enhanced survival of transplanted human induced pluripotent stem cell-derived cardiomyocytes by the combination of cell sheets with the pedicled omental flap technique in a porcine heart. Circulation. 2013;128(11 Suppl 1):S87–S94. doi:10.1161/circulationaha.112.000366
13. Yoshida Y, Yamanaka S. iPS cells: a source of cardiac regeneration. J Mol Cell Cardiol. 2011;50(2):327–332. doi:10.1016/j.yjmcc.2010.10.026
14. Burridge PW, Li YF, Matsa E, et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med. 2016;22(5):547–556. doi:10.1038/nm.4087
15. Hsu WT, Huang CY, Yen CYT, Cheng AL, Hsieh PCH. The HER2 inhibitor lapatinib potentiates doxorubicin-induced cardiotoxicity through iNOS signaling. Theranostics. 2018;8(12):3176–3188. doi:10.7150/thno.23207
16. Magdy T, Jouni M, Kuo HH, et al. Identification of drug transporter genomic variants and inhibitors that protect against doxorubicin-induced cardiotoxicity. Circulation. 2022;145(4):279–294. doi:10.1161/circulationaha.121.055801
17. Maillet A, Tan K, Chai X, et al. Modeling doxorubicin-induced cardiotoxicity in human pluripotent stem cell derived-cardiomyocytes. Sci Rep. 2016;6(25333). doi:10.1038/srep25333
18. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi:10.1093/nar/gks1193
19. Kattih B, Shirvani A, Klement P, et al. IDH1/2 mutations in acute myeloid leukemia patients and risk of coronary artery disease and cardiac dysfunction-A retrospective propensity score analysis. Leukemia. 2021;35(5):1301–1316. doi:10.1038/s41375-020-01043-x
20. Huang H, Christidi E, Shafaattalab S, et al. RARG S427L attenuates the DNA repair response to doxorubicin in induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Rep. 2022;17(4):756–765. doi:10.1016/j.stemcr.2022.03.002
21. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi:10.1093/bioinformatics/bts034
22. Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi:10.1186/gb-2004-5-10-r80
23. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
24. Breiman L. Random forests. Mach Learn. 2001;4:5–32.
25. Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Stat Soc Series B. 1996;58(1):267–288. doi:10.1111/j.2517-6161.1996.tb02080.x
26. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9(559). doi:10.1186/1471-2105-9-559
27. Kang J, Tang Q, He J, et al. RNAInter v4.0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic Acids Res. 2022;50(D1):D326–D332. doi:10.1093/nar/gkab997
28. Zhang Q, Liu W, Zhang HM, et al. hTFtarget: a comprehensive database for regulations of human transcription factors and their targets. Genomics Proteomics Bioinf. 2020;18(2):120–128. doi:10.1016/j.gpb.2019.09.006
29. Tokar T, Pastrello C, Rossos AEM, et al. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(D1):D360–D370. doi:10.1093/nar/gkx1144
30. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
31. Tian C, Yang Y, Li B, et al. Doxorubicin-Induced cardiotoxicity may be alleviated by bone marrow mesenchymal stem cell-derived exosomal lncRNA via inhibiting inflammation. J Inflamm Res. 2022;15:4467–4486. doi:10.2147/jir.S358471
32. Sumneang N, Tanajak P, Oo TT. Toll-like receptor 4 inflammatory perspective on doxorubicin-induced cardiotoxicity. Molecules. 2023;28(11). doi:10.3390/molecules28114294
33. Bhagat A, Shrestha P, Kleinerman ES. The innate immune system in cardiovascular diseases and its role in doxorubicin-induced cardiotoxicity. Int J Mol Sci. 2022;23(23). doi:10.3390/ijms232314649
34. Pan JA, Tang Y, Yu JY, et al. miR-146a attenuates apoptosis and modulates autophagy by targeting TAF9b/P53 pathway in doxorubicin-induced cardiotoxicity. Cell Death Dis. 2019;10(9):668. doi:10.1038/s41419-019-1901-x
35. Pawan K, Singal DS, Iliskovic N. Doxorubicin-induced cardiomyopathy. New Engl J Med. 1998;339(13):900–905.
36. Herrmann J. Adverse cardiac effects of cancer therapies: cardiotoxicity and arrhythmia. Nat Rev Cardiol. 2020;17(8):474–502. doi:10.1038/s41569-020-0348-1
37. Vandereyken K, Sifrim A, Thienpont B, Voet T. Methods and applications for single-cell and spatial multi-omics. Nat Rev Genet. 2023;24(8):494–515. doi:10.1038/s41576-023-00580-2
38. Barbulovic-Nad I, Lucente M, Sun Y, et al. Bio-microarray fabrication techniques--A review. Crit Rev Biotechnol. 2006;26(4):237–259. doi:10.1080/07388550600978358
39. Miyazaki S, Yamano H, Motooka D, et al. Zfp296 knockout enhances chromatin accessibility and induces a unique state of pluripotency in embryonic stem cells. Commun Biol. 2023;6(1):771. doi:10.1038/s42003-023-05148-8
40. Kloet SL, Karemaker ID, van Voorthuijsen L, et al. NuRD-interacting protein ZFP296 regulates genome-wide NuRD localization and differentiation of mouse embryonic stem cells. Nat Commun. 2018;9(1):4588. doi:10.1038/s41467-018-07063-7
41. Kjaer IM, Kahns S, Timm S, et al. Phase II trial of delta-tocotrienol in neoadjuvant breast cancer with evaluation of treatment response using ctDNA. Sci Rep. 2023;13(1):8419. doi:10.1038/s41598-023-35362-7
42. Matsuura T, Miyazaki S, Miyazaki T, Tashiro F, Miyazaki JI. Zfp296 negatively regulates H3K9 methylation in embryonic development as a component of heterochromatin. Sci Rep. 2017;7(1):12462. doi:10.1038/s41598-017-12772-y
43. Fujii Y, Kakegawa M, Koide H, Akagi T, Yokota T. Zfp296 is a novel Klf4-interacting protein and functions as a negative regulator. Biochem Biophys Res Commun. 2013;441(2):411–417. doi:10.1016/j.bbrc.2013.10.073
44. Gliddon HD, Kaforou M, Alikian M, et al. Identification of reduced host transcriptomic signatures for tuberculosis disease and digital PCR-based validation and quantification. Front Immunol. 2021;12(637164). doi:10.3389/fimmu.2021.637164
45. Gulberti S, Jacquinet JC, Chabel M, et al. Chondroitin sulfate N-acetylgalactosaminyltransferase-1 (CSGalNAcT-1) involved in chondroitin sulfate initiation: impact of sulfation on activity and specificity. Glycobiology. 2012;22(4):561–571. doi:10.1093/glycob/cwr172
46. Agnelli L, Forcato M, Ferrari F, et al. The reconstruction of transcriptional networks reveals critical genes with implications for clinical outcome of multiple myeloma. Clin Cancer Res. 2011;17(23):7402–7412. doi:10.1158/1078-0432.CCR-11-0596
47. Kruk D, Yeung ACY, Faiz A, et al. Gene expression profiles in mesenchymal stromal cells from bone marrow, adipose tissue and lung tissue of COPD patients and controls. Respir Res. 2023;24(1):22. doi:10.1186/s12931-023-02314-8
48. Yang J, Shen M, Wen H, et al. Recent advance in delivery system and tissue engineering applications of chondroitin sulfate. Carbohydr Polym. 2020;230(115650). doi:10.1016/j.carbpol.2019.115650
49. Zhao RR, Ackers-Johnson M, Stenzig J, et al. Targeting chondroitin sulfate glycosaminoglycans to treat cardiac fibrosis in pathological remodeling. Circulation. 2018;137(23):2497–2513. doi:10.1161/circulationaha.117.030353
50. Cen C, Chen J, Lin L, et al. Fancb deficiency causes premature ovarian insufficiency in mice†. Biol Reprod. 2022;107(3):790–799. doi:10.1093/biolre/ioac103
51. Du W, Amarachintha S, Erden O, et al. Fancb deficiency impairs hematopoietic stem cell function. Sci Rep. 2015;5(18127). doi:10.1038/srep18127
52. D’Andrea MGA. Fanconi anemia and DNA repair. Human Molecular Genetics. 2001;10(20):2253–2259.
53. Chung YH, Qian Q, Huang HY, et al. The nuclear function of IL-33 in desensitization to DNA damaging agent and change of glioma nuclear structure. Front Cell Neurosci. 2021;15(713336). doi:10.3389/fncel.2021.713336
54. Dong Y, Xu S, Liu J, et al. Non-coding RNA-linked epigenetic regulation in cardiac hypertrophy. Int J Biol Sci. 2018;14(9):1133–1141. doi:10.7150/ijbs.26215
55. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/s0092-8674(04)00045-5
56. Fa HG, Chang WG, Zhang XJ, Xiao DD, Wang JX. Noncoding RNAs in doxorubicin-induced cardiotoxicity and their potential as biomarkers and therapeutic targets. Acta Pharmacol Sin. 2021;42(4):499–507. doi:10.1038/s41401-020-0471-x
57. Sano S, Wang Y, Ogawa H, et al. TP53-mediated therapy-related clonal hematopoiesis contributes to doxorubicin-induced cardiomyopathy by augmenting a neutrophil-mediated cytotoxic response. JCI Insight. 2021;6(13). doi:10.1172/jci.insight.146076
58. Bhagat A, Shrestha P, Jeyabal P, et al. Doxorubicin-induced cardiotoxicity is mediated by neutrophils through release of neutrophil elastase. Front Oncol. 2022;12(947604). doi:10.3389/fonc.2022.947604
59. Kaczmarek A, Krysko O, Heyndrickx L, et al. TNF/TNF-R1 pathway is involved in doxorubicin-induced acute sterile inflammation. Cell Death Dis. 2013;4(12):e961. doi:10.1038/cddis.2013.496
60. Krysko DV, Kaczmarek A, Krysko O, et al. TLR-2 and TLR-9 are sensors of apoptosis in a mouse model of doxorubicin-induced acute inflammation. Cell Death Differ. 2011;18(8):1316–1325. doi:10.1038/cdd.2011.4
61. Obata Y, Ishimori N, Saito A, et al. Activation of invariant natural killer T cells by alpha-galactosylceramide ameliorates doxorubicin-induced cardiotoxicity in mice. Eur J Prev Cardiol. 2020;27(19):2358–2361. doi:10.1177/2047487319901208
62. Ye J, Huang Y, Que B, et al. Interleukin-12p35 knock out aggravates doxorubicin-induced cardiac injury and dysfunction by aggravating the inflammatory response, oxidative stress, apoptosis and autophagy in mice. EBioMedicine. 2018;35:29–39. doi:10.1016/j.ebiom.2018.06.009
63. Zhang H, Xu A, Sun X, et al. Self-maintenance of cardiac resident reparative macrophages attenuates doxorubicin-induced cardiomyopathy through the SR-A1-c-Myc axis. Circ Res. 2020;127(5):610–627. doi:10.1161/circresaha.119.316428
64. Inada R, Miyamoto K, Tanaka N, et al. Chondroitin sulfate N-acetylgalactosyltransferase-1 knockout shows milder phenotype in experimental autoimmune encephalomyelitis than in wild type. Glycobiology. 2021;31(3):260–265. doi:10.1093/glycob/cwaa072
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.